Abstract
Micropatterning techniques that control three-dimensional (3D) arrangement of biomolecules and cells at the microscale will allow development of clinically relevant tissues composed of multiple cell types in complex architecture. Although there have been significant developments to regulate spatial and temporal distribution of biomolecules in various materials, most micropatterning techniques are applicable only to two-dimensional patterning. We report here the use of two-photon laser scanning (TPLS) photolithographic technique to micropattern cell adhesive ligand (RGDS) in hydrogels to guide cell migration along pre-defined 3D pathways. The TPLS photolithographic technique regulates photo-reactive processes in microscale focal volumes to generate complex, free form microscale patterns with control over spatial presentation and concentration of biomolecules within hydrogel scaffolds. The TPLS photolithographic technique was used to dictate the precise location of RGDS in collagenase-sensitive poly(ethylene glycol-co-peptide) diacrylate hydrogels, and the amount of immobilized RGDS was evaluated using fluorescein-tagged RGDS. When human dermal fibroblasts cultured in fibrin clusters were encapsulated within the micropatterned collagenase-sensitive hydrogels, the cells underwent guided 3D migration only into the RGDS-patterned regions of the hydrogels. These results demonstrate the prospect of guiding tissue regeneration at the microscale in 3D scaffolds by providing appropriate bioactive cues in highly defined geometries.
Keywords: PEG, hydrogel, micropatterning, cell migration, tissue engineering
1. Introduction
Although numerous studies have suggested the promise of tissue engineering, clinical applications to regenerate architecture and functions of complex tissues, such as liver, kidney, and muscle, are yet to be demonstrated. To mimic tissue complexity and develop clinically relevant tissues, recent focus has been placed on techniques to control three-dimensional (3D) arrangement of biomolecules and cells at the microscale. 3D microfabrication techniques with microscale resolution developed up to date include sequential photolithographic patterning [1-3], soft-lithographic patterning [4,5], microfluidic patterning [6], electrochemical deposition [7], and 3D printing [8]. These micropatterning techniques generally employ techniques in which layers of material are serially patterned in the z direction to form the final 3D structures. Although this layer-by-layer approach provides a convenient method to pattern in 2D, it generally suffers from the loss of fidelity in z direction during the iterative procedures [9]. Departing from the traditional use of these layer-by-layer methods, this present study demonstrates application of two-photon laser scanning (TPLS) photolithography to create 3D micropatterning of biomolecules immobilized in preformed network of biomaterials, thereby guiding 3D cell migration.
We previously reported development of TPLS photolithographic technique to fabricate 3D micropatterns of biomolecules in poly(ethylene glycol) (PEG) hydrogels [10]. The TPLS photolithographic technique utilizes the simultaneous absorption of finely focused photons to achieve two-photon excitation in microscale focal volumes [11]. The laser focal point is the only location along the optical path where the two-photon excitation occurs, and thus the subsequent photo-reactive processes are confined to this microscale focal volume, leaving out of focus regions unaltered. Using this technique, 3D patterns and gradients of biochemical signals have been generated in PEG hydrogels [10].
In this present study, we use the TPLS photolithography to guide 3D cell migration within degradable, biomimetic hydrogels. Specifically, we used the TPLS photolithography to dictate the precise location of cell adhesive ligand, Arg-Gly-Asp-Lys (RGDSK), within proteolytically degradable hydrogels, and fibroblasts encapsulated in these hydrogels underwent guided 3D cell migration within the patterned regions of the hydrogels. In order to allow 3D cell migration within hydrogel scaffolds, poly(ethylene glycol) diacrylate (PEGDA) hydrogels, which are inherently biocompatible and non-degradable, were rendered biodegradable by incorporating collagenase-sensitive peptide sequence, Lys-Gly-Pro-Ala (LGPA) into the backbone of the precursor polymer chains [12-14]. Further modifications of the hydrogels with cell adhesive ligands can render otherwise bioinert hydrogels cell adhesive; we have reported modifications of PEGDA hydrogels with various peptides and proteins to support cell adhesion, proliferation, and protease-mediated migration of many different cell types [12,15-18]. Combining the 3D micropatterning technology with the ability to design any materials with controlled bioactivity may allow creation of engineered tissues with complex structures such as microvasculature.
