Abstract
Background
Sacoglossan sea slugs are well known for their unique ability among metazoans to incorporate functional chloroplasts (kleptoplasty) in digestive glandular cells, enabling the slugs to use these as energy source when starved for weeks and months. However, members assigned to the shelled Oxynoacea and Limapontioidea (often with dorsal processes) are in general not able to keep the incorporated chloroplasts functional. Since obviously no algal genes are present within three (out of six known) species with chloroplast retention of several months, other factors enabling functional kleptoplasty have to be considered. Certainly, the origin of the chloroplasts is important, however, food source of most of the about 300 described species is not known so far. Therefore, a deduction of specific algal food source as a factor to perform functional kleptoplasty was still missing.
Results
We investigated the food sources of 26 sacoglossan species, freshly collected from the field, by applying the chloroplast marker genes tufA and rbcL and compared our results with literature data of species known for their retention capability. For the majority of the investigated species, especially for the genus Thuridilla, we were able to identify food sources for the first time. Furthermore, published data based on feeding observations were confirmed and enlarged by the molecular methods. We also found that certain chloroplasts are most likely essential for establishing functional kleptoplasty.
Conclusions
Applying DNA-Barcoding appeared to be very efficient and allowed a detailed insight into sacoglossan food sources. We favor rbcL for future analyses, but tufA might be used additionally in ambiguous cases. We narrowed down the algal species that seem to be essential for long-term-functional photosynthesis: Halimeda, Caulerpa, Penicillus, Avrainvillea, Acetabularia and Vaucheria. None of these were found in Thuridilla, the only plakobranchoidean genus without long-term retention forms. The chloroplast type, however, does not solely determine functional kleptoplasty; members of no-retention genera, such as Cylindrobulla or Volvatella, feed on the same algae as e.g., the long-term-retention forms Plakobranchus ocellatus or Elysia crispata, respectively. Evolutionary benefits of functional kleptoplasty are still questionable, since a polyphagous life style would render slugs more independent of specific food sources and their abundance.
Keywords: DNA-barcoding, Food analyses, Kleptoplasty, rbcL, Sacoglossa, tufA
Background
Sacoglossa sea slugs are a relatively small group of heterobranch gastropods with currently about 300 species described [1], comprising the shelled Oxynoacea and the shell-less Plakobranchacea. The latter is further divided into the ceras-bearing Limapontioidea (although not monophyletic in phylogenetic analyses) and the parapodia bearing Plakobranchoidea [2-4]. Because of their ability to incorporate functional chloroplasts and subsequently use them for sustenance during starvation over weeks and months Sacoglossa fascinated scientists over decades. This feature is unique among metazoan life forms [3,5-8] and only known elsewhere in members of the Foraminifera (Rhizaria) [9], Ciliophora (Alveolata) [10] and in Dinoflagellata (Alveolata) [11]. Within Sacoglossa usually three states of kleptoplasty are differentiated, either based on measurements of functional chloroplasts with Pulse Amplitude Modulated (PAM) Fluorometry or on CO2 fixation experiments [3,12,13]. Following the classification of Händeler et al. [3], specimens that are not able to incorporate chloroplasts functional are considered as no-retention forms (NR); specimens that are able to incorporate functional chloroplasts for up to two weeks of starvation are called short-term-retention forms (StR) and those incorporating functional chloroplasts for over 20 days during starvation are called long-term-retention forms (LtR). The fact that chloroplasts survive for weeks and months in the slug’s digestive system and perform functional photosynthesis, despite the absence of the algal nucleus, has led to the intriguing hypothesis that a horizontal gene transfer must have occurred from the algal organisms into the metazoan life form [7,12,14]. Up to now, a few single genes amplified from Elysia chlorotica, Gould [15] were interpreted to encode proteins relevant for photosynthesis, such as Lhc (light harvesting complex), fcp (fucoxanthin protein), psbO (manganese stabilizing protein), and others [7,12,16,17].
Analyzing transcriptomic data for the first time, Wägele et al. [18] found no evidence for a horizontal gene transfer in two sacoglossan species known to maintain chloroplasts for several months, Elysia timida Risso [19] and Plakobranchus ocellatus van Hasselt [20]. Similar negative results were obtained later for Elysia chlorotica by transcriptomic and genomic data [21,22]. Conflicting results of Pierce et al. [23] that suggested a gene transfer based on few single reads in their transcriptomic data, have most recently been interpreted in a different way [24]. Thus, in our view the hypothesis that slugs are able to actively support the kleptoplasts via the translational products of transferred algal genes is rejected and the focus for understanding long-term incorporation of kleptoplasts has to include other factors, like properties of chloroplasts and their origin with regard to functional photosynthesis. We clearly demonstrated this recently by investigating Plakobranchus ocellatus during various starvation periods [25]: Despite a broad documented food range in fresh caught animals, only chloroplasts from the ulvophycean Halimeda macroloba Decaisne [26] remained in the digestive gland after a starvation period of two months. Unfortunately, our knowledge on sacoglossan food preferences is still scarce. In general the slugs are considered to be stenophagous and mainly sequester members of the Ulvophyceae sensu Floyd & O’Kelly [22,27-33]. Few species (Elysia crispata, E. clarki and P. ocellatus) are known to feed on a high variety of algae [18,25,30,34-37]. Only some species are recorded to feed on specific members of Rhodophyta (e.g., Hermaea bifida) [38,39], Heterokontophyta (e.g., Elysia chlorotica) or sea grasses (Elysia serca, Marcus [40]) [3,30,31,41]. Food sources were usually identified by observation, feeding experiments [28,33,42], or by electron microscopically studies of chloroplast types [43,44]. But it is obvious that, especially in potential polyphagous sea slugs, not every food alga may be detected by feeding experiments, in particular when not knowing, which ones to offer. On the other hand, slugs may feed on alternative food sources during food limitation rather than having these as host algae. Molecular barcoding has proved to be a high efficient method to identify algal food sources even when using a single barcoding-marker [30,34-37], instead of two [25]. This now well-established method opens the opportunity to study sacoglossan food sources and origin of kleptoplasts in a highly reliable mode. To find a pattern in chloroplast origin and functional kleptoplasty, a profound database on sequestered chloroplasts for all sacoglossan groups, and especially those taxa hardly studied at all, e.g., the genus Thuridilla[18,30], is needed. Following the methods introduced previously [30,34,35] we investigated chloroplast origin in 26 non-starved sacoglossan species, including NR, StR and LtR forms, by DNA-barcoding using the chloroplast markers tufA and rbcL to enlarge our insight in, and state more precisely, sacoglossan food spectrum. We combined the identified food sources with literature data of species for which retention ability is documented and analyzed in combination with this information if there is a correlation between food sources and retention-form. DNA-barcoding has become an important method in identifying plastid origin in Sacoglossa, but due to varying results and minor pitfalls in the application of rbcL, we compared reliability of this gene.
Results
Barcoding of tufA and rbcL
We successfully identified food sources for 30 sacoglossan specimens (overall 26 species) by at least applying one barcoding marker (Additional file 1). For 19 of the 26 species included here we were able to identify the food sources for the first time. For the remaining seven species, we could confirm literature data or even enlarged the food range (Additional file 1). Yet, no rbcL amplification products were obtained for four specimens and no tufA product was obtained for seven specimens (Additional file 1). TufA sequences obtained from Elysia amakusana (703) Baba [45], Elysia sp. (841), Cyerce nigra (860) Bergh [46] and Costasiella sp. (863) exclusively represented sequences of bacterial origin.
