Abstract
Background
Plasmodium vivax is a predominant species of malaria in parts of South America and there is increasing resistance to drugs to treat infections by P. vivax. The existence of latent hypnozoites further complicates the ability to classify recurrent infections as treatment failures due to relapse, recrudescence of hyponozoites or re-infections. Antigen loci are putatively under natural selection and may not be an optimal molecular marker to define parasite haplotypes in paired samples. Putatively neutral microsatellite loci, however, offer an assessment of neutral haplotypes. The objective here was to assess the utility of neutral microsatellite loci to reconcile cases of recurrent parasitaemia in Amazonian P. vivax populations in Peru.
Methods
Patient blood samples were collected from three locations in or around Iquitos in the Peruvian Amazon. Five putatively neutral microsatellite loci were characterized from 445 samples to ascertain the within and amongst population variation. A total of 30 day 0 and day of recurrent parasitaemia samples were characterized at microsatellite loci and five polymorphic antigen loci for haplotype classification.
Results
The genetic diversity at microsatellite loci was consistent with neutral levels of variation measured in other South American P. vivax populations. Results between antigen and microsatellite loci for the 30 day 0 and day of recurrent parasitaemia samples were the same for 80% of the pairs. The majority of non-concordant results were the result of differing alleles at microsatellite loci. This analysis estimates that 90% of the paired samples with the same microsatellite haplotype are unlikely to be due to a new infection.
Conclusions
A population-level approach was used to yield a better estimate of the probability of a new infection versus relapse or recrudescence of homologous hypnozoites; hypnozoite activation was common for this cohort. Population studies are critical with the evaluation of genetic markers to assess P. vivax biology and epidemiology. The additional demonstration of microsatellite loci as neutral markers capable of distinguishing the origin of the recurrent parasites (new infection or originating from the patient) lends support to their use in assessment of treatment outcomes.
Keywords: Plasmodium vivax, Malaria, Recrudescence, Relapse, Antigen locus, Microsatellite markers
Background
Plasmodium vivax contributes to a significant amount of morbidity in the Peruvian Amazon, accounting for 90% of all reported malaria cases in 2011 [1]. P. vivax is a predominant Plasmodium species in most areas of South America. The presence of the dormant hypnozoite stage along with reports of resistance to the most commonly used drugs to treat the blood stage, complicating treatment options for P. vivax complicate treatment options. Accordingly, the treatment and control of P. vivax remains an important issue to public health programmes.
P. vivax is characterized by the presence of a hypnozoite stage, which can persist in the liver for months to years, causing recurrent disease even though treatment and clearance of the initial blood stage infection were achieved. When a patient presents with recurrent P. vivax parasitaemia following treatment, there are several possible causes: 1) recrudescence of blood stage parasites; 2) relapse from latent hypnozoites; or, 3) a new infection (if the patient resides in an endemic area). Previous studies have shown that the risk and timing of relapse depends on the geographical origin of the initial infection. It is known that isolates of P. vivax from the tropics relapse sooner than those from more temperate areas, with 70% of relapses occurring within five months of initial infection [2]. The probability of relapse is higher than 20% for P. vivax patients when anti-hypnozoite therapy is not prescribed, and, thus, the ability to distinguish between relapse, recrudescence and re-infection is important in clinical efficacy studies [3].
Molecular genotype profiles of the parasites in an infection are critical to the assessment of clinical efficacy studies. These profiles may also be used to estimate the rate of new infections in populations that are continuously exposed. Increasingly, molecular genotyping has been used to differentiate treatment recrudescence from re-infection by comparing the profiles or haplotypes in a paired patient samples [4,5]. The probability that a given haplotype is identical in paired samples taken from two different time points greatly decreases with the use of a greater number of genetic loci and, also, with loci that have a high amount of polymorphism within a parasite population [6]. Using antigen-encoding genes is problematic as they are affected by immune selection. These genes alone or in concert are not able to distinguish recrudescence from re-infection with high levels of confidence [2].
Highly variable neutral microsatellite loci are ideal tools to assess population-level studies of Plasmodium falciparum and, more recently, P. vivax[7-16]. Complicating factors include the existence of multiple infections or clones within a single host and, in the case of P. vivax, the presence of dormant hypnozoites that persist for some time after the initial infection. However, studies that employ population level sampling and estimates of variation at each of the loci, in addition to paired patient samples taken at different points in time during an infection, can yield estimates of recurrence and drug failures [9,15]. An analysis of P. vivax populations and recurrent parasitemia was conducted. The haplotypes and the genetic variation allow for further assessment of paired patient samples as recrudescence/relapse or new infection.
Methods
Study sites and collection of samples
Samples were collected under a study aimed at assessing the efficacy of three doses of primaquine for the prevention of P. vivax relapses. The study was conducted from March 2006 to November 2007 in three sites, Padrecocha, Santa Clara and San Juan, located in or near the city of Iquitos, the largest city in the Peruvian Amazon (Figure 1). Patients included in the study were at least one year of age, had a fever or history of fever within 72 hours, had a mono-infection with P. vivax as assessed by microscopy and PCR [17], had parasitaemia levels ranging between 250 and 100,000 asexual parasites/ul, were not pregnant, did not present with symptoms of severe malaria, and had normal activities of glucose-6-phosphate dehydrogenase (G6PD). All patients were treated under direct observation with chloroquine (25 mg/kg) over three days and three regimens of primaquine, depending on the study arm. Prior to receiving treatment, a pre-treatment whole blood sample was obtained on day 0 (D-0). Patients who presented a second time in the subsequent six months with recurrent P. vivax parasitaemia were treated with a second round of primaquine-chloroquine according to the protocol of the Peruvian Ministry of Health, and a second sample of whole blood was collected prior to re-treatment on the day of recurrence (D-R). All samples were stored at -80°C until they were used. Informed consent was granted by the subject or guardian in the case of minors and assent was obtained from subjects between eight and 17 years of age. The study protocol was approved by the Ethical Review Committees of the Peruvian National Institute of Health and the US Naval Medical Research Center in compliance with all applicable Federal regulations governing the protection of human subjects.
Figure 1.
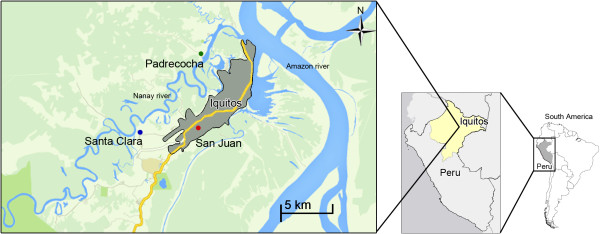
A map of the three populations used in this study, Padrecocha, Santa Clara, and San Juan.