2. Materials and Methods
Synthesis of crosslinkable and degradable multiblock (GGGLGPAGGK-PEG)
A collagenase-sensitive peptide sequence, GGGLGPAGGK, was synthesized by solid phase peptide synthesis based on standard Fmoc chemistry using an Apex396 peptide synthesizer (Aapptec, Louisville, KY, USA). Hydrogels with this peptide in the polymer backbone have been shown to be completely degraded by collagenases [14]. Following purification, synthesis of the peptide was confirmed with matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-ToF; Bruker Daltonics, Billerica, MA, USA). Synthesis of the polymer with the peptide linker was completed in a stepwise manner in 50 mM sodium bicarbonate buffer solution (pH 8.5) at room temperature as outlined in the Supplement Figure 1A. First, the peptide was reacted with succinimidyl α-methylbutanoate-PEG-succinimidyl α-methylbutanoate (SMB-PEG-SMB, 3400Da; Nektar, Huntsville, AL, USA) in a 2:1 (PEG:peptide) molar ratio, and then with a 2-fold excess of additional peptide. After reaction, the product was dialyzed (MWCO 10,000; Spectrum Laboratories, Inc., Rancho Dominguez, CA, USA) to remove unreacted peptide and PEG moieties. The resulting product was reacted with acrylate-PEG-N-hydroxysuccinimide (acrylate-PEG-NHS, 3400Da; Nektar) to introduce crosslinkable acrylate functional groups at both ends. The product (acrylate-PEG-(GGGLGPAGGK-PEG)n-acrylate, n≥3) with theoretical MW of ca. 15,700 g/mol was dialyzed, lyophilized, and stored frozen under argon until use. The products were analyzed by 1H-NMR (Advance 400, Bruker, Germany) and gel permeation chromatography (GPC, Polymer laboratories, Amherst, MA, USA) with UV/Vis and evaporative light scattering detectors.
Proteolytic degradation of collagenase-sensitive PEG hydrogels
100 μL of crosslinkable and degradable multi-block (acrylate-PEG-(GGGLGPAGGK-PEG)n-acrylate, n≥3) solution (0.1 g/mL) in 10 mM HEPES buffered saline (HBS, pH 7.4) was filtered (MILLEX GP filter, 0.22 μm PES membrane, Millpore corp. Bedford, MA) and then mixed with 10 μL/mL of a photoinitiator solution (2,2-dimethoxy-2-phenyl acetophenone in n-vinylpyrrolidone, 300 mg/mL). The prepolymer solution was poured into a 96-well plate and photopolymerized under long-wavelength UV light (365 nm, 10 mW/cm2, 20 sec). The resulting hydrogels were removed, weighed and then allowed to swell in HBS with 1 mM CaCl2 and 0.2 mg/mL sodium azide at 37 °C for 24 hr. Each hydrogel sample was then incubated at 37 °C with either HBS, 0.2 mg/mL collagenase from clostridium histolyticum (Sigma, St. Louis, MO, USA), 0.2 mg/mL plasmin (Sigma) or 0.2 mg/mL proteinase K (Invitrogen, Carlsbad, CA, USA). Degradation of the hydrogels was evaluated by monitoring changes in the wet weight of the hydrogels over time. The enzyme solution was refreshed every 24 hr.
