Abstract
Through advances in technology, the genetic basis of cancer has been investigated at the genomic level, and many fundamental questions have begun to be addressed. Among several key unresolved questions in cancer biology, the molecular basis for the link between nuclear deformation and malignancy has not been determined. Another hallmark of human cancer is aneuploidy; however, the causes and consequences of aneuploidy are unanswered and are hotly contested topics. We found that nuclear lamina proteins lamin A/C are absent in a significant fraction (38%) of human breast cancer tissues. Even in lamin A/C–positive breast cancer, lamin A/C expression is heterogeneous or aberrant (such as non-nuclear distribution) in the population of tumor cells, as determined by immunohistology and immunofluorescence microscopy. In most breast cancer cell lines, a significant fraction of the lamin A/C– negative population was observed. To determine the consequences of the loss of lamin A/C, we suppressed their expression by shRNA in non-cancerous primary breast epithelial cells. Down-regulation of lamin A/C in breast epithelial cells led to morphological deformation, resembling that of cancer cells, as observed by immunofluorescence microscopy. The lamin A/C–suppressed breast epithelial cells developed aneuploidy as determined by both flow Cytometry and fluorescence in situ hybridization. We conclude that the loss of nuclear envelope structural proteins lamin A/C in breast cancer underlies the two hallmarks of cancer aberrations in nuclear morphology and aneuploidy.
Keywords: Nuclear envelope, lamin A/C, aneuploidy, Polyploidy, nuclear grade, nuclear morphology, breast cancer
Advances in technology permitted the global and systematic investigation of the cancer genome to identify genetic changes at a molecular level[1],[2], yet several fundamental puzzles remain unsolved. A key unresolved question in cancer biology is the molecular basis of the link between nuclear deformation and malignancy[3],[4]. Most normal or benign cells have an oval shaped and smooth nuclear envelope, but an aberrantly shaped, large, and deformed nucleus is a hallmark of neoplastic cells[3]–[5]. Nuclear morphology is commonly used for diagnosis of malignancy and is referred to as nuclear grade[3],[6]. In the past five decades, the correlation of nuclear morphology with malignancy has been investigated[6]–[10]. Oncogenic signaling has been found to influence nuclear morphology[8]–[10], and alterations of nuclear matrix protein have also been suspected to play a role[5]. Nevertheless, the molecular changes associated with nuclear morphological transformation in cancer have not been established[3].
Another prominent hallmark of cancer is aneuploidy (or chromosomal numerical instability)[11]–[15], which was recognized by Boveri more than one hundred years ago as a factor closely associated with malignancies [11],[12]. Many oncogenic events are associated with aneuploidy [14],[16],[17], but the causes and significance of aneuploidy in cancer are not clear. It is commonly thought that in cancer cells, cytokinesis defects and frequent mitotic failure lead to the formation of tetraploid intermediates, which subsequently produce aneuploid cells following aberrant cell division[18],[19]. The role of aneuploidy in cancer is still a hotly debated subject[15],[16],[20]. On one hand, aneuploidy impairs cell growth and transformation[21]–[23]; however, aneuploidy, with aid from oncogenic mutations, may also enable the transformed cells to undergo changes and adaptation, evolution, and clonal selection to promote tumor progression[12],[15],[24].
We recently found that emerin, a nuclear envelope structural protein, is often lost or greatly reduced in ovarian cancer[25]. Based on experiments in cultured ovarian surface epithelial cells, we concluded that the loss of emerin may account for both nuclear deformation and aneuploidy in ovarian cancer [25]. Here, we investigated if emerin is also frequently absent in breast cancer cells and if the loss of other nuclear envelope structural proteins accounts for the defective nuclear envelope found in breast cancer. We present evidence that the loss of lamin A/C rather than emerin may be the cause of nuclear envelope defects in breast cancer.
Materials and Methods
Tumor specimens, human primary breast epithelial cells, and cell cultures
Breast cancer tissue microarrays were provided by the Tumor Bank of Fox Chase Cancer Center[26]. The microarray contained 56 invasive ductal carcinomas and 5 non-cancerous mammary tissues in duplicate cores. Primary human breast epithelial cells were prepared from fresh mammary tissues obtained from mammary reduction surgeries as detailed previously[26]. The cells were cultured in low calcium RPMI-1640 media containing 5% chelated horse serum in flasks coated with 0.1% swine skin gelatin. Seven independent preparations of primary human breast epithelial cells were used in this study for duplications and repetitions in the experiments. Six breast cancer cell lines (MDA-MB-468, MDA-MB-435, MDA-MB-231, SkBr3, T47D, and MCF-7) and 4 non-cancer cell lines (MCF-10A, MCF-10F, MCF-12A, and BT20) were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum. These cell lines were originally purchased from ATCC.
The study of human tissues and primary cells was reviewed by the Institutional Review Board of both Fox Chase Cancer Center and the University of Miami Miller School of Medicine and was approved and judged as non-human subject research.
Probes for Northern blot and antibodies for Western blot
Probes for lamin A/C and lamin B1 were obtained by reverse transcription-polymerase chain reaction (RT-PCR) using total RNA extracted from human ovarian surface epithelial (HOSE) cells at passage 2 as a template. The accession number, sense primer (SP), and anti-sense primer (ASP) are as follows: CGGTGTACATCGACAAGGTG (SP) and CAGCTGTTG-CTGCATTTGAT (ASP) for lamin B1 (BC078178); TCTG-CTGAGAGGAACAGCAA (SP) and GGTGATGGAG-CAGGTCATCT(ASP) for lamin A/C (NM_005572). The PCR products were sequenced to confirm the identity. The mouse emerin cDNA fragment was digested from an emerin expression construct, gel purified, and used as probe for Northern blot.