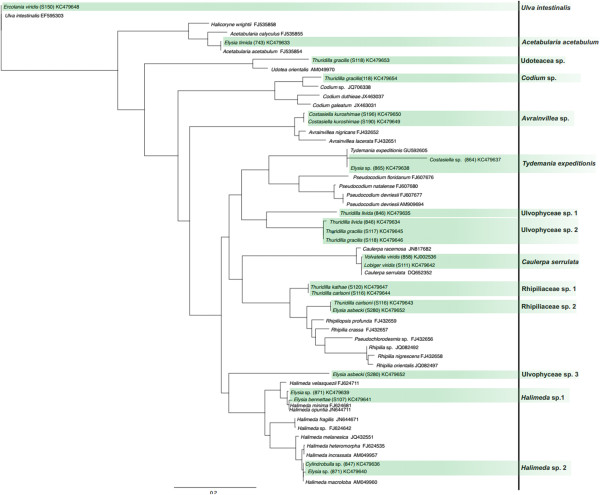
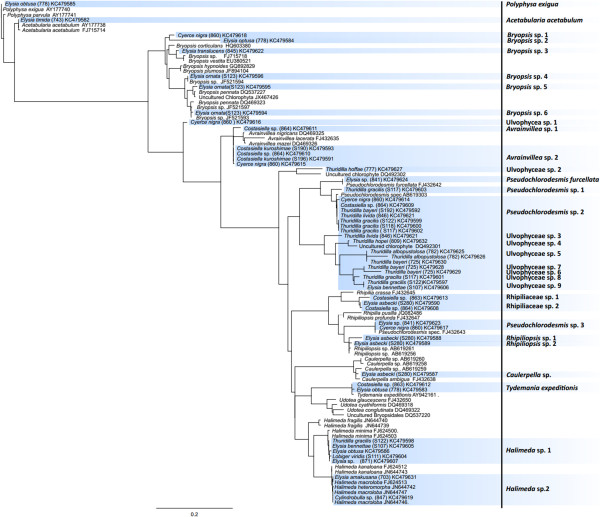
We were able to identify 14 different algal genera combining both markers: eleven genera can be assigned to Bryopsidophyceae, two to Dasycladophyceae and one to Ulvophyceae. RbcL revealed about two times more different haplotypes than tufA (30 for rbcL and 14 for tufA, respectively, Figures 1 and 2), though the number of genera is equal (9 for rbcL and 10 for tufA, respectively). Nine haplotypes for rbcL and three haplotypes for tufA could not be assigned to a certain algal genus, because of missing reference sequences in GenBank. We are not able to clarify if some of the unidentified haplotypes of rbcL and tufA actually represent the same algal species. Especially for rbcL almost always a higher number of algal haplotypes for a distinct sacoglossan species was revealed, with the highest number of haplotypes for an unknown sister clade of Pseudochlorodesmis (7) and for Bryopsis (6) (Additional file 1, Figures 1 and 2).
Figure 1.
Food sources identified using tufA as barcode marker. ML tree of identified algal haplotypes in Sacoglossa by using tufA (highlighted in green). Identified haplotype is noted on the right side. When sequence match was < 99%, higher taxon name of the algae that formed a monophyletic group with the corresponding haplotype was used. Haplotypes with no monophyletic grouping are named “Ulvophyceae sp.”.
Figure 2.
Food sources identified using rbcL as barcode marker. ML tree of identified algal haplotypes in Sacoglossa by using rbcL (highlighted in blue). Identified haplotype is noted on the right side. When sequence match was < 99%, higher taxon name of the algae that formed a monophyletic group with the corresponding haplotype was used. Haplotypes with no monophyletic grouping are named “Ulvophyceae sp.”.
Food sources and retention form
We combined the information obtained from barcoding and literature data, and analyzed this information with regard to the according retention-form. We found that the food of NR forms covers a broad spectrum of ulvophycean algae and includes items that are not consumed by StR or LtR forms: Boodlea, Chlorodesmis, Ulva, Urospora, red algae, angiosperms and eggs of other sacoglossans (Figure 3). In contrast, algal taxa like Poropsis, Rhipidosiphon, Rhipocephalus, Udotea and an unidentified member of the Ulvophyceae were not identified so far as food sources for NR-forms.
Figure 3.
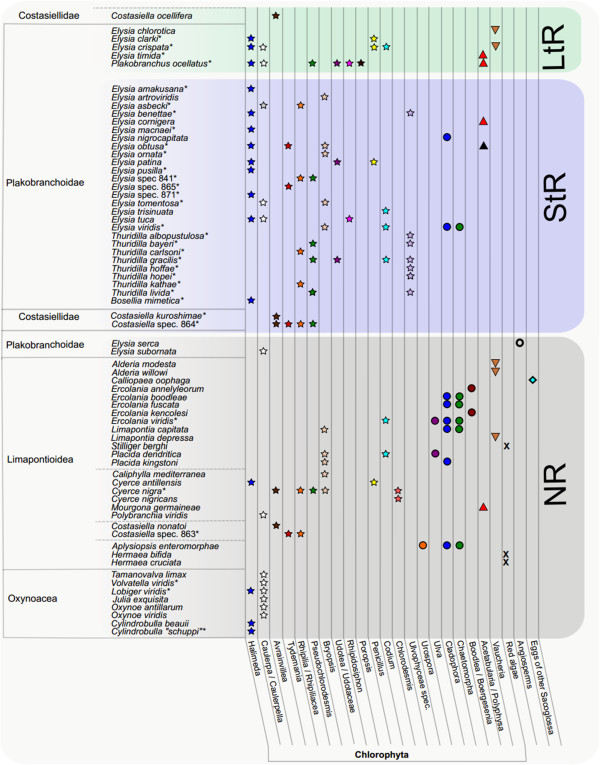
Food spectrum of Sacoglossa compared to functional retention form. Food sources of 68 species of Sacoglossa are shown. The classification of Sacoglossa is based on works of [3,4,46]. Asterisk indicates molecular identification of food (overall 33 species). Food sources were either obtained in this study or taken out of literature [3,4,18,25,30,34-37,80-83]. Classification of retention ability was done according to literature data [3,18,47,49,80-84]. LtR = Long-term-retention, StR = short-term-retention, NR = no retention. Stars indicate members of the Bryopsidales, filled circles members of the Ulvophyceae other than Bryopsidales; triangles represent Dasycladales, inverted triangles Heterokontophyta, X red algae, circle sea grass and diamond eggs of other Sacoglossa.
LtR forms consume various ulvophycean algal species, but prefer Halimeda, Caulerpa, Penicillus and Avrainvillea, the dasycladalen Acetabularia and the heterokontophyte Vaucheria (Figure 3). However, some NR and LtR forms belonging to different sacoglossan families [5] sequester the same algal species (e.g. Halimeda, Caulerpa, Avrainvillea, Vaucheria); therefore a correlation between food sources and functional kleptoplasty cannot be solely algal based. Interestingly, Thuridilla species that only comprise StR forms rarely feed on plastids sequestered by LtR forms of Elysia and Plakobranchus ocellatus (Figure 3).
Within sacoglossan clades, differences in preferred food items exist. All investigated members of the Oxynoacea are NR forms and stenophagous. This is in contrast to all other (non oxynoacean) sacoglossan genera that include polyphagous species (Figure 3). Cylindrobulla is the only member of the Oxynoacea that exclusively feeds on Halimeda, whereas the members of the remaining Oxynoacea specialized on Caulerpa species, with one exception that the rbcL analysis in Lobiger viridis revealed additional Halimeda as food source (Additional file 1, Figure 3). Within Limapontioidea, several species have a broad food spectrum, like Costasiella sp. (864) with at least four and Cyerce nigra Bergh [46] with at least five different food sources, respectively. However, Halimeda and Caulerpa, the major oxynoacean food, is scarcely represented amongst the food items of Limapontioidea and only found in Cyerce antillensis and Polybranchia viridis, respectively. Interestingly, Costasiella ocellifera Simroth [48], the only non-plakobranchoid LtR form ([49], unpublished data), feeds exclusively on Avrainvillea nigricans.