Synthesis of the fluorescein isothiocyanate tagged RGDSK-PEG (FITC-RGDSK-PEG-acrylate)
The N-α-Fmoc protected RGDSK peptide (Fmoc-RGDSK) was prepared using an Apex396 peptide synthesizer (Aapptec). Then, the free amine group on the lysine (K) side chain was conjugated to acrylate-PEG-NHS and purified as described previously [12]. To cleave Fmoc from Arginine (R), the Fmoc-RGDSK-PEG was dissolved in 20% piperidine in a mixture of dichloromethane (DCM) and N,N-dimethylformamide (DMF) and then reacted at room temperature for 5 h. After reaction, the sample was precipitated into diethyl ether and dried, and successful peptide synthesis was confirmed with MALDI-ToF (Bruker Daltonics). The Fmoc deprotected RGDSK-PEG was dissolved in DMF and then reacted with 1.5 molar excess of fluorescein isothiocyanate (Molecular Probes, Eugene, OR, USA) for 2 hr at room temperature. After the reaction, the sample was precipitated in diethyl ether and dried. The unbound fluorescein was removed by column chromatography with a Sephadex G-25 fine (Amersham Bioscience, Uppsala, Sweden), followed by dialysis (MWCO 3,500). 1H-NMR, FT-IR (660 plus, JASCO, Japan), HPLC (Varian, Inc. Walnut Creek, CA, USA) and GPC were used to confirm the synthesis of the peptides and conjugated products.
Cell culture
Human dermal fibroblasts (HDFs; Clonetics, San Diego, CA, USA) were maintained on Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS; Biowhittaker, Walkersville, MD, USA), 2 mM L-glutamine, 500 U penicillin and 100 mg/mL streptomycin. Fibroblasts were incubated at 37 °C in 5% CO2 environment. All experiments were conducted using fibroblasts at passages 3-6.
Preparation of cell clusters in fibrin gels
Fibrinogen solutions in 10 mg/mL were prepared from fibrinogen (from human plasma, Sigma) and dialyzed (MWCO 5,000; Spectrum Laboratories) for 24 h in Tris-buffered saline (28 mM Tris HCl (4.36 g/L), 5 mM Tris (0.64 g/L), 137 mM NaCl (8.0 g/L), 3 mM KCl (0.2 g/L); pH 7.4). The resulting solution was filtered through a syringe filter (MILLEX GP filter, 0.22 μm PES membrane). Concentration of the final fibrinogen solution was determined from the standard curve that was acquired by measuring the absorbance of the standard protein solution at 280 nm. The thrombin solution was prepared by dissolving thrombin at a concentration of 20 units/mL in phosphate-buffered saline (3 mM KCl (0.2 g/L), 2 mM KH2PO4(0.2 g/L), 8 mM Na2HPO4 (2.16 g/L), 137 mM NaCl (8.0 g/L), pH 7.4). To create cell cluster in fibrin gels, HDFs (106 cells) were suspended in 100 μL of fibrinogen solution (2 mg/mL) and mixed with 10 μL of thrombin solution (2 units/mL). 3 μL of cell suspended solution (2.7×104 cells) was dropped on glass slides coated with Sigmacote (Sigma) and then incubated at 37 °C for 20 min.
Preparation of pre-crosslinked hydrogel including cell cluster
The hydrogel precursor solution was prepared by dissolving multiblock (acrylate-PEG-(GGGLGPAGGK-PEG)n- acrylate, n≥3) (0.1 g/mL) in DMEM. The solution was filtered using a 0.22 μm syringe filter, and a photoinitiator solution (2,2-dimethoxy-2-phenyl acetophenone in n-vinylpyrrolidone, 300 mg/mL) was added at 10 μL/mL. To make hydrogels with homogenous distribution of RGDSK, 3.5 μmol/ml of RGDSK-PEG-acrylate was added in the precursor solution. 25 μL of the prepolymer solution was dropped on Lab-Tek ® II cover glass chambers (Nalge Nunc International, Naperville, IL, USA), and then the prepared fibroblast cluster in fibrin gel was placed inside the precursor solution. The sample solution with a fibroblast cluster suspended in the center was briefly exposed to long wavelength UV light (365 nm, 10 mW/cm2) for 20 sec. The same UV exposure condition has been used successfully to encapsulate cells in PEG hydrogels, and we have not seen any significant decrease in cell viability with this method of cell encapsulation [12,19]. The pre-crosslinked hydrogel was incubated in FITC-RGDSK-PEG-acrylate solution (30 μmol/mL) in the presence of a photoinitiator (10 μL/mL) for 30 min and subsequently patterned in 3D with the following procedure.