Mouse anti–lamin A/C antibodies and the lamin A–specific rabbit antibodies that do not recognize the lamin C spliced form were purchased from Santa Cruz Biotechnology Inc (Santa Cruz, CA, USA). Mouse anti-emerin, rabbit anti-p21, and goat anti–lamin B1 antibodies were also purchased from Santa Cruz. Monoclonal mouse anti–β-tubulin antibodies conjugated with Alexa were purchased from Sigma. The peroxidase-conjugated secondary antibodies were purchased from BioRad, Inc. A Super Signal Western Dura Extended Duration Substrate kit (Thermo Scientific, Rockford, IL, USA) was used for Western blot analysis.
Small interfering RNA (shRNA and siRNA) transfection
siRNA and shRNA directed against human lamin A/C, lamin B1, and scrambled controls were purchased from Santa Cruz Biotechnology and were transfected into cells using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen, CA). The siRNA and shRNA were provided as a mixture of oligos or selectable vectors, respectively, targeting multiple sequences of lamin A/C nucleotide sequences. Lamin A/C–specific shRNA plasmid (# sc-35776-SH) is a lentiviral vector expressing hairpins 19–25 nucleotides in length. Scrambled sequences were used as controls. For siRNA-mediated down-regulation, suppression seems to be optimal around 2–3 days. The shRNA-transfected cells were selected using puromycin (1.0 µg/mL for 5 days).
Flow Cytometry analysis
Cell sorting and flow analyses were performed on a FACS Vantage SE machine with Diva (Digital Vantage) software (Becton Dickinson, San Jose, CA), which is operated by the institutional core facility (University of Miami Miller School of Medicine). Cells were sorted according to DNA content and size based on their forward light scattering (FSC) value. The sideward light scattering (SSC) value obtained reflects the granularity or the irregularity of the cells. The sorted cells were either immediately used for analysis or placed in flasks for further Culturing.
Immunohistology of tissues and immunofluorescence microscopy of cultured cells
Immunostaining was performed using the mouse DAKO EnVision TM+ System and the Peroxidase (DAB) Kit (Dako) as previously described [25],[27]–[29]. Negative controls were performed by replacing the primary antibodies with non-immunized IgG.
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 for 5 min, blocked with 3% BSA in PBS containing 0.1% Tween-20 for 30 min, incubated for 1 h at 37°C with primary antibodies, and subsequently incubated with secondary antibodies conjugated with AlexaFluor 488 (green fluorescence) or AlexaFluor 594 (red fluorescence) (Molecular Probes). The nuclei were stained with Hoechst 33342 or DAPI (Molecular Probes). Cells were mounted/sealed in anti-fade reagent containing 0.1 mol/L n-propyl gallate (pH 7.4) and 90% glycerol in PBS. A Zeiss microscope and an AxioCam Camera and the AxioVision software 4.8 were used for image acquisition and analysis.
FISH and cytogenetic analysis
Chromosome number counting and fluorescence in situ hybridization (FISH) analysis were performed by the University of Miami Cytogenetic Core facility. Briefly, the cells were exposed to 15 µg/mL colcemid (demecolcine) for 12 h, resuspended in 10 mL hypotonic buffer (75 µmol/L KCI) for 20 min, and then kept in 1 mL of chilled fixative (methanol/acetic acid, 3:1). The cell suspension was dropped onto wet slides and heated at 70°C overnight, dyed with Wright's stain, and analyzed by counting 50 metaphases for each sample in duplicate. In a subset (5 to 10) of metaphase spreads, the chromosomes were identified by experienced cytogenetists, staff of the University of Miami Cytogenetic Core facility. For FISH analysis, DNA probes for chromosome X (red) and Y (green) were used for hybridization of unsynchronized cells.
Computer-assisted analysis of nuclear size
Following analysis with immunofluorescence microscopy, the images were scored for distribution of nuclear size by computer-assisted image analysis (AxioVision software 4).
Statistical analyses
Student's T-test was used to compare the difference between two groups. A P value less than 0.05 was considered significant.
Results
Lamin A/C expression is frequently lost in breast cancer cells and tissues
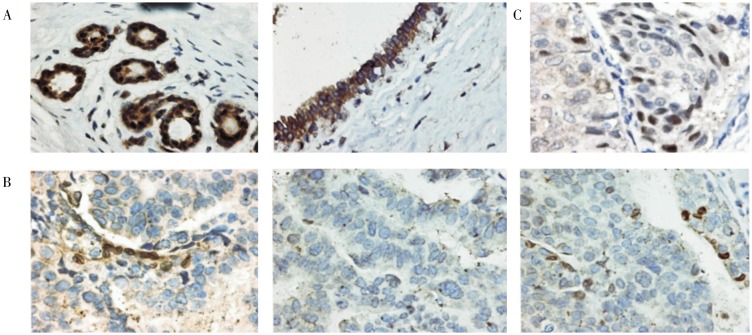
Since emerin is often lost in ovarian cancer[25], we examined whether emerin is also missing in breast cancer, or if the aberrant expression of other nuclear envelope proteins account for the nuclear morphological deformation of breast cancer cells. By immunohistochemistry, we found that lamin A/C, and not emerin, is frequently absent in breast carcinomas. In normal human mammary tissues, both globular and ductal epithelial cells exhibit strong lamin A/C staining around the nuclear envelope (Figure 1A). Lamin A/C staining was completely absent in 21 (38%) of the 56 invasive ductal carcinomas examined, as shown by three examples of breast carcinomas (Figure 1B). Among the lamin A/C–negative cancer cells, many exhibited irregular and aberrant nuclear morphology (Figure 1B). Fibroblast and some lymphocytic cells present in stroma stained strongly for lamin A/C, serving as an internal positive control. Additionally, in nearly all tumors cataloged as lamin A/C–positive, 10% to 80% of the tumor cells were often observed to be lamin A/C–negative, and the lamin A/C–positive and –negative tumor cells intermingled. Thus, heterozygous lamin A/C staining in tumor cell population was observed in most breast cancers, as shown by an example (Figure 1C).