DNA extraction
Parasitic DNA was extracted from 200 μl of whole blood following instructions from QIAamp DNA Blood Mini Kit (Qiagen). Genomic P. vivax DNA was used in order to amplify the polymorphic antigenic loci and also for the neutral microsatellite analysis.
Antigen genotyping of Plasmodium vivax
Thirty paired samples (D-0 and D-R from the same subject) were genotyped for five polymorphic antigenic loci of P. vivax: apical membrane antigen-1 (Pvama1) [18], circumsporozoite protein (Pvcsp) [19], merozoite surface protein 1 (Pvmsp-1) [20], merozoite surface protein 3 (Pvmsp3) [21], and duffy receptor binding protein (Pvdbp) [22]. Products of single PCR reactions were sequenced using an Applied Biosystems Prism 3130xl Avant Genetic Analyzer, and data analysis was performed using Sequencer software (Gene Codes Corporation).
Microsatellite characterization
Microsatellite characterization was conducted on 445 samples from 355 subjects (355 D-0 samples and 90 D-R samples) using five microsatellite loci in the P. vivax genome published previously by Imwong et al.: 14.185, 12.335, 7.67, 6.34, and 3.35 [10]. The full panel of eleven loci in the original study by Imwong et al., were tested, but these five loci were chosen because they reliably produced a product on a multitude of specimens and spurious peaks were rarely detected. Further, these loci amplified a polymorphic loci in the Imwong et al., study and also on a panel of test samples in the laboratory. The published PCR protocol was modified by using the PCR MasterMix (Promega) [10]. Fluorescently labelled PCR products were analysed on an Applied Biosystems Prism 3130xl Avant Genetic Analyzer and analysed using GeneMapper v4.0 (Applied Biosystems). A minimum peak height of 200 fluorescent units was used to define amplification products. For all analyses that use the microsatellite data, if multiple peaks were detected for a single sample, the value of the highest peak was used for the analysis.
Statistical analysis
The genetic variation for each microsatellite locus was measured by calculating the expected heterozygosity (H e ) and number of alleles per microsatellite locus (A). H e was calculated for each locus as H e = [n/(n - 1)][1 - ∑ p i 2], where n is the number of isolates sampled and p i is the frequency of the ith allele. The sampling variance for H e was calculated as 2(n - 1)/n3[2(n - 2)[∑ p i 3 - (∑ p i 2)2]] [23]. H e and the sampling variance were calculated using D-0 samples only. Arlequin ver 3.01 was used to compute H e [24], and the Excel Microsatellite Toolkit was used to format data [25].
Wright’s fixation index, FST, was used to test for genetic differentiation between two populations [26]. FST was calculated using Arlequin version 3.01 [24]. Linkage disequilibrium (LD) between microsatellite loci was assessed by using an exact test of LD [27]. Associations were tested between pairs of loci or sites by using 10,000 Monte Carlo steps in Arlequin; a Bonferroni-Holm correction was used to correct for multiple testing. For both FST and LD calculations, only microsatellite haplotypes from D-0 samples were used for the analyses. If a locus had multiple peaks, the allele represented by the highest peak was used for the analyses.
For paired D-0 and D-R samples, P(match), the probability of a second infection with a particular haplotype occurring purely by chance, was calculated using the population haplotype frequency. This calculation follows Brockman et al. and was conducted for the post-treatment sample pairs that had the same haplotype at each time point [28]. P(match) was calculated only for those sample pairs where the five-locus data was available; if data were missing from any of the loci then the paired samples were not considered for the P(match) analysis. Considering each geographic population separately, haplotype frequencies were determined using the five microsatellite loci and D-0 samples only. These frequencies were used as the ‘P(match)’ value for each of the haplotypes in question, and the P(match) value was considered significant if <0.05.
Results
Genetic relationships and differentiation among populations
In order to characterize the genetic variation overall and for each individual population, 355 D-0 samples were characterized at five neutral microsatellite loci. A total of 117, 54 and 184 samples were characterized for microsatellite loci from Padrecocha, Santa Clara, and San Juan, respectively. Approximately 2% of the samples in Padrecocha and San Juan and 3% in Santa Clara represented mixed infections of multiple P. vivax strains or haplotypes. H e , a measure of genetic variation per locus, was between 0.68 and 0.87 for each of the three populations individually and as a whole (Table 1). The loci 12.335, 7.67 and 3.35 had a higher number of alleles in each population and 14.185 and 6.34 had lower numbers of alleles.
Table 1.
Heterozygosity ( H e ) and number of alleles (A) per microsatellite locus
|
Locus |
14.185 |
|
12.335 |
|
7.67 |
|
6.34 |
|
3.35 |
|
|---|---|---|---|---|---|---|---|---|---|---|
| H e | A | H e | A | H e | A | H e | A | H e | A | |
| Padrecocha |
0.6833 ± 0.000 |
4 |
0.8430 ± 0.000 |
12 |
0.7149 ± 0.001 |
11 |
0.7711 ± 0.000 |
7 |
0.7998 ± 0.000 |
12 |
| Santa Clara |
0.7279 ± 0.001 |
6 |
0.8714 ± 0.000 |
11 |
0.7072 ± 0.002 |
9 |
0.7973 ± 0.000 |
7 |
0.8197 ± 0.001 |
10 |
| San Juan |
0.7282 ± 0.000 |
7 |
0.8665 ± 0.000 |
14 |
0.7227 ± 0.001 |
15 |
0.7775 ± 0.001 |
9 |
0.8071 ± 0.000 |
14 |
| All | 0.7173 ± 0.000 | 7 | 0.8665 ± 0.000 | 17 | 0.7168 ± 0.000 | 18 | 0.7812 ± 0.000 | 11 | 0.8149 ± 0.000 | 15 |
There was a significant amount of genetic differentiation (measured by FST) between Padrecocha and San Juan P. vivax populations (0.0118, p < 0.01). Additionally, the Padrecocha and San Juan P. vivax populations showed pairwise LD at all five loci. The Santa Clara population exhibited less LD (Figure 2).
Figure 2.
Linkage disequilibrium between microsatellite loci at the three sites, Padrecocha (A), Santa Clara (B), and San Juan (C). Each box represents one comparison between polymorphic pairs of loci. Shaded cells represent significance at 0.01.