Patterning of RGDS in pre-crosslinked PEG hydrogels using TPLS photolithography
The pre-crosslinked hydrogel submerged in FITC-RGDSK-PEG-acrylate solution was positioned on a stage in LSM 510 META NLO confocal microscope (Carl Zeiss Inc.; Oberkochen, Germany). Virtual pattern image was drawn by the Region of Interest (ROI) function in the LSM software. The initial and final planes were also selected, enclosing the fibroblast cluster in the middle. In typical experiments, z planes covering 300 μm thick sections of hydrogels were chosen for irradiation. A two-photon titanium:sapphire laser tuned to 720 nm was scanned across an x-y plane within the hydrogel according to the pre-defined ROI using at 203 mW/μm2 and 240 μsec/μm2, using a 10X Plan-Apochromat objective lens. The same procedure was repeated across the z planes in 3 μm increments. The patterned gel was washed with DMEM to remove any unbound FITC-RGDSK-PEG-acrylate and photoinitiator and incubated at 37 °C with DMEM in 5% CO2 environment. The degradable PEG hydrogels ubiquitously conjugated with RGDS without any patterns were fabricated and used as positive controls in the cell migration studies.
Quantification of patterned RGDK in hydrogel
Successful patterning of FITC-RGDSK-PEG-acrylate was confirmed by visualization of spatially restricted FITC fluorescence within the hydrogels using confocal microscope. To quantify the concentration of FITC-RGDSK-PEG-acrylate patterned in the hydrogels, control hydrogels were first prepared with the known concentrations of FITC-RGDSK-PEG-acrylate (0.45, 0.9 and 1.8 μmol/mL). Then, their fluorescence images were acquired using the same exposure settings in a confocal microscope, and intensity of each fluorescence image was measured with Scion image to generate a standard calibration curve. Hydrogels used in cell migration studies were patterned with FITC-RGDSK-PEG-acrylate as described above, and fluorescence images were obtained from these hydrogels. The concentration of the FITC-RGDSK-PEG-acrylate in the patterned regions of the hydrogels was extrapolated from the calibration curve using fluorescence intensity of the fluorescence images.
Visualization of 3D cell migration in the patterned RGDS regions
3D cell migration within the hydrogels was observed every 24 hr using a phase-contrast microscope. On day 10, cell migration was visualized by fluorescent staining and confocal microscopy. The hydrogel samples were fixed in 3.7% formaldehyde in PBS for 1 hr, followed by cell permeabilization with 0.5% Triton X-100 in PBS for 30 min. Nonspecific adsorption was blocked with 3% bovine serum albumin (BSA; Sigma) in PBS for 1 hr and washed with PBS. F-actin and nuclei were stained with rhodamine-conjugated phalloidin (5 U/ml, Probes) for 2 hr and DAPI (300 nM, Invitrogen) for 20 min. Confocal imaging was performed on a Zeiss LSM 510 META system.
3. Results
Characterization of the synthesized materials
Synthesis of the peptides (GGGLGPAGGK and Fmoc-RGDSK) was confirmed by MALDI-TOF and 1H-NMR. After PEG modification of these peptides, 1H-NMR analysis demonstrated that acrylate-PEG moieties were successfully conjugated to GGGLGPAGGK and Fmoc-RGDSK. After purification, these PEG conjugated products showed the methylene protons of PEG as a triplet at 3.6-3.7 ppm as well as the acrylate protons at 6.0-6.5 ppm. In addition, cleavage of the Fmoc group from Fmoc-RGDSK-PEG-acrylate and the subsequent FITC conjugation were confirmed by the presence of characteristic protons of aromatic ring at 8.5-9.5 ppm. Changes in the molecular weight of each product after PEG or FITC modification were confirmed with MALDI-TOF and GPC equipped an evaporative light scattering detector. This analysis also confirmed that unreacted species (PEG and peptide) were effectively removed by the dialysis procedure.