Figure 1. Loss of lamin A/C expression in breast cancer.
Breast normal and tumor tissues were analyzed by immunostaining for lamin A/C. A, representative examples show strong staining of lamin A/C around the nuclei of normal mammary lobular and ductal epithelial cells. The nuclei are generally smooth, oval shaped in all normal mammary epithelial cells. B, three examples show loss of lamin A/C expression in breast carcinomas. A few cells show strong lamin A/C staining around the nuclei, serving as internal positive controls. The nuclear morphology is heterogeneous, and some cells with large and deformed nuclei are present. C, a representative example of breast carcinomas shows heterogeneous expression of lamin A/C among tumor cells. The images were obtained with 400x magnification and were processed identically.
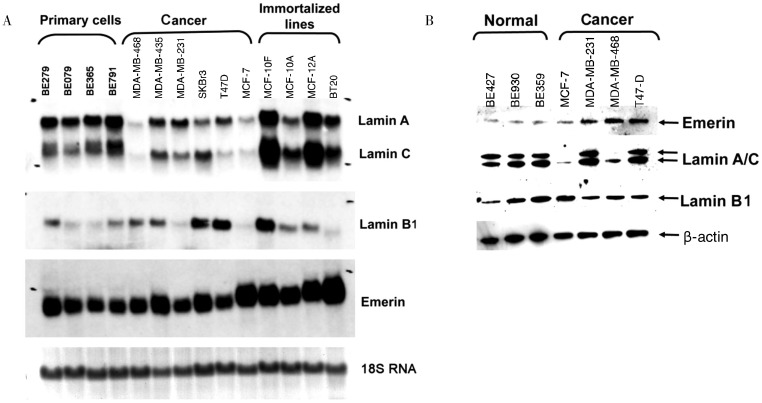
We also investigated the expression of several nuclear envelope proteins in a panel of cell lines. Emerin and lamin B mRNA levels were increased in immortalized and cancer cells compared with primary mammary epithelial cells (Figure 2A), and the expression of the emerin protein also increased in breast cancer cells (Figure 2B). Among several nuclear envelope proteins examined, lamin A/C were absent or greatly reduced in two breast cancer cell lines (MCF-7 and MDA-MB-468), but were present in all primary normal breast epithelial cells as shown by Western blot (Figure 2B).
Figure 2. Expression of nuclear envelope proteins in breast epithelial and cancer cell lines.
Primary non-cancer cells and cancer cell lines were analyzed for mRNA and protein expression of nuclear envelope proteins lamin A/C, lamin B, and emerin. A, total RNA was isolated and analyzed by Northern blot for lamin A/C, lamin B1, and emerin expression in human primary breast epithelial cells (BE279, BE079, BE365, and BE791), non-cancer immortalized breast epithelial lines (BT20, MCF-12A, MCF-10A, and MCF-10F), and breast cancer cells (MDA-MB-468, MDA-MD-435, MDA-MB-231, SKBr3, T47D, and MCF-7). Staining of 18S RNA on the gel was used as total RNA loading control. B, Western blot analysis was performed on total cell lysate from human primary breast epithelial cells (BE427, BE930, and BE359) and breast cancer cells (T47-D, MCF-7, MDA-MB231, and MDA-MB-468) to analyze the expression of lamin A/C, lamin B1, and emerin. Beta-actin was used as protein loading control.