Analysis of paired samples
A total of 30 paired samples collected at the D-0 and D-R visits were analysed for P. vivax strain identity using five polymorphic antigenic loci (PAL): Pvama, Pvcsp, Pvmsp1, Pvmsp3, and Pvdbp. Twelve (40%) paired samples had identical antigenic haplotypes and 18 (60%) had at least one locus that differed between the paired samples (Table 2).
Table 2.
Summary of polymorphic antigen loci (PAL) and microsatellite haplotypes from paired D-0 and D-R samples
| Padrecocha | Santa Clara | San Juan | Total | |
|---|---|---|---|---|
| Number pairs with differenta PAL haplotype (%) |
13 (65) |
2 (100) |
3 (37.5) |
18 (60) |
| Number pairs with sameb PAL haplotype (%) |
7 (35) |
0 (0) |
5 (62.5) |
12 (40) |
| Number pairs with differenta microsatellite haplotype (%) |
19 (55.9) |
5 (50) |
21 (45.7) |
45 (50) |
| Number pairs with sameb microsatellite haplotype (%) |
15 (44.1) |
5 (50) |
25 (54.3) |
45 (50) |
| Number pairs where p(match) <0.05 (%) | 9 (69)c | 5 (100) | 23 (100)c | 37 (90.2) |
aDifferent is defined as having at least one out of five loci different between paired samples.
bSame is defined as having identical alleles at all loci from which a viable PCR product was obtained.
cp (match) was only calculated for samples where there was complete data; two paired samples in both Padrecocha and San Juan populations had incomplete data.
Ninety paired D-0 and D-R samples were analysed for P. vivax strain identity by using the five loci microsatellite haplotypes (Table 3). Fifty percent of all pairs had identical alleles at all loci and the same proportion had a different allele at a minimum of one locus (Table 2). These proportions were similar for each of the three populations individually. A total of 37/41 paired samples (90.2%) had a significant P(match) < 0.05. Thus, it was unlikely that 90.2% of the paired samples were the result of a new infection. It should be noted that P(match) was only calculated for 41 out of 45 total pairs where the haplotypes were the same. Four pairs had incomplete data, and thus were removed from P(match) calculations.
Table 3.
Microsatellite analysis of paired D-0 and D-R samples
| Population | Case | Treatment a | Day b | 14.185 c | 12.335 | 7.67 | 6.34 | 3.35 | MS final d | p(match) e |
|---|---|---|---|---|---|---|---|---|---|---|
| Padrecocha |
1 |
5 |
0 |
269 |
162 |
102 |
150 |
125 |
S |
0.01 |
| |
|
|
70 |
269 |
162 |
102 |
150 |
125 |
|
|
| |
2 |
5 |
0 |
269 |
160 |
100 |
135 |
127 |
D |
|
| |
|
|
183 |
269 |
166 |
100 |
160 |
127 |
|
|
| |
3 |
5 |
0 |
|
|
|
135 |
|
D |
|
| |
|
|
141 |
|
|
|
160 |
|
|
|
| |
4 |
7 |
0 |
265 |
171 |
102 |
150 |
|
S |
|
| |
|
|
84 |
265 |
171 |
102 |
150 |
|
|
|
| |
5 |
14 |
0 |
265 |
171 |
|
135 |
|
D |
|
| |
|
|
83 |
265 |
166 |
|
135 |
|
|
|
| |
6 |
5 |
0 |
269 |
158 |
127 |
146 |
125 |
D |
|
| |
|
|
154 |
269 |
158 |
127 |
150 |
151 |
|
|
| |
7 |
5 |
0 |
282 |
162 |
100 |
146 |
|
D |
|
| |
|
|
105 |
265 |
171 |
102 |
150 |
|
|
|
| |
8 |
14 |
0 |
282 |
166 |
100 |
135 |
135 |
D |
|
| |
|
|
81 |
265 |
171 |
102 |
150 |
151 |
|
|
| |
9 |
7 |
0 |
282 |
166 |
|
160 |
|
D |
|
| |
|
|
28 |
282 |
166 |
|
135 |
|
|
|
| |
10 |
7 |
0 |
282 |
162 |
101 |
150 |
151 |
D |
|
| |
|
|
43 |
282 |
166 |
101 |
135 |
133 |
|
|
| |
11 |
5 |
0 |
282 |
163 |
98 |
150 |
151 |
D |
|
| |
|
|
120 |
282 |
163 |
98 |
150 |
153 |
|
|
| |
12 |
14 |
0 |
282 |
166 |
|
135 |
133 |
D |
|
| |
|
|
55 |
282 |
166 |
|
160 |
127 |
|
|
| |
13 |
5 |
0 |
269 |
158 |
126 |
146 |
125 |
D |
|
| |
|
|
154 |
269 |
158 |
126 |
135 |
127 |
|
|
| |
14 |
5 |
0 |
269 |
158 |
125 |
145 |
125 |
S |
0.051 |
| |
|
|
210 |
269 |
158 |
125 |
145 |
125 |
|
|
| |
15 |
14 |
0 |
282 |
166 |
|
160 |
125 |
D |
|
| |
|
|
43 |
282 |
166 |
|
135 |
133 |
|
|
| |
16 |
7 |
0 |
282 |
166 |
102 |
135 |
133 |
D |
|
| |
|
|
105 |
269 |
162 |
102 |
160 |
125 |
|
|
| |
17 |
5 |
0 |
282 |
166 |
122 |
150 |
117 |
D |
|
| |
|
|
53 |
282 |
166 |
101 |
135 |
125 |
|
|
| |
18 |
5 |
0 |
269 |
166 |
101 |