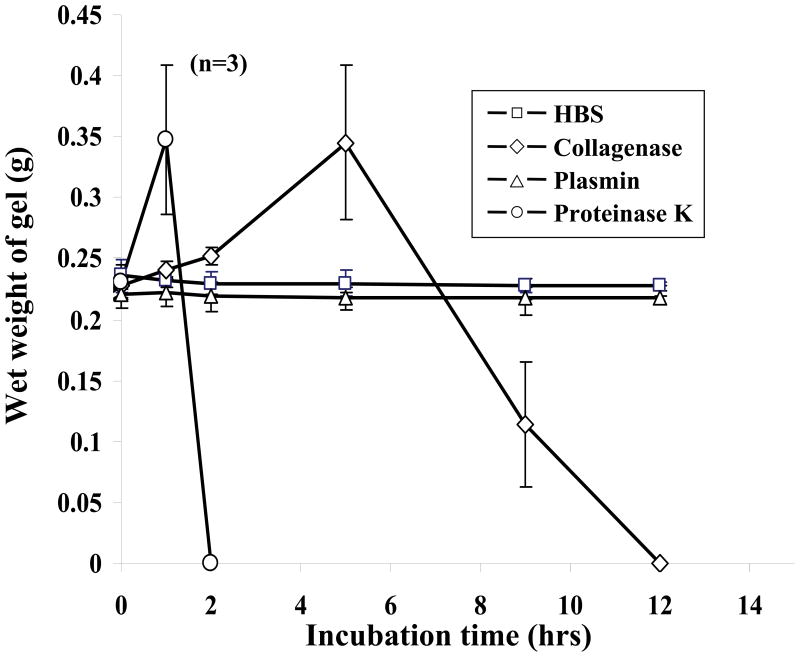
Incorporation of protease-specific peptide sequence into the backbone of the PEG-based hydrogel precursor renders the resulting hydrogels degradable by specific proteases [14]. Degradation profiles of the hydrogels modified with collagenease sensitive peptide sequence, GGGLAPAGGK with a proteolytic site between glycine (G) and leucine (L) [20], were obtained by measuring changes in the wet weight of the hydrogels incubated in collagenase solutions (Figure 1). In the initial degradation phase of hydrogels, the wet weight increased as proteases cleaved the peptides, loosening the hydrogel network and allowing more water to penetrate. Eventually, hydrogels were completely degraded as additional sequences were cleaved and degraded moieties diffused away from the hydrogel. The hydrogels were completely degraded by collagenase (0.2 mg/mL) after 10 hr of incubation. Proteinase K (0.2 mg/mL), a potent protease known to degrade most peptide sequences non-specifically [21], served as a positive control and completely degraded the hydrogels in 2 hr. Plasmin or the buffer solution used as negative controls did not degrade the hydrogels, demonstrating the specificity of the LGPA sequence to collagenase-mediated degradation.
Figure 1.
Degradation profiles of GGGLGPAGGK-derivative PEG hydrogels in protease solutions. Each hydrogel sample was swollen in HBS with 1 mM CaCl2 and 0.2 mg/mL sodium azide at 37 °C for 24 hr, and its weight was monitored in 0.2 mg/mL protease solution at 37 °C: (□) HBS, (◇) collagenase, (△) plasmin, (○) proteinase K. Hydrogels incubated with either collagenase or proteinase K underwent protease-mediated degradation.
The overall patterning strategy
Figure 2 illustrates the overall methodology of guiding cell migration in the preformed hydrogels by rapid 3D TPLS photolithography. First, the hydrogel precursor solution with a fibroblast-seeded fibrin cluster was pre-crosslinked in the presence of a photoinitiator by brief exposure to long wavelength UV light (365 nm, 10 mW/cm2, 20 sec) to form the base hydrogel without adhesion ligands. Although the bulk of acrylate groups is crosslinked at this point, the hydrogel matrices still have free acrylate groups that are available for subsequent conjugation to acrylate-containing moieties [10]. The RGDS peptide modified with an acrylated PEG and a fluorescein was then allowed to diffuse into the hydrogels, and specific regions to be scanned within each focal plane of the hydrogels were drawn as virtual masks in the confocal microscope software. The hydrogels were then irradiated according to the virtual masks by scanning with Ti:sapphire two-photon laser tuned to 720 nm within each focal plane and then incrementing focal planes to create freeform 3D patterns of the cell-adhesive peptide within the hydrogels. This resulted in 3D patterns of covalently immobilized biomolecules within the hydrogel networks.