Most breast cancer cell lines and tissues have a heterogeneous lamin A/C protein expression pattern
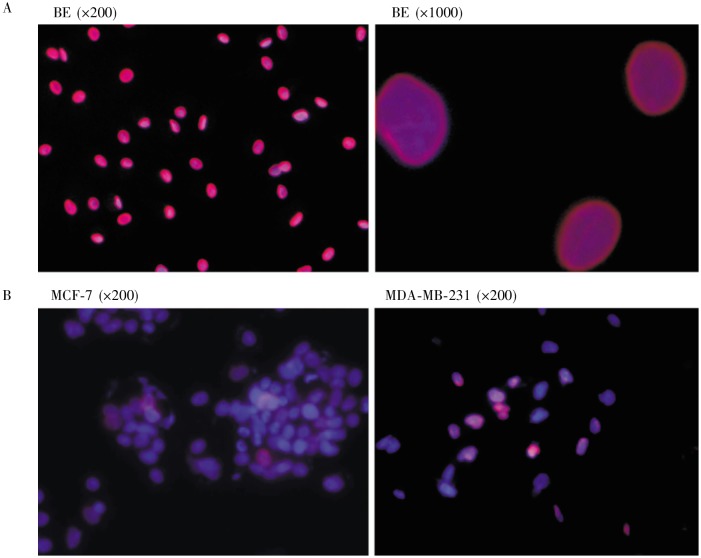
Using immunofluorescence microscopy, we examined the presence of lamin A/C in individual cells. In primary human breast epithelial cells, all nuclei stained strongly for lamin A/C and showed a smooth and oval shaped morphology (Figure 3A). In cancer cell lines, lamin A/C was either lost or expressed heterogeneously (Figure 3B). For example, in MDA-MB-231 breast cancer cells, lamin A/C were positive as detected by Western blot (Figure 1C); however, the cells contained both lamin A/C–positive and –negative populations, as detected by immunofluorescence microscopy (Figure 3B). Heterogeneous expression of lamin A/C was also observed in MCF-7 cells (Figure 3B). Thus, the reduced and heterogeneous expression of lamin A/C observed in tumor tissues by immunohistochemistry was also found in cell lines by Northern blot, Western blot, and immunofluorescence microscopy.
Figure 3. Heterogeneous lamin A/C expression in breast cancer cells.
Lamin A/C expression in normal and cancer breast epithelial cells was analyzed by immunofluorescence microscopy. A, primary human breast epithelial (BE) cells were stained for DAPI (blue) and lamin A/C (red), and representative images, with a magnification of 200x and 1000x, are shown. Five primary breast epithelial lines were examined. B, all breast cancer cell lines show heterogeneous staining of lamin A/C within the cell population. Examples (MCF-7 and MDA-MB-231) are shown.
Lamin A/C suppression in human mammary epithelial cells causes nuclear morphological deformation
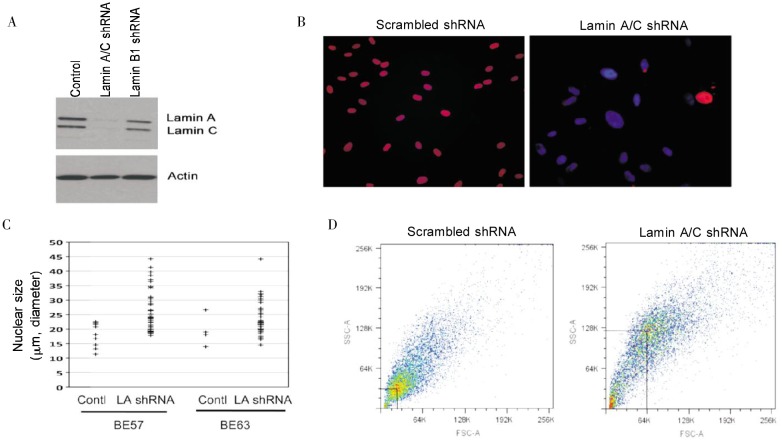
To investigate the potential functional significance of lamin A/C loss in breast cancer, we studied the effect of reduced lamin A/C expression in primary mammary epithelial cells. These human mammary epithelial cells were obtained from breast reduction surgeries of non-cancer patients, and the primary cells can be cultured for about 5–7 passages, with a culture life span of 2–3 months[26]. Suppression of lamin A/C proteins with shRNA was achieved as indicated by Western blot analysis (Figure 4A). Staining of lamin A/C in immunofluorescence microscopy indicated that lamin A/C expression was reduced in more than 90% of the cells (Figure 4B). We noticed a dramatic proportion of cells with large and irregularly shaped nuclei starting three days after targeting lamin A/C with shRNA but did not observe similar phenotypes in control cells treated with scrambled shRNA (Figure 4B). In several experiments using multiple breast epithelial cell preparations, lamin A/C down-regulation consistently resulted in 30%–70% of the cells having large and atypical nuclear morphology. Conversely, cells transfected with control siRNA exhibited mostly smooth, round or oval shaped nuclei, with fewer than 4% of cells having atypical nuclei. In comparison, down-regulation of emerin caused nuclear morphological deformation in approximately 20% of cells (not shown), which is consistent with our previous report[25]. The effect of lamin A/C down-regulation on nuclear size (diameter of the long axis) was quantified by computer-assisted imaging analysis, and the results indicated that lamin A/C down-regulation caused a general increase in nuclear size (Figure 4C). In two independent studies using two preparations of primary breast epithelial cells (BE57 and BE63), the average nuclear size increased from 18 to 32 µm (P < 0.001) and from 20 to 28 µm (P < 0.005), respectively.
Figure 4. Suppression of lamin A/C expression in breast epithelial cells leads to an enlarged and deformed nuclear shape.
Primary breast epithelial cells were treated with shRNA against lamin A/C and analyzed for the effect on nuclear size and morphology. A, Western blot shows the specific suppression of lamin A/C by shRNA in primary breast epithelial cells. B, immunofluorescence microscopy shows that lamin A/C expression (red) was suppressed in the majority of cells, whereas lamin A/C are expressed strongly and evenly in all the nuclei of control cells. The nuclei (DAPI staining, blue) of the control cells are largely smooth and oval shaped, and nuclei in many lamin A/C–suppressed cells are large and irregular. C, two breast epithelial cell lines (BE57 and BE63) were analyzed for nuclear size following shRNA treatment for 3 days. The nuclear size was scored by computer-assisted image analysis (AxioVision software 4) for randomly selected 50 nuclei in the image fields from each group. The length/diameter of the long axis from the 50 nuclei was plotted. The difference was found statistically significant by Student's T test. D, the cell size and granularity (reflecting nuclear size and shape, respectively) were assessed by flow Cytometry and the results were plotted as forward light scattering-area (FSC-A) verses sideward light scattering-area (SSC-A).