160 |
127 |
D |
|
| |
|
|
63 |
282 |
162 |
101 |
146 |
125 |
|
|
| |
19 |
14 |
0 |
265 |
171 |
102 |
150 |
153 |
S |
0.02 |
| |
|
|
91 |
265 |
171 |
102 |
150 |
153 |
|
|
| |
20 |
5 |
0 |
265 |
171 |
102 |
150 |
153 |
D |
|
| |
|
|
84 |
265 |
171 |
102 |
146 |
125 |
|
|
| |
21 |
5 |
0 |
269 |
158 |
125 |
145 |
125 |
S |
0.031 |
| |
|
|
111 |
269 |
158 |
125 |
145 |
125 |
|
|
| |
22 |
5 |
0 |
269 |
158 |
125 |
145 |
125 |
S |
0.031 |
| |
|
|
126 |
269 |
158 |
125 |
145 |
125 |
|
|
| |
23 |
14 |
0 |
269 |
159 |
100 |
135 |
127 |
S |
0.01 |
| |
|
|
73 |
269 |
159 |
100 |
135 |
127 |
|
|
| |
24 |
5 |
0 |
265 |
170 |
102 |
150 |
153 |
D |
|
| |
|
|
70 |
265 |
170 |
102 |
150 |
125 |
|
|
| |
25 |
7 |
0 |
265 |
170 |
101 |
150 |
153 |
S |
0.01 |
| |
|
|
53 |
265 |
170 |
101 |
150 |
153 |
|
|
| |
26 |
7 |
0 |
265 |
170 |
102 |
149 |
153 |
S |
0.051 |
| |
|
|
76 |
265 |
170 |
102 |
149 |
153 |
|
|
| |
27 |
14 |
0 |
265 |
|
|
149 |
153 |
D |
|
| |
|
|
52 |
265 |
|
|
134 |
131 |
|
|
| |
28 |
5 |
0 |
265 |
170 |
102 |
149 |
153 |
S |
0.051 |
| |
|
|
112 |
265 |
170 |
102 |
149 |
153 |
|
|
| |
29 |
7 |
0 |
265 |
170 |
102 |
149 |
153 |
S |
0.051 |
| |
|
|
80 |
265 |
170 |
102 |
149 |
153 |
|
|
| |
30 |
14 |
0 |
265 |
170 |
102 |
149 |
153 |
S |
0.051 |
| |
|
|
67 |
265 |
170 |
102 |
149 |
153 |
|
|
| |
31 |
7 |
0 |
265 |
170 |
102 |
149 |
153 |
S |
0.051 |
| |
|
|
120 |
265 |
170 |
102 |
149 |
153 |
|
|
| |
32 |
5 |
0 |
265 |
|
|
136 |
125 |
D |
|
| |
|
|
63 |
265 |
|
|
134 |
|
|
|
| |
33 |
5 |
0 |
265 |
170 |
102 |
150 |
153 |
S |
0.01 |
| |
|
|
202 |
265 |
170 |
102 |
150 |
153 |
|
|
| |
34 |
7 |
0 |
265 |
|
|
149 |
|
S |
|
| |
|
|
121 |
265 |
|
|
149 |
153 |
|
|
| Santa Clara |
35 |
7 |
0 |
265 |
162 |
98 |
150 |
151 |
D |
|
| |
|
|
52 |
265 |
164 |
102 |
135 |
127 |
|
|
| |
36 |
5 |
0 |
265 |
162 |
100 |
135 |
119 |
D |
|
| |
|
|
126 |
265 |
162 |
100 |
135 |
127 |
|
|
| |
37 |
5 |
0 |
265 |
171 |
102 |
150 |
153 |
D |
|
| |
|
|
87 |
265 |
171 |
102 |
150 |
151 |
|
|
| |
38 |
14 |
0 |
265 |
164 |
101 |
150 |
135 |
D |
|
| |
|
|
134 |
265 |
164 |
101 |
150 |
125 |
|
|
| |
39 |
5 |
0 |
265 |
170 |
102 |
149 |
153 |
S |
0.019 |
| |
|
|
30 |
265 |
170 |
102 |
149 |
153 |
|
|
| |
40 |
5 |
0 |
265 |
|
|
160 |
125 |
D |
|
| |
|
|
38 |
265 |
|
|
160 |
153 |
|
|
| |
41 |
14 |
0 |
265 |
158 |
125 |
145 |
125 |
S |
0.019 |
| |
|
|
178 |
265 |
158 |
125 |
145 |
125 |
|
|
| |
42 |
5 |
0 |
265 |
158 |
126 |
145 |
125 |
S |
0.019 |
| |
|
|
117 |
265 |
158 |
126 |
145 |
125 |
|
|
| |
43 |
5 |
0 |
265 |
170 |
102 |
149 |
127 |
S |
0.019 |
| |
|
|
110 |
265 |
170 |
102 |
149 |
127 |
|
|
| |
44 |
14 |
0 |
265 |
164 |
115 |
149 |
125 |
S |
0.019 |
| |
|
|
32 |
265 |
164 |
115 |
149 |
125 |
|
|
| San Juan |
45 |
7 |
0 |
265 |
|
|
150 |
|
D |
|
| |
|
|
28 |
265 |
|
|
135 |
|
|
|
| |
46 |
5 |
0 |
265 |
162 |
102 |
150 |
127 |
D |
|
| |
|
|
43 |
265 |
171 |
102 |
150 |
151 |
|
|
| |
47 |
5 |
0 |
265 |
166 |
122 |
150 |
127 |
S |
0.006 |
| |
|
|
71 |
265 |
166 |
122 |
150 |
127 |
|
|
| |
48 |
7 |
0 |
265 |
166 |
122 |
150 |
117 |
D |
|
| |
|
|
40 |
265 |
164 |
104 |
135 |
125 |
|
|
| |
49 |
5 |
0 |
265 |
164 |
102 |
135 |
|
D |
|
| |
|
|
62 |
265 |
164 |
102 |
135 |
|
|
|
| |
50 |
14 |
0 |
265 |
158 |
102 |
135 |
127 |
D |
|
| |
|
|
84 |
265 |
171 |
102 |
150 |
127 |
|
|
| |
51 |
5 |
0 |
265 |
171 |
102 |
150 |
127 |
S |
0.006 |
| |
|
|
70 |
265 |
171 |
102 |
150 |
127 |
|
|
| |
52 |
5 |
0 |
265 |
171 |
102 |
150 |
151 |
S |
0.006 |
| |
|
|
80 |
265 |
171 |
102 |
150 |
151 |
|
|
| |
53 |
14 |
0 |
265 |
164 |
101 |
160 |
125 |
S |
0.012 |
| |
|
|
99 |
265 |
164 |
101 |
160 |
125 |
|
|
| |
54 |
14 |
0 |
265 |
158 |
125 |
149 |
125 |
D |
|
| |
|
|
144 |
265 |
158 |
125 |
145 |
125 |
|
|
| |
55 |
5 |
0 |
265 |
160 |
102 |
145 |
151 |
D |
|
| |
|
|
86 |
265 |
160 |
102 |
145 |
151 |
|
|
| |
56 |
5 |
0 |
265 |
162 |
102 |
135 |
151 |
S |
0.006 |
| |
|
|
51 |
265 |
162 |
102 |
135 |
151 |
|
|
| |
57 |
5 |
0 |
265 |
170 |
100 |
150 |
127 |
S |
0.006 |
| |
|
|
53 |