Four genera of the Plakobranchoidea that all include functional retention forms, are investigated. Bosellia mimetica is stenophagous, feeding on Halimeda, contrary to the polyphagous Plakobranchus ocellatus that feeds on seven different algae (including Halimeda, Caulerpa and Udotea) exhibiting the most diverse food spectrum of all investigated sacoglossans so far (Figure 3); however, these data are obtained from different specimens of different localities [25,37]. The geographically wide distributed taxon Plakobranchus might show geographic differences in the consumed food items. Furthermore, cryptic speciation with already ecological differences in feeding can also not be ruled out [50]. Based on the molecular phylogeny of Händeler et al. [3], Thuridilla is the sister taxon to Plakobranchus, its members principally feeding on a variety of food sources, though Thuridilla species neither consume Halimeda, nor Caulerpa or Poropsis (food sources of Plakobranchus), but a bryopsidophycean genus related to Pseudochlorodesmis (Figure 2). This peculiar food item could not be identified more specifically due to lack of reference sequences in GenBank. Members of the fourth investigated genus, Elysia, consume food items recorded also for the shelled sacoglossans, Halimeda and Caulerpa. Additionally, many Elysia species feed on the same bryopsidophycean genera similar to those observed in Thuridilla (exceptions see above) or Plakobranchus. Some species became very specialized. Elysia chlorotica feeds on Vaucheria (like the limapontioidean genus Alderia), whereas its sister taxon, Elysia serca, is reported to feed solely on higher plants (i.e., sea grasses [31]), a unique feature within Sacoglossa. Several species are confirmed in their specific narrow food spectrum, e.g., Elysia timida, which exclusively feeds on the dasycladophycean Acetabularia acetabulum P.C. Silva [51].
Discussion
Food sources and retention form
We analyzed a wide spectrum of sacoglossan sea slugs with regard to their food preferences and plastid origin by using a combination of two barcoding markers, rbcL and tufA, instead of one [3,18,34-37]. This method proved to be more rapid and more precise in identification of food sources of a large set of Sacoglossa sea slugs, compared to former feeding experiments or feeding observations. Although not every algal food source is identified on species level, the information we provide here increases our knowledge considerably and can now be used for further ecological or behavioral studies.
We found that the ability to perform photosynthesis for at least several days and weeks is established with certain food sources (members of the genera Elysia, Bosellia and Plakobranchus, see Figure 3 and [3]). At least one of the following six algae, the bryopsidophycean Halimeda, Caulerpa, Penicillus and Avrainvillea, the heterokontophyte Vaucheria, or the dasycladophycean Acetabularia, seem to be essential for establishing long-term-retention. However, non-retention forms also feed on these specific algae and are not able to perform photosynthesis. Both aspects indicate that a special type of chloroplast is not sufficient to establish functional kleptoplasty and intrinsic factors of the slugs also contribute to a functional photosynthesis. Here, we pave the way for further research on these specific algae consumed by LtR forms to better understand which genetically and physiological commonalities these target plastids may have. Pelletreau et al. [21] recently considered special abilities of chloroplasts from respective host algae as relevant for long-term incorporation, an option already mentioned earlier [52,53]. A factor now found to possible enhance plastid longevity is ftsH, a protein important for the repair of Photosystem II. It is plastid encoded in the food algae of the two LtR forms Elysia chlorotica and Plakobranchus ocellatus[54]. With the information of this study we are now able to specifically examine the presence of ftsH in plastids of the identified food sources of other retention forms and correlate plastid origin, photosynthetic capability and plastid longevity. Interestingly, Thuridilla species – as close relatives of Plakobranchus and Elysia – and which exhibit extremely short chloroplast retention [3,55], did not reveal any of these six algal species mentioned above (Additional file 1, Figure 3).
Besides plastid origin, several other factors surely influence survival of chloroplasts and render them nutritional at least in sacoglossans in one way or the other. A horizontal gene transfer as a factor was excluded not only for the sea slugs Plakobranchus ocellatus, Elysia timida and E. chlorotica[18,22], but also for the Foraminifera Elphidium margaritaceum and most likely for the dinoflagellate Dinophysis acuminate[56,57]. Common to all three different systems is that there is no mechanical grinding before chloroplast uptake. E.g., the sacoglossans pierce the cell wall with their teeth and then suck out the cytoplasm of the algae, which is then processed in the digestive gland. Second, the intracellular digestion of plastids seems to be stopped or regulated in a way yet to be discovered.
Händeler et al. [3] have shown in their phylogenetic analysis by using ancestral character state reconstruction, that the ancestor of the Plakobranchoidea most likely developed an unknown mechanism, which hinders direct digestion of chloroplasts. However, the genus Costasiella was not included in their analysis, which is now verified in comprising both, LtR and StR species ([49], [58], unpublished data). Thus, at least in Costasiella, this mechanism must be developed as well. Evertsen & Johnsen [59] showed that in Elysia viridis starch grains persist throughout degradation of plastids and are not broken up, as was the case in the NR form Placida dendritica (Alder & Hancock) [60]. If the slugs cannot access the photosynthetic produced starch of functional chloroplasts in the first place, the benefit, under starvation conditions, has to arise from another process. In this case the accumulated photosynthates would only become accessible for slug metabolism after the complete degradation of the chloroplast. The nutritional benefit of the chloroplasts would lie in the presence of an additional food reservoir (starch) that becomes available only after degradation or digestion of the chloroplasts. This is in contrast to former hypotheses that usually assume a continuous supply of photosynthates and therefore a continuous exchange between chloroplast and slug’s cytosol [61,62].
General food source of Sacoglossa
Our results in combination with literature data show that sacoglossans generally feed on a high variety of algal species, although the majority prefers bryopsidophycean taxa. Jensen [4] assumed Halimeda as the ancestral food of Sacoglossa based on her observations on Cylindrobulla. We can confirm here by molecular analyses of the gut content that this species exclusively feeds on Halimeda, whereas all other oxynoacean species feed on Caulerpa, though Lobiger viridis seems also to at least feed additional on Halimeda. The position of Cylindrobulla within the Oxynoacea is not resolved yet, thus the ancestral food source of this clade cannot be deduced [3,63-65]. Future phylogenetic studies are needed to address these questions. All higher sacoglossan taxa switched to other food sources than Halimeda (especially members of the Limapontioidea), or broadened their food spectrum (many Elysia species).
Why some species feed on several different algal species and others specialized on just one prey species, is difficult to explain. Further information on biology has to be considered, e.g., incorporation of defensive compounds obtained via sequestration of toxic algae, or de novo synthesis (rendering the slug independent from algal toxins). We have to emphasize that we were not able to study all species on a broad scale by including specimens from various geographic areas or seasons. Food availability and seasonality, and/or intraspecific or interspecific competition might also force specimens from the same species to switch to other food items and thus influences the finding of certain food items in sacoglossan sea slugs. Further starvation studies on polyphagous species (see e.g. [25]) will verify if these species have a preferred food source, as was shown for Plakobranchus ocellatus and these investigations will certainly contribute to our understanding of polyphagous strategies [37].