To further assess cell size and shape, we used a flow cytometer, which can analyze these properties in a large population of cells in suspension culture. Compared with control cells, the lamin A/C–suppressed cell population shifted to a larger and irregular shape (Figure 4D), as indicated by the increase in FSC-A values and SSC-A values, respectively. Presumably, the size and shape of cells in suspension reflect the changes in size and shape of the nuclei. These experiments were performed over 2 years with four repetitions and independent preparations of primary breast epithelial cells.
Suppression of lamin A/C leads to Polyploidy and aneuploidy in breast epithelial cells
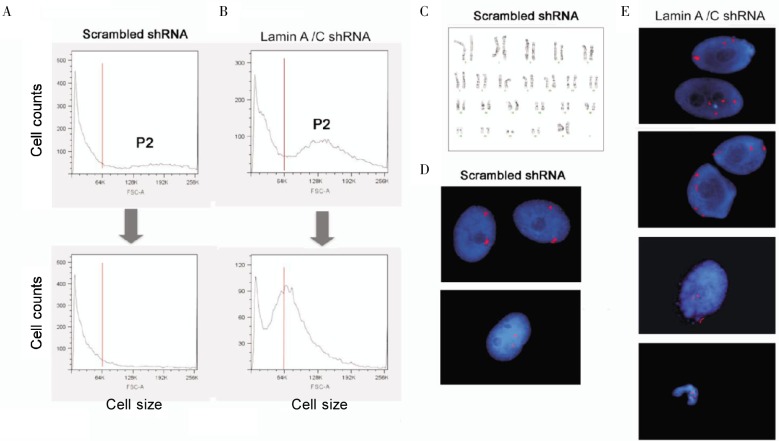
Next, we determined if the increased cell or nuclear size following lamin A/C suppression can be accounted for by increased nuclear DNA content, or Polyploidy and aneuploidy. In subsequent experiments, primary breast epithelial cells were transfected with shRNA vectors and selected with puromycin for 15 days. We collected the larger cells (the P2 population) from both control and lamin A/C–suppressed populations by flow Cytometry. The collected cells were cultured further for 2 weeks and were then re-analyzed by flow Cytometry. In control cells, most of the P2 population re-distributed back into a pattern with mostly smaller (2n) population; however, most (79%) of the P2 population of the lamin A/C– suppressed cells remained larger and only a small fraction of the P2 cells re-distributed back to smaller, 2n cells (Figure 5A, B). We determined chromosomal number by cytogenetic approaches in P2 cells from both the first and second flow sorting. The control cells readily formed metaphase spreads with 46 chromosomes per nucleus (Figure 5C). However, in two attempts with different lines of primary breast epithelial cells, we failed to obtain a sufficient number of metaphase spreads to allow a valid determination of ploidy of lamin A/C–suppressed P2 populations. These results suggest that the P2 cells underwent mitotic arrest and did not complete metaphase.
Figure 5. Lamin A/C suppression in primary breast epithelial cells results in aneuploidy and Polyploidy.
Primary breast epithelial cells were transfected with control or shRNA to suppress lamin A/C. The pools of transfected cells were selected using puromycin for 15 days. Following selection, the control (A) and lamin A/C (B) shRNA-treated cells (BE57) were sorted by flow Cytometry to collect the P2 fraction (by size, FSC-A value) of live G2/M cells. The collected cells of the P2 fraction were cultured for another 15 days and then re-analyzed by flow Cytometry. C, the sorted and re-cultured cells were subjected to cytogenetic analysis of metaphase. Metaphase spreads containing 46 chromosomes from the control cells treated with scrambled shRNA were readily observed, as shown in the example. Metaphase spreads were not observed in cells treated with shRNA targeting lamin A/C. The cells were also subjected to FISH analysis using X (red) and Y (green) chromosome probes in 50 nuclei examined, as shown in 2 examples for controls (D) and 4 examples for lamin A/C–suppressed cells (E).
Nevertheless, we were able to determine the ploidy of the interphase cells using fluorescence in situ hybridization (FISH) with X (red) and Y (green) chromosome probes. In control cells, all 50 nuclei analyzed were diploid as shown in two fields (Figure 5D); however, 12% of the lamin A/C–suppressed cells contained 3 to 4 X-chromosomes in each nucleus as shown in four different fields of examples (Figure 5E), suggesting these cells are polyploid or aneuploid. In the examples, a nucleus containing a third X-chromosome in a micronuclear or nuclear protrusion (third panel down, Figure 5E) and a condensed nucleus containing three X-chromosomes (lowest panel, Figure 5E) were observed. The result of the FISH technique is likely an underestimation of the percentage of aneuploid cells because the other 2-X-chromosome–containing nuclei may gain or lose chromosome (s) other than the X-chromosome. Thus, suppression of lamin A/C leads to the formation of tetraploid and aneuploid breast epithelial cells.