265 |
170 |
100 |
150 |
127 |
|
|
| |
58 |
5 |
0 |
265 |
174 |
100 |
134 |
119 |
S |
0.006 |
| |
|
|
53 |
265 |
174 |
100 |
134 |
119 |
|
|
| |
59 |
7 |
0 |
265 |
160 |
|
160 |
125 |
S |
|
| |
|
|
120 |
265 |
160 |
|
160 |
125 |
|
|
| |
60 |
5 |
0 |
265 |
162 |
101 |
150 |
149 |
S |
0.006 |
| |
|
|
72 |
265 |
162 |
101 |
150 |
149 |
|
|
| |
61 |
7 |
0 |
265 |
|
|
134 |
|
D |
|
| |
|
|
99 |
265 |
|
|
134 |
|
|
|
| |
62 |
7 |
0 |
265 |
170 |
100 |
159 |
153 |
S |
0.006 |
| |
|
|
129 |
265 |
170 |
100 |
159 |
153 |
|
|
| |
63 |
5 |
0 |
265 |
174 |
102 |
134 |
125 |
S |
0.006 |
| |
|
|
42 |
265 |
174 |
102 |
134 |
125 |
|
|
| |
64 |
5 |
0 |
265 |
|
|
149 |
121 |
D |
|
| |
|
|
122 |
265 |
|
|
159 |
121 |
|
|
| |
65 |
14 |
0 |
265 |
166 |
101 |
134 |
121 |
S |
0.006 |
| |
|
|
98 |
265 |
166 |
101 |
134 |
121 |
|
|
| |
66 |
14 |
0 |
265 |
174 |
100 |
149 |
133 |
D |
|
| |
|
|
56 |
265 |
170 |
102 |
149 |
133 |
|
|
| |
67 |
14 |
0 |
265 |
166 |
151 |
134 |
127 |
S |
0.006 |
| |
|
|
100 |
265 |
166 |
151 |
134 |
127 |
|
|
| |
68 |
5 |
0 |
265 |
158 |
122 |
149 |
127 |
D |
|
| |
|
|
113 |
265 |
158 |
122 |
149 |
127 |
|
|
| |
69 |
5 |
0 |
265 |
|
|
159 |
121 |
D |
|
| |
|
|
85 |
265 |
|
|
149 |
121 |
|
|
| |
70 |
14 |
0 |
265 |
166 |
121 |
134 |
121 |
S |
0.006 |
| |
|
|
76 |
265 |
166 |
121 |
134 |
121 |
|
|
| |
71 |
5 |
0 |
265 |
|
101 |
145 |
|
D |
|
| |
|
|
210 |
265 |
164 |
105 |
150 |
|
|
|
| |
72 |
5 |
0 |
265 |
162 |
100 |
160 |
153 |
S |
0.006 |
| |
|
|
162 |
265 |
162 |
100 |
160 |
153 |
|
|
| |
73 |
7 |
0 |
265 |
166 |
99 |
135 |
|
D |
|
| |
|
|
71 |
265 |
162 |
121 |
149 |
|
|
|
| |
74 |
5 |
0 |
265 |
164 |
101 |
160 |
125 |
S |
0.012 |
| |
|
|
103 |
265 |
164 |
101 |
160 |
125 |
|
|
| |
75 |
5 |
0 |
265 |
166 |
101 |
134 |
|
D |
|
| |
|
|
197 |
265 |
162 |
101 |
134 |
|
|
|
| |
76 |
5 |
0 |
265 |
170 |
102 |
150 |
125 |
S |
0.006 |
| |
|
|
72 |
265 |
170 |
102 |
150 |
125 |
|
|
| |
77 |
5 |
0 |
265 |
166 |
100 |
160 |
125 |
S |
0.006 |
| |
|
|
56 |
265 |
166 |
100 |
160 |
125 |
|
|
| |
78 |
5 |
0 |
265 |
|
|
135 |
133 |
D |
|
| |
|
|
168 |
265 |
|
|
135 |
148 |
|
|
| |
79 |
14 |
0 |
265 |
158 |
125 |
134 |
127 |
D |
|
| |
|
|
80 |
265 |
154 |
125 |
134 |
151 |
|
|
| |
80 |
7 |
0 |
265 |
170 |
101 |
149 |
125 |
D |
|
| |
|
|
51 |
265 |
166 |
121 |
149 |
125 |
|
|
| |
81 |
5 |
0 |
265 |
|
|
134 |
|
S |
|
| |
|
|
136 |
265 |
|
|
134 |
121 |
|
|
| |
82 |
5 |
0 |
265 |
|
|
145 |
|
D |
|
| |
|
|
59 |
265 |
|
|
135 |
151 |
|
|
| |
83 |
5 |
0 |
265 |
174 |
102 |
149 |
133 |
D |
|
| |
|
|
163 |
265 |
170 |
102 |
149 |
148 |
|
|
| |
84 |
7 |
0 |
269 |
162 |
103 |
145 |
153 |
S |
0.006 |
| |
|
|
168 |
269 |
162 |
103 |
145 |
153 |
|
|
| |
85 |
14 |
0 |
269 |
162 |
100 |
149 |
148 |
S |
0.006 |
| |
|
|
71 |
269 |
162 |
100 |
149 |
148 |
|
|
| |
86 |
14 |
0 |
265 |
167 |
101 |
149 |
153 |
S |
0.006 |
| |
|
|
98 |
265 |
167 |
101 |
149 |
153 |
|
|
| |
87 |
5 |
0 |
269 |
162 |
121 |
149 |
127 |
S |
0.006 |
| |
|
|
136 |
269 |
162 |
121 |
149 |
127 |
|
|
| |
88 |
5 |
0 |
269 |
166 |
101 |
135 |
153 |
S |
0.006 |
| |
|
|
79 |
269 |
166 |
101 |
135 |
153 |
|
|
| |
89 |
14 |
0 |
271 |
166 |
101 |
134 |
121 |
D |
|
| |
|
|
183 |
276 |
162 |
100 |
140 |
148 |
|
|
| |
90 |
14 |
0 |
282 |
162 |
100 |
160 |
153 |
S |
0.006 |
| 73 | 282 | 162 | 100 | 160 | 153 |
aTreatment given to the patient on D-0. ‘5’ represents 30 mg of primaquine for 5 days; ‘7’ represents 30 mg of primaquine for 7 days.
‘14’ represents 15 mg of primaquine for 14 days.
bDay of sample collection, where ‘0’ is Day-0 (D-0) and the second day is the day of recurrence (D-R) respective to D-0.
cAllele for each microsatellite loci are reported as PCR product size.
dThe final determination of concordance (S = same) or disconcordance (D = different) between the two microsatellite haplotypes (D-0 and D-R). Different alleles between a pair of samples are highlighted in red.
eP(match) values were calculated separately for each population and only for complete 5-locus haplotypes.