Figure 2.
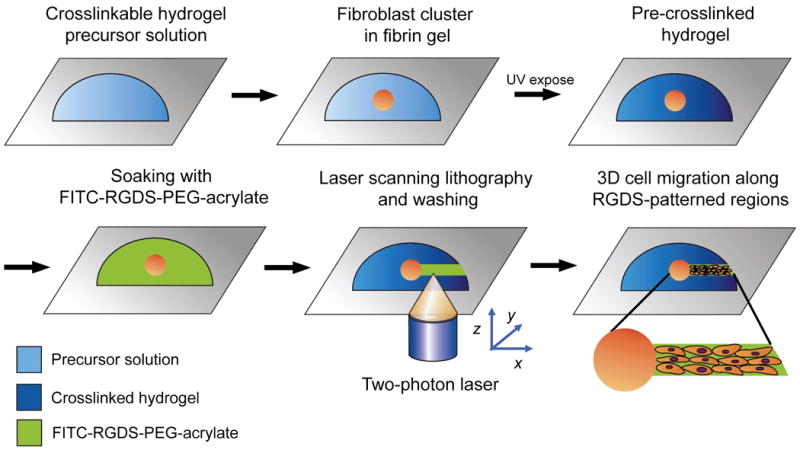
The overall methodology for three-dimensional RGDS patterning by two-photon laser scanning (TPLS) photolithography. First, HDFs encapsulated in fibrin clusters were photopolymerized into collagenase-sensitive PEG hydrogels by exposure to long wavelength UV light. The hydrogels were soaked in PEG-RGDS solution, allowing its diffusion into the bulk of materials. The TPLS photolithographic technique was used to irradiate the hydrogels according to the pre-designed virtual patterns, conjugating PEG-RGDS in 3D network of hydrogels in pre-determined patterns. After washing steps, cell migration was subsequently monitored over time.
Fidelity of the patterned RGDSK in hydrogel
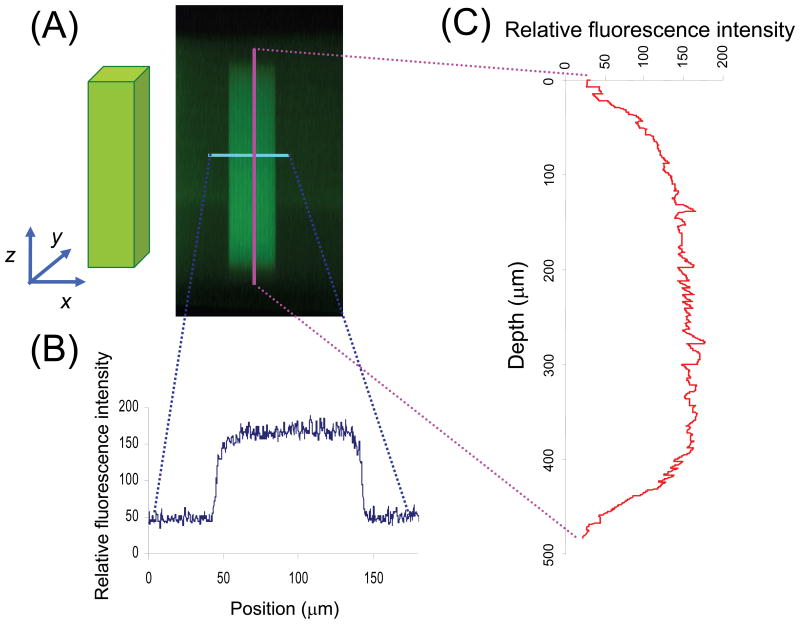
To demonstrate the high fidelity of 3D TPLS photolithographic patterning, cubical regions were patterned in hydrogels, and their fluorescence intensity profiles were examined in the horizontal and vertical cross-sections of the patterned regions using confocal microscopy (Figure 3). A fluorescent marker, FITC-RGDSK-PEG-acrylate, was patterned in the cubical region with d = 100 × 100 × 500 μm and visualized (Figure 3A). The fluorescence intensity profile in the horizontal cross-section displays a sharp contrast between the irradiated and non-irradiated regions of the hydrogel, demonstrating minimal lateral scattering in hydrogels during TPLS irradiation (Figure 3B). The vertical cross-section profile shows relatively consistent fluorescence intensity in the depths ranging from 80 to 430 μm from the surface of the hydrogel (Figure 3C), demonstrating superior capability of TPLS to immobilize biomolecules in selective volumes in the 3D hydrogel networks.