Jensen [22,53] studied morphological differentiation of the radula teeth in connection with algal food. Based on her findings she suggested that certain radula teeth are correlated with the polysaccharide of cell walls of the respective food sources. However, Händeler & Wägele [23] mapped radula teeth forms on their molecular based cladogram and were not able to confirm this previously suggested correlation. We therefore compared three unusual food switches within the Sacoglossa with regard to a possible correlation of food source and radula tooth shape i) The NR form Hermaea bifida (Montagu) [66] feeds on the red algae Griffithsia with cell walls composed of cellulose. However, the radula of H. bifida is not different from species feeding on the green alga Caulerpa with cell walls composed of xylan [67]. ii) Elysia serca, also a NR form [68], presents a unique switch to sea grasses with cell walls composed of cellulose [28,29], but the radula is similar to species feeding on Halimeda with cell walls composed of xylan. iii) The NR forms Alderia modesta and A. willowi, as well as the LtR form Elysia chlorotica, feed on the heterokontophyte genus Vaucheria composed of cellulose-based cell walls. The radulae of Alderia species and Elysia chlorotica are similar to species feeding on the green alga Codium composed of mannan-based cell walls. Thus, we think that different radula shapes did not force or influence a host switch in these three examples, though it cannot be ruled out for others that are not examined here.
Whether functional kleptoplasty is of higher evolutionary benefit than e.g., feeding on a wide spectrum of algae may be reconsidered. A polyphagous strategy –in contrast to stenophagous strategies – would render the slugs more independent of seasonality or general availability of their host algae. LtR forms like Costasiella ocellifera, Elysia chlorotica and Elysia timida feed on one algal species. Acetabularia acetabulum, the only natural food of E. timida, calcifies in fall, and therefore probably forces Elysia timida to rely on its incorporated chloroplasts [69]. Unfortunately we have no similar information on seasonality of Avrainvillea and Vaucheria, the major food items of C. ocellifera and E. chlorotica, respectively. Plakobranchus ocellatus on the other side has a broad food spectrum. As Maeda et al. [37] showed in a subtle experiment, the algae show a high seasonality and the slugs combine here functional kleptoplasty and multiple food sources that certainly enhances survival.
Usefulness of barcoding markers
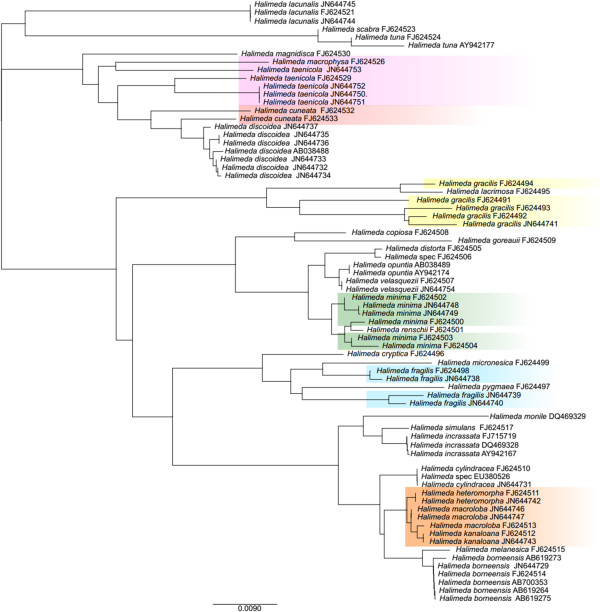
RbcL and tufA are now state of the art markers for barcoding algal species [70] and are therefore intensively used for identification of sequestered chloroplasts in Sacoglossa [25,30,34-37]. So far it is not possible to identify the food of every sample we screened, and reasons are unknown. Christa et al. [25] showed that rbcL almost always revealed more haplotypes for one sacoglossan specimen than tufA, especially within the genus Halimeda. In the present study we see exactly the same pattern. We therefore compared several rbcL sequences of randomly chosen Halimeda species in order to reveal possible problems by using rbcL as barcoding marker for this genus (Figure 4). Our analyses show that for several Halimeda species various rbcL sequences exist. p-distances between haplotypes of the same species exceeded in some cases those of rbcL sequences between different species. For example, the p-distance of haplotypes of H. cuneata FJ624533 and FJ624532 is 2.5%, the p-distance of Halimeda gracilis FJ624494 and Halimeda lacrimosa FJ624495 1%. Assuming that there are no misidentifications (which is important for DNA-Barcoding) with regard to the sequences obtained from GenBank, this problem can lead to an overestimated number of haplotypes in cases where no reference sequences are available. With regard to our study, this might be the case for several algal sequences obtained from Thuridilla, which are now assigned as seven different unidentified haplotypes. It is very likely that these problems do not only occur in the taxon Halimeda, but also in other ulvophyceaen taxa where detailed studies are still missing. Händeler et al. [30] demonstrated for Caulerpa that identification and annotation using tufA on species level could also be problematic due to sequence similarities. Hanyuda et al. [71] reported introns in rbcL sequences from members of the Caulerpaceae, a problem we faced in Caulerpa sequences obtained from Volvatella viridis. These sequences cannot be aligned with the remaining rbcL sequences, and were therefore not included in our analyses here.
Figure 4.
ML tree of rbcL sequences of Halimeda species. ML tree of randomly chosen rbcL sequences of Halimeda. Multiple haplotypes of a single species are highlighted in colored boxes.
An additional problem for identifying food organisms in Sacoglossa is connected to the lack of references for both genes in the databases. Available information does not cover many of the obtained haplotypes; therefore an extension of algal taxon sampling in the future by algal specialists is absolutely necessary. Here we show that the use of two markers for plastid identification in Sacoglossa only leads to a more detailed identification of ingested plastids in some species. Yet, we suggest that, based on the higher reliability and despite possible multiple haplotypes for some genera, rbcL should be used preferentially. TufA may be added in cases where an identification of rbcL failed or sequence variability of rbcL hinders unambiguous identification. Unfortunately both primer pairs are still not applicable at the moment for Cladophoraceae [71,72]. Constructing new primers for both genes based on chloroplast genomic data probably could solve this problem as soon as a cladophoraceaen plastid genome is available. Until then it has to be kept in mind that barcoding results may cover only a reduced food range. Even with the limitations mentioned above, we consider DNA-Barcoding a more accurate and effective method than feeding observations, especially in polyphagous specimens.
Conclusions
DNA-Barcoding of sequestered chloroplasts in Sacoglossa led to a more specific insight in food sources of sacoglossan sea slugs than by direct observation and is certainly much more efficient. RbcL should be used primarily because of higher amplification and sequencing success, while tufA should be added in analyses with ambiguous results.
Our results revealed food items for many species not investigated before and additionally confirmed literature data. We could show that food sources are highly variable in some species and morphological features such as radula shape have probably only little impact on food preferences or food switches. There seem to be certain food sources that are essential for functional kleptoplasty. LtR forms preferably consume algal species belonging to the genera Halimeda, Caulerpa, Avrainvillea, Acetabularia or Vaucheria. Kleptoplasty is only established in slugs feeding on green algae and heterokontophytes, but not in slugs feeding on rhodophytes and sea grasses. NR forms may also feed on algae that are known to be the sole food of LtR forms. Factors like physiology of food items, genetic and physiological properties of the plastids and digestion properties in slugs need to be more investigated in future studies to reveal principles of establishing functional kleptoplasty. The evolutionary benefit of kleptoplasty is still enigmatic, since a polyphagous life style may lead to more independence from specific food source’s seasonality and abundance.
Methods
Sacoglossan specimens used in this study were identified by morphological examination using a taxonomic species file based on original literature ([73] on Thuridilla), as well as identification books (e.g., [74]) and the sea slug forum (http://www.seaslugforum.net). These identified specimens, covering NR, StR and LtR forms, are listed in Additional file 1 and represent those for which DNA-Barcoding of food source was successful, in order to determine the minimal food items of these species. Specimens were fixed in 96%-EtOH immediately after collection. Slug parts containing digestive glandular tissue were cut off and subsequently DNA extracted using the DNeasy® Blood & Tissue Kit (Qiagen, Germany) following manufacturer’s instructions and stored at -20°C.