The microsatellite data were compared to the PAL data for 30 paired samples. Concordance between the paired samples was examined for both PAL and microsatellite data (both having the criteria of no differing loci for a D-0 and D-R pair). Twenty-four pairs (24/30, 80%) had concordant results between the two methods and six pairs (six/30, 20%) were non-concordant. Of the six that were non-concordant, one pair was determined as the same haplotype by microsatellite analysis at both time points and a different haplotype by antigen loci (the difference was at one of five loci, Pvcsp). Five pairs were found to have the same haplotype by antigen loci but a different haplotype by using microsatellite loci.
Discussion
The high levels of genetic diversity seen in these samples from the Peruvian Amazon are consistent with neutral levels of microsatellite and tandem repeat variation measured in P. vivax from multiple sites in Brazil, Colombia, and Peru [7,13,15,29-32]. This pattern is curious because it is entirely different from South American P. falciparum populations, as they show much lower levels of variation [33]. These differences could be explained by a number of factors, including differences in demography, natural selection due to drug pressure or differences in microsatellite dynamics between the two Plasmodium species. A low number of multiple clone infections in individual patients were observed in this study. This is an area where there is relatively lower transmission of parasites, and it is expected that these patients would experience less inoculations than patients in a hyperemic area of disease, and, thus a reduced chance for multiple clones existing within a single patient. Further, the specific composition of loci examined (i.e. the microsatellite motifs) and the effects of natural selection (i.e. strong immune selection on PALs) on these loci, may affect the detection of multiple infections [34].
There was evidence of population differentiation between Padrecocha and San Juan; this differentiation was not observed between Santa Clara and either of the two sites. This result could be due to a sample size bias, as only 54 samples were available from Santa Clara. Extensive LD among microsatellite markers has been noted in Colombian and some (but not all) Brazilian P. vivax populations. The existence of LD in South American populations is not surprising, given the low rate of parasite transmission and high levels of genetic inbreeding documented in many areas of the continent [13,29,35]. The Santa Clara population had less observed LD, but this population was represented by a smaller number of samples, which could affect the calculation of LD as it is dependent on the frequency of allele or loci pairs in a population. An alternative explanation could be that the Santa Clara population has experienced a larger amount of genetic recombination or mutation. Additional studies would be needed to better understand this observation.
There have been several studies that have utilized population-based estimates of genetic variation to calculate the probability that a pair of samples from the same patient would have the same genetic profile or haplotype by chance. P (identical), the probability that a random pair of haplotypes is identical, is affected by low frequency haplotypes in a population [36]. There are many low frequency haplotypes in each of the three Peruvian populations evaluated here. Another estimate, p (maximum), the maximum probability of two clones carrying the same haplotype by chance, requires independence of loci [9]. There was extensive LD between loci in the populations from Padrecocha and San Juan; thus, non-independence of loci was found. The third method, P(match), utilizes the population haplotype frequencies to assess whether the paired haplotype frequency occurs above or below a defined frequency value (for example, 0.05) [28]. Given the before mentioned limitations of the populations, P(match) was chosen as a method to assess the maximum probability of two clones or parasites carrying the same haplotype by chance in a population. P(match) has the limitation of requiring a population-level assessment of parasite diversity prior to assigning probability values. In areas with low genetic diversity, as is observed in South America, sample sizes must be relatively large in order to appropriately define the extent of diversity in the parasite populations [28].
Eighty percent of the paired samples examined by both microsatellite and antigen loci methods obtained the same overall result. Five out of six non-concordant paired samples (83%) were found to have different haplotypes at the two sampling times by microsatellite loci, but not by using antigen loci. This result is not unexpected, as microsatellite loci are putatively neutral with respect to selection, and, thus, should exhibit a greater amount of variation. Loci under strong selection, such as antigen loci, exhibit lower levels of variation in the population. These results emphasize the utility of using neutral loci to determine the P. vivax strain characteristics, and this can be illustrated by considering the application of these tests to assess a treatment regimen. If, for example, this study assessed drug failures amongst these 30 paired samples, results from antigen-encoding loci would lead one to believe that 12 (40%) patients potentially had a relapse of liver-stage parasites, whereas microsatellite loci would assess this rate at eight (26.7%) patients. It should be emphasized, though, that this type of simple interpretation has many caveats. Of note, the microsatellite methods used here employed a population-based level of sampling and analysis - such an approach is critical when assessing and interpreting genetic variation. Although use of PAL and microsatellite loci can be used in conjunction, researchers should use caution in the interpretation of the data and consider the effect of selection or neutral mechanisms on each of type of loci.
Interpretation of recurrent parasitaemia and drug treatment regimens in P. vivax is complicated by the presence of hypnozoites. Although the transmission of P. vivax in the Iquitos area of Peru is relatively low, the possibility of re-infection for the subjects in this study cannot be eliminated. Paired samples with different initial and recurrent haplotypes are the result of relapse with a heterologous hypnozoite or a new infection; here, 50% of the paired samples had different parasite lineages. Paired samples with the same lineage (here, 50%) are the result of recrudescence of the blood stage parasite (potentially a chloroquine treatment failure), relapse with a homologous hypnozoite (potentially a primaquine treatment failure), or a new infection with the same parasite strain. Furthermore, the population-level analysis here indicates that 90.2% of the paired samples that had the same haplotype were unlikely due to a new infection. These results indicate a potential large frequency of recurrent infections resulting from hypnozoite activation, either heterologous or homologous. This could arise from either an initial infection or multiple infections over time with different P. vivax strains, all resulting in dormant hypnozoites. The activation of heterologous hypnozoites has been noted before in patients from Asia [9].
There are limitations to microsatellite, tandem repeats, and PCR-based techniques to discern treatment outcomes. PCR bias, potential artifacts, and the inability to discriminate multiple clones are significant hurdles with these methods [14]. Indeed, microsatellite diversity is associated with the underlying repeat motif structure of each loci, and, thus, there may be an inherent bias in the results based on the loci that are chosen [32,34,37]. Careful consideration of the underlying motif structure of these loci is an important factor for researchers to consider. Further, the ability to rapidly screen many infections and assess genetic variation of the larger parasite population is key to understanding and interpreting these assays, irrespective of the locus. Population-based approaches have an advantage over smaller sampling strategies in both analyses and assessments [15].