Suppression of lamin A/C leads to reduced mitosis and retarded cell growth
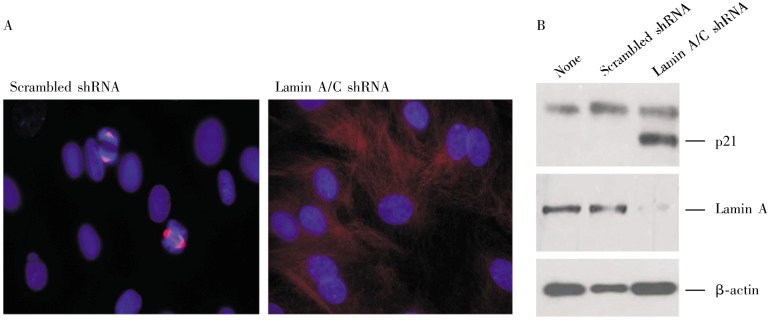
The observation of the lack of metaphase spread suggests that suppression of lamin A/C may interfere with cell proliferation and normal mitosis of primary breast epithelial cells. Although primary breast cells grow slower than cancer cells in culture, we observed that about 9% of the cells contained mitotic spindles as shown by staining for β-tubulin (Figure 6A). In contrast, mitotic spindles were rarely observed (less than 1%) in lamin A/C–suppressed cells (Figure 6A), indicating a reduced rate of cell division following the lamin A/C suppression and acquiring of Polyploidy and aneuploidy.
Figure 6. Lamin A/C–suppressed primary breast epithelial cells fail to undergo mitosis.
Primary breast epithelial cells were transfected with scrambled or lamin A/C shRNA. The pools of transfected cells were selected using puromycin for 15 days. Following selection, the cells were sorted by flow Cytometry to collect the P2 fraction (by size, FSC-A value) of live G2/M cells. The cells collected from the P2 fraction were cultured and analyzed. A, the cells were analyzed by immunofluorescence microscopy of β-tubulin (red) to detect mitotic cells. Beta-tubulin-stained mitotic spindles in cells treated with control shRNA were observed. Mitotic spindles were rarely observed in lamin A/C–suppressed cells. B, the cells were subjected to Western blot analysis for lamin A/C and p21 expression, with β-actin as a protein loading control. Cells without shRNA transfection or flow Cytometry sorting were used as controls.
Aneuploidy and polypoidy were reported to trigger cell growth arrest through the p53/p21 pathway[30]. Thus, we analyzed the expression of the cyclin inhibitor p21 by Western blot (Figure 6B). We observed the induction of p21 in lamin A/C–suppressed cells but not in untreated or vector-transfected control cells. Thus, we reason that the occurrence of aneuploidy and Polyploidy in lamin A/C–suppressed cells induces p21 and subsequent cell growth arrest, providing an explanation for the inability to obtain metaphase spreads in lamin A/C–suppressed cells.
Discussion
Unlike in ovarian cancer in which emerin expression is lost[25], emerin is expressed abundantly in breast cancer cells; instead, lamin A/C proteins are often reduced or absent in breast cancer. Lamin A/C have been suggested as a biomarker for various types of cancer[31]. Lamin A/C are absent in embryonic stem cells and some adult stem cell types but are expressed in the majority of cell types in adults, thereby suggesting that their expression is an indicator of differentiation[32],[33]. Loss of lamin A/C expression has been reported for colon cancer[34],[35], lung cancer[36], prostate cancer[10], gastric cancer[37], and leukemia and lymphoma[38],[39]. The current study is the first report on a loss of lamin A/C expression in breast cancer. In the analysis of cell lines, it appears that loss of lamin A/C occurs primarily at protein level, instead of mRNA level, which may explain why loss or reduction of lamin A/C has not been recognized or reported previously based on expression array studies.
The loss of lamin A/C in breast cancer provides an obvious explanation for the deformed nuclear morphology of cancer, including breast carcinoma cells. Previously, the correlation between nuclear size and shape and degree of malignancy has been appreciated[3],[6],[7], but a molecular mechanism has not been well defined[3]. In basic cell biology studies, the role of lamin A/C in determining cell shape has been well established[40]. The current investigation makes this translational link, where loss of the lamin A/C protein accounts for the deformed nuclear morphology in a significant percentage of breast cancer (38% of the 56 invasive ductal carcinomas examined are lamin A/C–negative). Furthermore, most breast cancers examined exhibit a heterogeneous pattern of lamin A/C expression in the population of cancer cells, which may account for the variation of nuclear shapes of the cancer cells within a tumor.
Another important conclusion from the current study is that loss of lamin A/C may account for aneuploidy and chromosomal numerical instability of breast cancer cells. Thus, in addition to non-disjunction in chromosome segregation, a defective nuclear envelope may account for the generation of aneuploidy. Simply, inefficiency in the formation of new nuclear envelope to cage the newly formed nuclei likely leads to furrow regression and cytokinesis failure. In C. elegans, which has only one lamin gene, lamin plays a critical role in chromatin segregation and is essential for mitosis[41]. In mammals, although lamin A/C may be dispensable for cytokinesis and even for development, as shown in lamin A/C knockout mice[42], a role for lamin A/C in mitosis and the formation of new nuclear envelope immediately following chromatin separation is recognized [43],[44]. It may be speculated that, though non-essential, lamin A/C loss may reduce the efficiency of nuclear envelope formation in mitosis. Thus, mitotic failure and furrow regression may be more frequent in the absence of lamin A/C in the breast epithelial cells, resulting in the formation of tetraploid intermediates and subsequent aneuploid cell generation.