DNA amplification
Standard PCR reactions for rbcL and tufA as reported elsewhere [25,30,34-37] were performed using a touchdown protocol and ulvophycean specific primers. 2.5 μl of genomic DNA was used as template in a 20 μl final volume reaction supplied with 5.5 μl sterilized water, 2 μl Qiagen® Q-Solution, 10 μl of double concentrated QIAGEN® Multiplex PCR Master Mix and 1 μl of 5 pmol/μl concentrated primer each. PCR for amplification of rbcL was performed with primer pairs rbcL 1 ([36] and rbcL R, 5′-CCA WCG CAT ARA NGG TTG HGA-3′ ([25] modifid after [36]) by an initially denaturation for 15 min at 95°C, followed by 9 touch-down cycles at 94°C for 45 s, 53°C (-1°C per cycle) for 45 s, 72°C for 90 s, followed by 25 standard cycles (94°C for 45 s, 45°C for 45 s and 72°C for 90 s.) and a final extension at 72°C for 10 min. tufA amplification was performed with primer pair tufAF and tufAR [75] by an initially denaturation for 15 min at 95°C, followed by 9 touch-down cycles at 94°C for 45 s, 57°C (-1°C per cycle) for 45 s, 72°C for 90 s, followed by 25 standard cycles (94°C for 45 s, 48°C for 45 s and 72°C for 90 s.) and a final extension at 72°C for 10 min. PCR products were size-fractionated in a 1.5% agarose gel for 90 min at 70 V and bands according to desired gene-fragment length and subsequently gel-extracted using Machery-Nagel NucleoSpin® Extract II (Düren, Germany) kit following manufacturer’s instructions. Isolated fragments were ligated into pGEM T-easy Vector (Promega, Germany) and cloned into competent E. coli XL1-blue cells from Stratagene (Heidelberg, Germany). For each specimen 12 clones were sequenced by Macrogen Inc, Amsterdam.
Sequence analysis
Sequence identity of tufA and rbcL of every clone was verified by BLAST search using Geneious (Biomatters Ltd, New Zealand, v. 6.0.3). Consensus sequences of one slug specimen were created when sequence divergence of chloroplast genes was lower than 1%, as introduced by Händeler et al. [30] (see Additional files 2, 3 and 4). All gained sequences were again verified by BLAST search and the first top 5 BLAST results of each sequence were taken to create a dataset of overall 54 sequences for tufA (688 bp) and 94 sequences for rbcL (561 bp). Both datasets were aligned with MAFFT plugin as implemented in Geneious (v6.814b[76]). A maximum likelihood tree for each dataset was calculated using PhyML plugin [77] implemented in Geneious with GTR + I + R as substitution model. For final identifying sequence origin we used a combination of similarity-based and tree-based DNA-barcoding approaches [78,79]: Sequences were assigned to a certain algal species by a similarity-based approach using BLAST analysis when sequence similarity was 99-100%. We included a 99% match as positive identification of an algal species, thus allowing a small amount of ambiguity nucleotides within our consensus sequences which are not identified as similarity by the BLAST search in NCBI (http://www.ncbi.nlm.nih.gov). When multiple positive identifications were obtained, sequences were assigned to the corresponding genus based on the position within the ML tree. When not able to define on species level (BLAST similarity <99%), sequences were assigned to higher taxa based on their position within monophyletic algal groups in the ML tree analysis. When sequences obtained in our study did not group within distinct algal taxa, we assigned them to the taxon level Ulvophyceae.
Sequence analysis of rbcL sequences of Halimeda
Randomly chosen rbcL sequences of Halimeda species were downloaded from Genbank to analyze sequences variability. Overall 72 sequences were used, representing 32 species. Dataset was aligned with MAFFT plugin as implemented in Geneious (v6.814b[76]). A maximum likelihood tree for the dataset was calculated using PhyML plugin [77] implemented in Geneious with GTR + I + R as substitution model. P-distances of sequences were calculated using HyPhy (v 2.1). On the basis of these p-distances the sequence variability of haplotypes of the same algal species were then analyzed.
Comparison of literature data and new barcoding data
DNA-Barcoding results of the present study were combined with results taken from literature to compare food sources of different retention forms (LtR, StR, NR). Literature data on food sources and retention form were taken from [3,4,18,25,30,34-37,80-84] (Additional files 1 and 5). The retention ability of the majority of the species investigated here was measured by means of a PAM. Therefore, we used the classification scheme of Händeler et al. [3]. Results of Clark et al. [68] based on 14C methods were re-assigned, when new data were available. For example, Mourgona germaineae Marcus, 1970 is classified as StR form by Clark et al. [68], however PAM measurements revealed no fluorescence at all (unpublished data). We therefore set 14C obtained by values similar to M. germaineae as “no retention”, e.g. Caliphylla mediterranea Costa [85], Hermaea cruciata, Placida kingstoni Thompson [86] and Elysia serca. Most recently Schmitt et al. [87] could show that E. timida can be artificially raised on Acetabularia peniculus, an algal species that naturally does not co-occur with the slugs. Similar retention of plastids was observed as from the natural food A.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HW, KH and GC conceived and designed the experiment. GC, TS and KH performed the experiments. HW, GC and KH analyzed the data. GC, TS, GK and HW wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Origin of specimen and identified food sources with comparison of literature data. In Table S1 we provide a list of Sacoglossa specimens and species analyzed with regard to food items. Table S2 displays the identified food sources in Sacoglossa specimens by analyzing the chloroplast markers tufA and rbcL. The number of haplotypes per gene obtained from each slug specimen is listed in the last two columns. * tufA sequences of Thuridilla hopei (809), Vérany were published previously [18]. Table S3 shows a comparison of chloroplast origin by feeding observations (4th column) and identified by DNA-Barcoding (this study and literature data – last column). When not specified specifically, information on retention-form is taken from Händeler et al. [3]. Food sources identified by using tufA are indicated with 1, by rbcL with 2 in the last column. Literature data on food sources base on the review of Händeler & Wägele [29] or is indicated otherwise.
Sequence affinity based on BLAST search.
Alignment of tufA sequences used for consensus sequences.
Alignment of rbcL sequences used for consensus sequences.
Classification on retention form in Sacoglossa.
Contributor Information
Gregor Christa, Email: gchrista@uni-bonn.de.
Katharina Händeler, Email: k.haendeler@gmx.de.
Till F Schäberle, Email: till.schaeberle@uni-bonn.de.
Gabriele M König, Email: g.koenig@uni-bonn.de.
Heike Wägele, Email: h.waegele@zfmk.de.
Acknowledgments
We thank all collectors Ingo Burghardt, Yvonne Grzymbowski, Annette Klussmann-Kolb, Patrick Krug, Wolfgang Loch, Tane Sinclair, Kristina Stemmer, Carmen Zinßmeister, and the directors of the Lizard Island Research Station, Anne Hoggett and Lyle Vail and the director of the Marine Laboratory, Guam, Peter Schupp, providing us with material. Emilie Goralski and Claudia Etzbauer helped with the molecular work. Anonymous reviewers provided valuable information and helped considerably in improving the first version of this manuscript. We thank the German Science Foundation (DFG) for financial support to HW (Wa 618/8 and Wa 618/12). GC was partly financed by an ERC grant to W. Martin (Düsseldorf, Germany): Networkorigins Proj. Ref. 232975.