In conclusion, the P. vivax parasite population in the Peruvian Amazon has considerable diversity and population differentiation. Antigen and microsatellite-based P. vivax genotyping methods can be useful tools to evaluate drug treatment regimens and characterize genetic profiles of parasite populations; however, researchers should be careful when interpreting data. Antigen-encoding loci do incorporate a bias due to immune selection. The description of the extent of hypnozoite activation in this population emphasizes the importance of treatments that target this stage. The P. vivax dormant hypnozoite stage poses challenges in the assessment of paired patient samples; nevertheless, population-based studies with appropriate methods can provide insights into the biology and epidemiology of P. vivax populations. The further development and assessment of molecular tools that allow a broad analysis of an individual parasite’s genetic construction will be immensely useful to drug treatment and resistance studies.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AMM, VU, AAE, and DJB designed the study. AMM, VS, CS, and MS conducted the molecular genetics studies and data analysis. AMM, PCFG and DJB drafted the manuscript. SD, CC and DJB participated in the field study design and coordination of sample collection. All authors read and approved the final manuscript.
Contributor Information
Andrea M McCollum, Email: amccollum@cdc.gov.
Valeria Soberon, Email: valeria.soberon@gmail.com.
Carola J Salas, Email: Carola.Salas@med.navy.mil.
Meddly L Santolalla, Email: Meddly.Santolalla@med.navy.mil.
Venkatachalam Udhayakumar, Email: vxu0@cdc.gov.
Ananias A Escalante, Email: Ananias.Escalante@asu.edu.
Paul CF Graf, Email: Paul_Graf@URMC.Rochester.edu.
Salomon Durand, Email: salomondurand@gmail.com.
Cesar Cabezas, Email: ccabezas@ins.gob.pe.
David J Bacon, Email: david.bacon@nrl.navy.mil.
Acknowledgements
The authors would like to acknowledge the contribution of the patients to this study, the microscopy performed by Delores Rimarachin, Gerson Guedes and Greys Braga, and the map prepared for publication by Hugo Valdivia. The authors greatly appreciate comments by G Christain Baldeviano and Willy Lescano on this manuscript.
This work was developed and sponsored jointly by the Instituto Nacional de Salud of the Peruvian Ministry of Health and the US Naval Medical Research Unit Six (NAMRU-6). Funding for this project was received from the Global Emerging Infections Surveillance and Response System of the US Department of Defense (AFHSC/GEIS). This work was supported and funded by work unit number 847705.82000.256B.B0016. Additional financial support from the CDC Drug Resistance Working Group and support from the Atlanta Research and Education Foundation (Atlanta, GA, USA) are appreciated.
Disclaimer
The views expressed in this article are those of the author and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, Centers for Disease Control and Prevention, nor the US Government.
Copyright statement
Some of the authors are a military service members or employees of the U.S. Government. This work was prepared as part of their official duties. Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
References
- Peru MS. Sala de Situacion de Salud. Ministry of Health Publication; 2011. [Google Scholar]
- Baird JK. Chloroquine resistance in Plasmodium vivax. Antimicrob Agents Chemother. 2004;48:4075–4083. doi: 10.1128/AAC.48.11.4075-4083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird JK, Hoffman SL. Primaquine therapy for malaria. Clin Infect Dis. 2004;39:1336–1345. doi: 10.1086/424663. [DOI] [PubMed] [Google Scholar]
- Hwang J, Alemayehu BH, Reithinger R, Tekleyohannes SG, Takele T, Birhanu SG, Demeke L, Hoos D, Melaku Z, Kassa M, Jima D, Malone JL, Nettey H, Green M, Poe A, Akinyi S, Udhayakumar V, Kachur SP, Filler S. In vivo efficacy of artemether-lumefantrine and chloroquine against Plasmodium vivax: a randomized open label trial in central Ethiopia. PLoS One. 2013;8:e63433. doi: 10.1371/journal.pone.0063433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaukat AM, Gilliams EA, Kenefic LJ, Laurens MB, Dzinjalamala FK, Nyirenda OM, Thesing PC, Jacob CG, Molyneux ME, Taylor TE, Plowe CV, Laufer MK. Clinical manifestations of new versus recrudescent malaria infections following anti-malarial drug treatment. Malar J. 2012;11:207. doi: 10.1186/1475-2875-11-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snounou G, Beck HP. The use of PCR genotyping in the assessment of recrudescence or reinfection after antimalarial drug treatment. Parasitol Today. 1998;14:462–467. doi: 10.1016/S0169-4758(98)01340-4. [DOI] [PubMed] [Google Scholar]
- Ferreira MU, Karunaweera ND, da Silva-Nunes M, da Silva NS, Wirth DF, Hartl DL. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J Infect Dis. 2007;195:1218–1226. doi: 10.1086/512685. [DOI] [PubMed] [Google Scholar]
- Gomez JC, McNamara DT, Bockarie MJ, Baird JK, Carlton JM, Zimmerman PA. Identification of a polymorphic Plasmodium vivax microsatellite marker. Am J Trop Med Hyg. 2003;69:377–379. [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Snounou G, Pukrittayakamee S, Tanomsing N, Kim JR, Nandy A, Guthmann JP, Nosten F, Carlton J, Looareesuwan S, Nair S, Sudimack D, Day NP, Anderson TJ, White NJ. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J Infect Dis. 2007;195:927–933. doi: 10.1086/512241. [DOI] [PubMed] [Google Scholar]
- Imwong M, Sudimack D, Pukrittayakamee S, Osorio L, Carlton JM, Day NP, White NJ, Anderson TJ. Microsatellite variation, repeat array length, and population history of Plasmodium vivax. Mol Biol Evol. 2006;23:1016–1018. doi: 10.1093/molbev/msj116. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Karunaweera ND, Ferreira MU, Phone-Kyaw M, Pollack RJ, Alifrangis M, Rajakaruna RS, Konradsen F, Amerasinghe PH, Schousboe ML, Galappaththy GN, Abeyasinghe RR, Hartl DL, Wirth DL. Geographic structure of Plasmodium vivax: microsatellite analysis of parasite populations from Sri Lanka, Myanmar, and Ethiopia. Am J Trop Med Hyg. 2010;82:235–242. doi: 10.4269/ajtmh.2010.09-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjuela-Sanchez P, da Silva NS, da Silva-Nunes M, Ferreira MU. Recurrent parasitemias and population dynamics of Plasmodium vivax polymorphisms in rural Amazonia. Am J Trop Med Hyg. 2009;81:961–968. doi: 10.4269/ajtmh.2009.09-0337. [DOI] [PubMed] [Google Scholar]