We found that lamin A/C–suppressed, aneuploid and/or polyploid cells were not able to form metaphase spreads and had a reduced mitotic population. This observation suggests that the cells were under growth arrest. This is consistent with the report that aneuploidy and Polyploidy activate the p53/p21 pathway to induce growth arrest[30]. Aneuploid cells may have a reduced growth and survival ability initially[21],[23]. However, clones with optimal chromosomal composition or oncogenic mutation(s) may be selected and ultimately expand and transform. Our finding that a reduction of lamin A/C in cancer cells may be an important mechanism for aneuploidy and chromosomal numerical instability provides rationale to design and test the hypothesis that the loss of lamin A/C, with additional oncogenic mutation(s) to overcome growth impairment, may enable breast epithelial cells to transform into aneuploid and neoplastic cells resembling human mammary carcinomas.
The loss of lamin A/C expression or mutations in both humans and mice are not known to link with cancer. A possible explanation is that the severity of diseases or phenotypes related to muscular dystrophy and cardiomyopathy does not allow the presentation of cancer that usually occurs in older age. We also observed that lamin A/C expression is reduced or heterogeneously expressed instead of completely absent in breast cancer. It is possible that a certain event (such as degradation) leads to loss of lamin A/C at one stage, and aneuploidy develops. Subsequently, the return of lamin A/C expression may benefit the mitotic machinery of the proliferating cancer cells.
In summary, our study suggests that reduction/loss of lamin A/C may be the cause of both nuclear morphological deformation and aneuploidy, two prominent characteristics of cancer cells. Further elucidation of this molecular mechanism will provide a basis for additional study to answer the question on the role of aneuploidy in breast cancer development.
Acknowledgments
We thank Lisa Vanderveen Carolyn Slater, and Malgorzata Rula at the Fox Chase Cancer Center for their excellent technical assistance in preparing and Culturing primary human mammary epithelial cells and their analysis. We acknowledge Shannon Opiela, Jay Enten, and James Phillips (Flow Cytometry Core), and Carmen Casas and Ana Prada (Cytogenetic Core) at the University of Miami Miller School of Medicine for the assistance and service from their core facility. We thank our colleague, Drs. Elizabeth Smith and Robert Moore, as well as our laboratory members and colleagues for reading, suggestions, and editing during the course of the experiments and the preparation of the manuscript.
These studies were supported by funds from concept awards BC097189 and BC076832 from Department of Defense (USA). Grants R01 CA095071, R01 CA099471, and CA79716 to Xiang-Xi Xu from NCI, NIH (USA) also contributed to the studies.
References
- 1.Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes [J] Nature. 2009;462(7276):1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers [J] Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 3.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells [J] Nat Rev Cancer. 2004;4(9):677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 4.Papanicolaou GN. A new procedure for staining vaginal smears [J] Science. 1942;95(2469):438–439. doi: 10.1126/science.95.2469.438. [DOI] [PubMed] [Google Scholar]
- 5.Nickerson JA. Nuclear dreams: the malignant alteration of nuclear architecture [J] J Cell Biochem. 1998;70(2):172–180. [PubMed] [Google Scholar]
- 6.Pienta KJ, Coffey DS. Correlation of nuclear morphometry with progression of breast cancer [J] Cancer. 1991;68(9):2012–2016. doi: 10.1002/1097-0142(19911101)68:9<2012::aid-cncr2820680928>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 7.Hsu CY, Kurman RJ, Vang R, et al. Nuclear size distinguishes low- from high-grade ovarian serous carcinoma and predicts outcome [J] Hum Pathol. 2005;36(10):1049–1054. doi: 10.1016/j.humpath.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Boyd J, Pienta KJ, Getzenberg RH, et al. Preneoplastic alterations in nuclear morphology that accompany loss of tumor suppressor phenotype [J] J Natl Cancer Inst. 1991;83(12):862–866. doi: 10.1093/jnci/83.12.862. [DOI] [PubMed] [Google Scholar]
- 9.Fischer AH, Taysavang P, Jhiang SM. Nuclear envelope irregularity is induced by RET/PTC during interphase [J] Am J Pathol. 2003;163(3):1091–1100. doi: 10.1016/S0002-9440(10)63468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debes JD, Sebo TJ, Heemers HV, et al. p300 modulates nuclear morphology in prostate cancer [J] Cancer Res. 2005;65(3):708–712. [PubMed] [Google Scholar]
- 11.Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris [J] J Cell Sci. 2008;121(Suppl 1):1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- 12.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis [J] Nat Rev Mol Cell Biol. 2009;10(7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roschke AV, Tonon G, Gehlhaus KS, et al. Karyotypic complexity of the NCI-60 drug-screening panel [J] Cancer Res. 2003;63(24):8634–8647. [PubMed] [Google Scholar]
- 14.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability [J] Curr Biol. 2010;20(6):R285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajagopalan H, Lengauer C. Aneuploidy and cancer [J] Nature. 2004;432(7015):338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 16.Duesberg P. Does aneuploidy or mutation start cancer? [J] Science. 2005;307(5706):41. doi: 10.1126/science.307.5706.41d. [DOI] [PubMed] [Google Scholar]