References
- Jensen KR. Biogeography of the Sacoglossa (Mollusca, Opisthobranchia) Bonn Zool Beitr. 2007;55:255–281. [Google Scholar]
- Kohnert P, Brenzinger B, Jensen KR, Schrödl M. 3D- microanatomy of the semiterrestrial slug Gascoignella aprica Jensen, 1985—a basal plakobranchacean sacoglossan (Gastropoda, Panpulmonata) Org Divers Evol. 2013;13:583–603. doi: 10.1007/s13127-013-0142-6. [DOI] [Google Scholar]
- Händeler K, Grzymbowski YP, Krug PJ, Wägele H. Functional chloroplasts in metazoan cells - a unique evolutionary strategy in animal life. Front Zool. 2009;6:28. doi: 10.1186/1742-9994-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KR. Evolution of the Sacoglossa (Mollusca, Opisthobranchia) and the ecological associations with their food plants. Evol Ecol. 1997;11:301–335. doi: 10.1023/A:1018468420368. [DOI] [Google Scholar]
- Muscatine L, Greene RW. Chloroplasts and algae as symbionts in molluscs. Int Rev Cytol. 1973;36:137–169. doi: 10.1016/s0074-7696(08)60217-x. [DOI] [PubMed] [Google Scholar]
- Trench RK. Of “leaves that crawl”: functional chloroplasts in animal cells. Sym Soc Exp Biol. 1975;29:229–265. [PubMed] [Google Scholar]
- Rumpho ME, Summer EJ, Manhart JR. Solar-powered sea slugs. Mollusc/algal chloroplast symbiosis. Plant Physiol. 2000;123:29–38. doi: 10.1104/pp.123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wägele H, Johnson G. Observations on the histology and photosynthetic performance of “solar-powered” opisthobranchs (Mollusca, Gastropoda, Opisthobranchia) containing symbiotic chloroplasts or zooxanthellae. Org Divers Evol. 2001;1:193–210. doi: 10.1078/1439-6092-00016. [DOI] [Google Scholar]
- Lee JJ. Algal symbiosis in larger Foraminifera. Symbiosis. 2006;42:63–75. [Google Scholar]
- McManus GB. Chloroplast symbiosis in a marine ciliate: ecophysiology and the risks and rewards of hosting foreign organelles. Front Microbiol. 2012;3:1–9. doi: 10.3389/fmicb.2012.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gast RJ, Moran DM, Dennett MR, Caron DA. Kleptoplasty in an Antarctic dinoflagellate: caught in evolutionary transition? Environ Microbiol. 2007;9:39–45. doi: 10.1111/j.1462-2920.2006.01109.x. [DOI] [PubMed] [Google Scholar]
- Rumpho ME, Pelletreau KN, Moustafa A, Bhattacharya D. The making of a photosynthetic animal. J Exp Biol. 2011;214:303–311. doi: 10.1242/jeb.046540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wägele H, Martin WF. In: Endosymbiosis. Löffelhardt W, editor. Vienna: Springer Vienna; 2013. Endosymbioses in sacoglossan seaslugs: plastid-bearing animals that keep photosynthetic organelles without borrowing genes; pp. 291–324. [Google Scholar]
- Pierce S, Biron R, Rumpho M. Endosymbiotic chloroplasts in molluscan cells contain proteins synthesized after plastid capture. J Exp Biol. 1996;199:2323–2330. doi: 10.1242/jeb.199.10.2323. [DOI] [PubMed] [Google Scholar]
- Gould AA. Report on the Invertebrata of Massachusetts. 2. Boston: Wright and Potter, State Printers; 1870. [Google Scholar]
- Schwartz JA, Curtis NE, Pierce SK. Using algal transcriptome sequences to identify transferred genes in the sea slug, Elysia chlorotica. Evol Biol. 2010;37:29–37. doi: 10.1007/s11692-010-9079-2. [DOI] [Google Scholar]
- Rumpho ME, Worful JM, Lee J, Kannan K, Tyler MS, Bhattacharya D, Moustafa A, Manhart JR. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc Natl Acad Sci USA. 2008;105:17867–17871. doi: 10.1073/pnas.0804968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wägele H, Deusch O, Händeler K, Martin R, Schmitt V, Christa G, Pinzger B, Gould SB, Dagan T, Klussmann-Kolb A, Martin W. Transcriptomic evidence that longevity of acquired plastids in the photosynthetic slugs Elysia timida and Plakobranchus ocellatus does not entail lateral transfer of algal nuclear genes. Mol Biol Evol. 2011;28:699–706. doi: 10.1093/molbev/msq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso A. Mémoire sur quelques gastéropodes nouveaux, nudibranches et tectibranches observés dans la Mer de Nice. J Phys Chim Hist Nat Arts. 1818;87:368–377. [Google Scholar]
- Hasselt JCV. Extrait d’une lettre du Dr JC van Hasselt au Prof. van Swinderen, sur mollusques de Java. Bull Sci Nat Géol. 1824;2:237–245. [Google Scholar]
- Pelletreau KN, Bhattacharya D, Price DC, Worful JM, Moustafa A, Rumpho ME. Sea slug kleptoplasty and plastid maintenance in a metazoan. Plant Physiol. 2011;155:1561–1565. doi: 10.1104/pp.111.174078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Pelletreau KN, Price DC, Sarver KE, Rumpho ME. Genome analysis of Elysia chlorotica egg DNA provides no evidence for horizontal gene transfer into the germ line of this kleptoplastic mollusc. Mol Biol Evol. 2013;8:1843–1852. doi: 10.1093/molbev/mst084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SK, Fang X, Schwartz JA, Jiang X, Zhao W, Curtis NE, Kocot KM, Yang B, Wang J. Transcriptomic evidence for the expression of horizontally transferred algal nuclear genes in the photosynthetic sea slug, Elysia chlorotica. Mol Biol Evol. 2012;29:1545–1556. doi: 10.1093/molbev/msr316. [DOI] [PubMed] [Google Scholar]
- Christa G, Zimorski V, Woehle C, Tielens AGM, Wägele H, Martin WF, Gould SB. Plastid-bearing sea slugs fix CO2 in the light but do not require photosynthesis to survive. Proc R Soc B. 2014;281:20132493. doi: 10.1098/rspb.2013.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christa G, Wescott L, Schäberle TF, König GM, Wägele H. What remains after 2 months of starvation? Analysis of sequestered algae in a photosynthetic slug, Plakobranchus ocellatus (Sacoglossa, Opisthobranchia), by barcoding. Planta. 2013;237:559–572. doi: 10.1007/s00425-012-1788-6. [DOI] [PubMed] [Google Scholar]
- Decaisne J. Plantes de l’Arabie Heureuse, receuillies par MP-E. Botta. Archs Mus. natn. Hist nat Paris. 1842;2:89–193. [Google Scholar]
- Taylor DL. Chloroplasts as symbiotic organelles in the digestive gland of Elysia viridis (Gastropoda, Opisthobranchia) J Mar Biol Ass. 1968;48:1–15. doi: 10.1017/S0025315400032380. [DOI] [Google Scholar]
- Jensen KR. Morphological adaptations and plasticity of radular teeth of the Sacoglossa (= Ascoglossa) (Mollusca: Opisthobranchia) in relation to their food plants. Biol J Linn Soc. 1993;48:135–155. doi: 10.1111/j.1095-8312.1993.tb00883.x. [DOI] [Google Scholar]
- Händeler K, Wägele H. Preliminary study on molecular phylogeny of Sacoglossa and a compilation of their food organisms. Bonn Zool Beitr. 2007;55:231–254. [Google Scholar]
- Händeler K, Wägele H, Wahrmund U, Rüdinger M, Knoop V. Slugs’ last meals: molecular identification of sequestered chloroplasts from different algal origins in Sacoglossa (Opisthobranchia, Gastropoda) Mol Ecol Resour. 2010;10:968–978. doi: 10.1111/j.1755-0998.2010.02853.x. [DOI] [PubMed] [Google Scholar]