- Rezende AM, Tarazona-Santos E, Couto AD, Fontes CJ, De Souza JM, Carvalho LH, Brito CF. Analysis of genetic variability of Plasmodium vivax isolates from different Brazilian Amazon areas using tandem repeats. Am J Trop Med Hyg. 2009;80:729–733. [PubMed] [Google Scholar]
- Vab den Eede P, Erhart A, Van der Auwera G, Van Overmeir C, Thang ND, Hung Le X, Anne J, D'Alessandro U. High complexity of Plasmodium vivax infections in symptomatic patients from a rural community in central Vietnam detected by microsatellite genotyping. Am J Trop Med Hyg. 2010;82:223–227. doi: 10.4269/ajtmh.2010.09-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo E, Imwong M, Rojas W, Carmona-Fonseca J, Maestre A. High genetic polymorphism of relapsing P. vivax isolates in northwest Colombia. Acta Trop. 2011;119:23–29. doi: 10.1016/j.actatropica.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Araujo FC, de Rezende AM, Fontes CJ, Carvalho LH, Alves De Brito CF. Multiple-clone activation of hypnozoites is the leading cause of relapse in Plasmodium vivax infection. PLoS One. 2012;7:e49871. doi: 10.1371/journal.pone.0049871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–292. doi: 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- Han ET, Park JH, Shin EH, Choi MH, Oh MD, Chai JY. Apical membrane antigen-1 (AMA-1) gene sequences of re-emerging Plasmodium vivax in South Korea. Korean J Parasitol. 2002;40:157–162. doi: 10.3347/kjp.2002.40.3.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Pukrittayakamee S, Gruner AC, Renia L, Letourneur F, Looareesuwan S, White NJ, Snounou G. Practical PCR genotyping protocols for Plasmodium vivax using Pvcs and Pvmsp1. Malar J. 2005;4:20. doi: 10.1186/1475-2875-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A, Vicini J, Patarroyo ME, Murillo LA, Patarroyo MA. Plasmodium vivax: polymorphism in the merozoite surface protein 1 gene from wild Colombian isolates. Exp Parasitol. 2000;95:215–219. doi: 10.1006/expr.2000.4534. [DOI] [PubMed] [Google Scholar]
- Bruce MC, Galinski MR, Barnwell JW, Snounou G, Day KP. Polymorphism at the merozoite surface protein-3alpha locus of Plasmodium vivax: global and local diversity. Am J Trop Med Hyg. 1999;61:518–525. doi: 10.4269/ajtmh.1999.61.518. [DOI] [PubMed] [Google Scholar]
- Ampudia E, Patarroyo MA, Patarroyo ME, Murillo LA. Genetic polymorphism of the Duffy receptor binding domain of Plasmodium vivax in Colombian wild isolates. Mol Biochem Parasitol. 1996;78:269–272. doi: 10.1016/S0166-6851(96)02611-4. [DOI] [PubMed] [Google Scholar]
- Nash D, Nair S, Mayxay M, Newton PN, Guthmann JP, Nosten F, Anderson TJ. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc Biol Sci. 2005;272:1153–1161. doi: 10.1098/rspb.2004.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Park SDE. PhD thesis. University of Dublin; 2001. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. [Google Scholar]
- Wright S. The interpretation of population structure by F-statistics with special regard to systems of mting. Evolution. 1965;19:395–420. doi: 10.2307/2406450. [DOI] [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.2307/2410454. [DOI] [PubMed] [Google Scholar]
- Brockman A, Paul RE, Anderson TJ, Hackford I, Phaiphun L, Looareesuwan S, Nosten F, Day KP. Application of genetic markers to the identification of recrudescent Plasmodium falciparum infections on the northwestern border of Thailand. Am J Trop Med Hyg. 1999;60:14–21. doi: 10.4269/ajtmh.1999.60.14. [DOI] [PubMed] [Google Scholar]
- Imwong M, Nair S, Pukrittayakamee S, Sudimack D, Williams JT, Mayxay M, Newton PN, Kim JR, Nandy A, Osorio L, Carolton JM, White NJ, Day NP, Anderson TJ. Contrasting genetic structure in Plasmodium vivax populations from Asia and South America. Int J Parasitol. 2007;37:1013–1022. doi: 10.1016/j.ijpara.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Van den Eede P, Van der Auwera G, Delgado C, Huyse T, Soto-Calle VE, Gamboa D, Grande T, Rodriguez H, Llanos A, Anne J, Erhart A, D'Alessandro U. Multilocus genotyping reveals high heterogeneity and strong local population structure of the Plasmodium vivax population in the Peruvian Amazon. Malar J. 2010;9:151. doi: 10.1186/1475-2875-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosek M, Yori PP, Gilman RH, Calderon M, Zimic M, Chuquiyauri R, Jeri C, Pinedo-Cancino V, Matthias MA, Llanos-Cuentas A, Vinetz JM. High degree of Plasmodium vivax diversity in the Peruvian Amazon demonstrated by tandem repeat polymorphism analysis. Am J Trop Med Hyg. 2012;86:580–586. doi: 10.4269/ajtmh.2012.11-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton PL, Neyra V, Hernandez JN, Branch OH. Plasmodium falciparum and Plasmodium vivax infections in the Peruvian Amazon: propagation of complex, multiple allele-type infections without super-infection. Am J Trop Med Hyg. 2009;81:950–960. doi: 10.4269/ajtmh.2009.09-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- Sutton PL. A call to arms: on refining Plasmodium vivax microsatellite marker panels for comparing global diversity. Malar J. 2013;12:447. doi: 10.1186/1475-2875-12-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezende AM, Tarazona-Santos E, Fontes CJ, Souza JM, Couto AD, Carvalho LH, Brito CF. Microsatellite loci: determining the genetic variability of Plasmodium vivax. Trop Med Int Health. 2010;15:718–726. doi: 10.1111/j.1365-3156.2010.02535.x. [DOI] [PubMed] [Google Scholar]
- Conway DJ, McBride JS. Population genetics of Plasmodium falciparum within a malaria hyperendemic area. Parasitology. 1991;103(Pt 1):7–16. doi: 10.1017/s0031182000059229. [DOI] [PubMed] [Google Scholar]
- Chuquiyauri R, Penataro P, Brouwer KC, Fasabi M, Calderon M, Torres S, Gilman RH, Kosek M, Vinetz JM. Microgeographical differences of Plasmodium vivax relapse and re-infection in the Peruvian Amazon. Am J Trop Med Hyg. 2013;89:326–338. doi: 10.4269/ajtmh.13-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]