- 17.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer [J] Curr Opin Genet Dev. 2007;17(2):157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines [J] Nature. 2005;437(7061):1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 19.Margolis RL. Tetraploidy and tumor development [J] Cancer Cell. 2005;8(5):353–354. doi: 10.1016/j.ccr.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? [J] Curr Opin Cell Biol. 2006;18(6):658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Williams BR, Prabhu VR, Hunter KE, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells [J] Science. 2008;322(5902):703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver BA, Cleveland DW. The aneuploidy paradox in cell growth and tumorigenesis [J] Cancer Cell. 2008;14(6):431–433. doi: 10.1016/j.ccr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance [J] Genetics. 2008;179(2):737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pihan G, Doxsey SJ. Mutations and aneuploidy: co-conspirators in cancer? [J] Cancer Cell. 2003;4(2):89–94. doi: 10.1016/s1535-6108(03)00195-8. [DOI] [PubMed] [Google Scholar]
- 25.Capo-chichi CD, Cai KQ, Testa JR, et al. Loss of GATA6 leads to nuclear deformation and aneuploidy in ovarian cancer [J] Mol Cell Biol. 2009;29(17):4766–4777. doi: 10.1128/MCB.00087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith ER, Cai KQ, Smedberg JL, et al. Nuclear entry of activated MAPK is restricted in primary ovarian and mammary epithelial cells [J] PLoS One. 2010;5(2):e9295. doi: 10.1371/journal.pone.0009295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai KQ, Klein-Szanto A, Karthik D, et al. Age-dependent morphological alterations of human ovaries from populations with and without BRCA mutations [J] Gynecol Oncol. 2006;103(2):719–28. doi: 10.1016/j.ygyno.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 28.Cai KQ, Caslini C, Capo-chichi CD, et al. Loss of GATA4 and GATA6 expression specifies ovarian cancer histological subtypes and precedes neoplastic transformation of ovarian surface epithelia [J] PLoS One. 2009;4(7):e6454. doi: 10.1371/journal.pone.0006454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Capo-chichi CD, Roland IH, Vanderveer L, et al. Anomalous expression of epithelial differentiation-determining GATA factors in ovarian tumorigenesis [J] Cancer Res. 2003;63(16):4967–4977. [PubMed] [Google Scholar]
- 30.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism [J] J Cell Biol. 2010;188(3):369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster CR, Przyborski SA, Wilson RG, et al. Lamins as cancer biomarkers [J] Biochem Soc Trans. 2010;38(Pt 1):297–300. doi: 10.1042/BST0380297. [DOI] [PubMed] [Google Scholar]
- 32.Rober RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study [J] Development. 1989;105(2):365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- 33.Lin F, Worman HJ. Expression of nuclear lamins in human tissues and cancer cell lines and transcription from the promoters of the lamin A/C and B1 genes [J] Exp Cell Res. 1997;236(2):378–384. doi: 10.1006/excr.1997.3735. [DOI] [PubMed] [Google Scholar]
- 34.Willis ND, Cox TR, Rahman-Casafls SF, et al. Lamin A/C is a risk biomarker in colorectal cancer [J] PLoS One. 2008;3(8):e2988. doi: 10.1371/journal.pone.0002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moss SF, Krivosheyev V, de Souza A, et al. Decreased and aberrant nuclear lamin expression in gastrointestinal tract neoplasms [J] Gut. 1999;45(5):723–729. doi: 10.1136/gut.45.5.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machiels BM, Broers JL, Raymond Y, et al. Abnormal A-type lamin organization in a human lung carcinoma cell line [J] Eur J Cell Biol. 1995;67(4):328–335. [PubMed] [Google Scholar]
- 37.Wu Z, Wu L, Weng D, et al. Reduced expression of lamin A/C correlates with poor histological differentiation and prognosis in primary gastric carcinoma [J] J Exp Clin Cancer Res. 2009;28(1):8. doi: 10.1186/1756-9966-28-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadelmann B, Khandjian E, Hirt A, et al. Repression of nuclear lamin A and C gene expression in human acute lymphoblastic leukemia and non-Hodgkin's lymphoma cells [J] Leuk Res. 1990;14(9):815–821. doi: 10.1016/0145-2126(90)90076-l. [DOI] [PubMed] [Google Scholar]
- 39.Agrelo R, Setien F, Espada J, et al. Inactivation of the lamin A/C gene by CpG island promoter hypermethylation in hematologic malignancies, and its association with poor survival in nodal diffuse large B-cell lymphoma [J] J Clin Oncol. 2005;23(7):3940–3947. doi: 10.1200/JCO.2005.11.650. [DOI] [PubMed] [Google Scholar]
- 40.Lammerding J, Fong LG, Ji JY, et al. Lamins A and C but not lamin B1 regulate nuclear mechanics [J] J Biol Chem. 2006;281(35):25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Rolef Ben-Shahar T, Riemer D, et al. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes [J] Mol Biol Cell. 2000;11(11):3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan T, Escalante-Alcalde D, Bhatt H, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy [J] J Cell Biol. 1999;147(5):913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao K, Capell BC, Erdos MR, et al. A lamin A protein isoform overexpressed in Hutchinson-Gilford progena syndrome interferes with mitosis in progeria and normal cells [J] Proc Natl Acad Sci USA. 2007;104(12):4949–4954. doi: 10.1073/pnas.0611640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dechat T, Shimi T, Adam SA, et al. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging [J] Proc Natl Acad Sci USA. 2007;104(12):4955–4960. doi: 10.1073/pnas.0700854104. [DOI] [PMC free article] [PubMed] [Google Scholar]