- Jensen KR. A review of sacoglossan diets, with comparative notes on radular and buccal anatomy. Malacol Rev. 1980;13:55–77. [Google Scholar]
- Thompson TE, Jarman GM. Nutrition of Tridachia crispata (Mörch) (Sacoglossa) J Moll Stud. 1989;55:239–244. doi: 10.1093/mollus/55.2.239. [DOI] [Google Scholar]
- Williams SI, Walker DI. Mesoherbivore–macroalgal interactions: feeding ecology of sacoglossan sea slugs (Mollusca, Opisthobranchia) and their effects on their food algae. Oceanogr Mar Biol Annu Rev. 1999;37:87–138. [Google Scholar]
- Curtis NE, Massey SE, Schwartz JA, Maugel TK, Pierce SK. The intracellular, functional chloroplasts in adult sea slugs (Elysia crispata) come from several algal species, and are also different from those in juvenile slugs. Microsc Microanal. 2005;11:1195–1196. [Google Scholar]
- Curtis NE, Massey SE, Pierce SK. The symbiotic chloroplasts in the sacoglossan Elysia clarki are from several algal species. Invertebr Biol. 2006;125:336–345. doi: 10.1111/j.1744-7410.2006.00065.x. [DOI] [Google Scholar]
- Pierce SK, Curtis NE, Massey SE, Bass AL, Karl SA, Finney CM. A morphological and molecular comparison between Elysia crispata and a new species of kleptoplastic sacoglossan sea slug (Gastropoda: Opisthobranchia) from the Florida Keys, USA. Mollusc Res. 2006;26:23–38. [Google Scholar]
- Maeda T, Hirose E, Chikaraishi Y, Kawato M, Takishita K, Yoshida T, Verbruggen H, Tanaka J, Shimamura S, Takaki Y, Tsuchiya M, Iwai K, Maruyama T. Algivore or phototroph? Plakobranchus ocellatus (Gastropoda) continuously acquires kleptoplasts and nutrition from multiple algal species in nature. PLoS ONE. 2012;7:e42024. doi: 10.1371/journal.pone.0042024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DL. Photosynthesis of symbiotic chloroplasts in Tridachia crispata (Bergh) Comp Biochem Physiol. 1971;38:233. doi: 10.1016/0300-9629(71)90111-3. [DOI] [Google Scholar]
- Vogel RM. The biology and a redescription of the opisthobranch mollusk Hermaea cruciata Gould, from Chesapeake Bay (Maryland) Veliger. 1971;14:155–157. [Google Scholar]
- Marcus E. Opisthobranchia from Brazil. Bol Fac Filos, Cienc Letr. 1955;20:89–261. [Google Scholar]
- Krug PJ. Bet-hedging dispersal strategy of a specialist marine herbivore: a settlement dimorphism among sibling larvae of Alderia modesta. Mar Ecol Prog Ser. 2001;213:177–192. [Google Scholar]
- Jensen KR. Observations on feeding methods in some Florida Ascoglossans. J Moll Stud. 1981;47:190–199. [Google Scholar]
- Kawaguti S, Yamasu T. Electron microscopy on the symbiosis between an elysioid gastropod and chloroplasts of a green alga. Biol J Okayama Univ. 1965;11:57–65. [Google Scholar]
- Hirose E. Digestive system of the sacoglossan Plakobranchus ocellatus (Gastopoda: Opishtobranchia): Light and electron-microscopic observations with remarks on chloroplast retention. Zool Sci. 2005;22:905–916. doi: 10.2108/zsj.22.905. [DOI] [PubMed] [Google Scholar]
- Baba K. Opisthobranchia of Kii, Middle Japan. J Dep Agriculture, Kyushu Imp Univ. 1938;6:1–19. [Google Scholar]
- Bergh L. In: Reisen im Archipel der Philippinen. Semper CG, editor. Wissenschaftliche Resultate; 1870. Malacologische Untersuchungen. 1:1–30. [Google Scholar]
- Christa G, Gould SB, Franken J, Vleugels M, Karmeinski D, Händeler K, Martin WF, Wägele H, Functional kleptoplasty in a limapontioid genus. Phylogeny, food preferences and photosynthesis in Costasiella with focus on C. ocellifera (Gastropoda, Sacoglossa) J Mollusc Stud. (in press)
- Simroth H. Opisthobranchia Gymnobranchia. Die Gastropoden der Plankton Expedition. 1895;2:1–206. [Google Scholar]
- Clark KB, JK R, Hugh SM, Fermin C. Chloroplast symbiosis in a non-elysiid mollusc, Costasiella lilianae Marcus (Hermaeidae: Ascoglossa (= Sacoglossa): Effects of temperature, light intensity, and starvation on carbon fixation rate. Biol Bull. 1981;160:43–54. doi: 10.2307/1540899. [DOI] [Google Scholar]
- Krug PJ, Händeler K, Vendetti J. Genes, morphology, development and photosynthetic ability support the resurrection of Elysia cornigera (Heterobranchia:Plakobranchoidea) as distinct from the “solar-powered” sea slug, E. timida. Invertebr Syst. 2011;25:477. doi: 10.1071/IS11026. [DOI] [Google Scholar]
- Silva PC. A review of nomenclatural conservation in the algae from the point of view of the type method. Univ Calif Publ Botany. 1952;25:241–323. [Google Scholar]
- Wright SW, Grant BR. Properties of chloroplasts isolated from siphonous algae: effects of osmotic shock and detergent treatment on intactness. Plant Physiol. 1978;61:768–771. doi: 10.1104/pp.61.5.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trench RK, Boyle JE, Smith DC. The association between chloroplasts of Codium fragile and the mollusc Elysia viridis. I. Characteristics of isolated Codium chloroplasts. Proc R Soc B. 1973;184:51–61. doi: 10.1098/rspb.1973.0030. [DOI] [Google Scholar]
- de Vries J, Habicht J, Woehle C, Changjie H, Christa G, Wägele H, Nickelsen J, Martin WF, Gould SB. Is ftsH the key to plastid longevity in sacoglossan slugs? Genome Biol Evol. 2013;5:2540–2548. doi: 10.1093/gbe/evt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura P, Calado G, Jesus B. Photosynthetic efficiency and kleptoplast pigment diversity in the sea slug Thuridilla hopei (Vérany, 1853) J Exp Mar Biol Ecol. 2013;441(C):105–109. [Google Scholar]
- Pillet L, Pawlowski J. Transcriptome analysis of foraminiferan Elphidium margaritaceum questions the role of gene transfer in kleptoplastidy. Mol Biol Evol. 2013;30:66–69. doi: 10.1093/molbev/mss226. [DOI] [PubMed] [Google Scholar]
- Wisecaver JH, Hackett JD. Transcriptome analysis reveals nuclear-encoded proteins for the maintenance of temporary plastids in the dinoflagellate Dinophysis acuminata. BMC Genomics. 2010;11:366. doi: 10.1186/1471-2164-11-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Händeler K. Evolution of Sacoglossa (Opisthobranchia) with Emphasis on their Food (Ulvophyceae) and the Ability to Incorporate Kleptoplasts. University Bonn; 2011. pp. 1–134. (Doctoral thesis). [Google Scholar]
- Evertsen J, Johnsen G. In vivo and in vitro differences in chloroplast functionality in the two north Atlantic sacoglossans (Gastropoda, Opisthobranchia) Placida dendritica and Elysia viridis. Mar Biol. 2009;156:847–859. doi: 10.1007/s00227-009-1128-y. [DOI] [Google Scholar]
- Alder J, Hancock A. A Monograph of the British Nudibranchiate Mollusca with Figures of all the Species. London: The Ray Society; 1845. [Google Scholar]
- Trench RK, Gooday GW. Incorporation of [H-3]-leucine into protein by animal tissues and by endosymbiotic chloroplasts in Elysia viridis Montagu. Comp Biochem Physiol. 1973;44:321. doi: 10.1016/0300-9629(73)90485-4. [DOI] [Google Scholar]
- Gallop A. Evidence for the presence of a “factor” in Elysia viridis which stimulates photosynthate release from its symbiotic chloroplasts. New Phytol. 1974;73:1111–1117. doi: 10.1111/j.1469-8137.1974.tb02140.x. [DOI] [Google Scholar]
- Maeda T, Kajita T, Maruyama T, Hirano Y. Molecular phylogeny of the sacoglossa, with a discussion of gain and loss of kleptoplasty in the evolution of the group. Biol Bull. 2010;219:17–26. doi: 10.1086/BBLv219n1p17. [DOI] [PubMed] [Google Scholar]
- Jörger KM, Stöger I, Kano Y, Fukuda H, Knebelsberger T, Schrödl M. On the origin of Acochlidia and other enigmatic euthyneuran gastropods, with implications for the systematics of Heterobranchia. BMC Evol Biol. 2010;10:323. doi: 10.1186/1471-2148-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neusser TP, Fukuda H, Jörger KM, Kano Y, Schrödl M. Sacoglossa or Acochlidia? 3D reconstruction, molecular phylogeny and evolution of Aitengidae (Gastropoda: Heterobranchia) J Moll Stud. 2011;77:332–350. doi: 10.1093/mollus/eyr033. [DOI] [Google Scholar]
- Montagu A. An account of some new and rare British shells and animals. Transac Linn Soc Lond. 1815;11:179–204. [Google Scholar]
- Thompson TE. Eastern Mediterranean Opisthobranchia: oxynoidae, Polybranchiidae, Stiligeridae (Sacoglossa) J Moll Stud. 1988;54:157–172. doi: 10.1093/mollus/54.2.157. [DOI] [Google Scholar]
- Clark KB, Jensen KR, Stirts HM. Survey for functional kleptoplasty among west Atlantic Ascoglossa (= Sacoglossa) (Mollusca: Opisthobranchia) Veliger. 1990;33:339–345. [Google Scholar]
- Marìn A, Ros J. Ultrastructural and ecological aspects of the development of chloroplast retention in the sacoglossan gastropod Elysia timida. J Moll Stud. 1993;59:95–104. doi: 10.1093/mollus/59.1.95. [DOI] [Google Scholar]
- Saunders GW. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philos T R Soc B. 2005;360:1879–1888. doi: 10.1098/rstb.2005.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyuda T, Arai S, Ueda K. Variability in the rbcL introns of Caulerpalean algae (Chlorophyta, Ulvophyceae) J Plant Res. 2000;113:403–413. doi: 10.1007/PL00013948. [DOI] [Google Scholar]
- Verbruggen H, Ashworth M, LoDuca ST, Vlaeminck C, Cocquyt E, Sauvage T, Zechman FW, Littler DS, Littler MM, Leliaert F. A multi-locus time-calibrated phylogeny of the siphonous green algae. Mol Phylogenet Evol. 2009;50:642–653. doi: 10.1016/j.ympev.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Gosliner TM. The genus Thuridilla (Opisthobranchia: Elysiidae) from the tropical Indo-Pacific, with a revision of the phylogeny of the Elysiidae. Proceedings of the California Academy of Sciences, Volume 49. 1995. pp. 1–54.
- Gosliner TM, Behrens DW, Valdés Á. Indo-Pacific Nudibranchs and Sea Slugs: a Field Guide to the World’s Most Diverse Fauna. San Francisco: California Academy of Sciences/Sea Challengers Natural History Books; 2008. pp. 1–426. [Google Scholar]
- Famà P, Wysor B, Kooistra WH, Zuccarello GC. Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene. J Phycol. 2002;38:1040–1050. doi: 10.1046/j.1529-8817.2002.t01-1-01237.x. [DOI] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Tanabe AS, Toju H. Two New computational methods for universal DNA barcoding: a benchmark using barcode sequences of bacteria, archaea, animals, fungi, and land plants. PLoS ONE. 2013;8:e76910. doi: 10.1371/journal.pone.0076910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little DP. DNA barcode sequence identification incorporating taxonomic hierarchy and within taxon variability. PLoS ONE. 2011;6:e20552. doi: 10.1371/journal.pone.0020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Yusa Y, Yamamoto S, Hirano Y, Hirano Y, Motomura T, Tanemura T, Obokata J. Identification of photosynthetic sacoglossans from Japan. Endocyt Cell Res. 2009;19:112–119. [Google Scholar]
- Klochkova TA, Han J-W, Kim J-H, Kim K-Y, Kim G-H. Feeding specificity and photosynthetic activity of Korean sacoglossan mollusks. Algae. 2010;25:217–227. doi: 10.4490/algae.2010.25.4.217. [DOI] [Google Scholar]
- Klochkova TA, Han J-W, Chah K-H, Kim RW, Kim J-H, Kim K-Y, Kim G-H. Morphology, molecular phylogeny and photosynthetic activity of the sacoglossan mollusc, Elysia nigrocapitata, from Korea. Mar Biol. 2012;160:155–168. [Google Scholar]
- Wägele H, Stemmer K, Burghardt I, Haendeler K. Two new sacoglossan sea slug species (Opisthobranchia, Gastropoda): Ercolania annelyleorum sp. nov. (Limapontioidea) and Elysia asbecki sp. nov. (Plakobranchoidea), with notes on anatomy, histology and biology. Zootaxa. 2010;2676:1–28. [Google Scholar]
- Evertsen J, Burghardt I, Johnsen G, Wägele H. Retention of functional chloroplasts in some sacoglossans from the Indo-Pacific and Mediterranean. Mar Biol. 2007;151:2159–2166. doi: 10.1007/s00227-007-0648-6. [DOI] [Google Scholar]
- Costa A. Illustrazione di due generi di Molluschi Nudibranchi. Rendi Accad Sci Fis Mat Napoli. 1867;6:136–137. [Google Scholar]
- Thompson TE. Jamaican opisthobranch molluscs I. J Moll Stud. 1977;43:93–140. [Google Scholar]
- Schmitt V, Händeler K, Gunkel S, Escande ML, Menzel D, Gould SB, Martin WF, Wägele H. Chloroplast incorporation and long-term photosynthetic performance through the life cycle in laboratory cultures of Elysia timida (Sacoglossa, Heterobranchia) Front Zool. 2014;11:5. doi: 10.1186/1742-9994-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Origin of specimen and identified food sources with comparison of literature data. In Table S1 we provide a list of Sacoglossa specimens and species analyzed with regard to food items. Table S2 displays the identified food sources in Sacoglossa specimens by analyzing the chloroplast markers tufA and rbcL. The number of haplotypes per gene obtained from each slug specimen is listed in the last two columns. * tufA sequences of Thuridilla hopei (809), Vérany were published previously [18]. Table S3 shows a comparison of chloroplast origin by feeding observations (4th column) and identified by DNA-Barcoding (this study and literature data – last column). When not specified specifically, information on retention-form is taken from Händeler et al. [3]. Food sources identified by using tufA are indicated with 1, by rbcL with 2 in the last column. Literature data on food sources base on the review of Händeler & Wägele [29] or is indicated otherwise.
Sequence affinity based on BLAST search.
Alignment of tufA sequences used for consensus sequences.
Alignment of rbcL sequences used for consensus sequences.
Classification on retention form in Sacoglossa.