Figure 3.
The characterization of RGDS-patterned region by confocal microscopy (A) XZ cross-sectional image of hydrogel (green fluorescence represents RGDS channel) was used to obtain (B) horizontal and (C) longitudinal fluorescence intensity profile across the RGDS channel. The horizontal and longitudinal cross-sections show sharp contrast between irradiated and non-irradiated regions, demonstrating high fidelity of the TPLS photolithographic technique.
Quantification of the patterned RGDSK in hydrogels
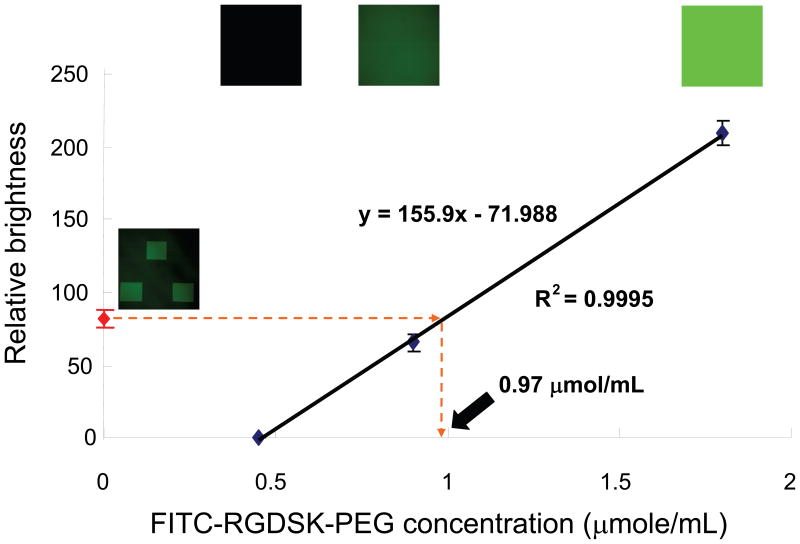
The concentration of the bound cell adhesive peptide (RGDSK) moiety in the patterned regions was quantified by comparing its fluorescence intensity with a standard curve generated from hydrogels photopolymerized with the same moiety in known concentrations. Various concentrations of FITC-RGDSK-PEG-acrylate were mixed with PEGDA precursor solution and photopolymerized to fabricate hydrogels with homogenous distribution of the fluorescent marker. The hydrogels with varying concentrations of FITC-labeled RGDSK were visualized with a confocal microscope using the same settings between the samples. In each hydrogel, planes distant from either the top or bottom surface were used for measurement of fluorescence intensity to avoid any artifacts from interfaces. Fluorescence intensity from each image was measured and plotted against the FITC-RGDSK-PEG-acrylate concentrations to generate a calibration curve as shown in Figure 4. We assumed that 100% of the FITC-RGDSK-PEG-acrylate precursors added and thoroughly mixed with the precursor solution was conjugated to the PEGDA hydrogel networks after photopolymerization as this has been experimentally demonstrated previously using radiolabeled moieties [22]. The calibration curve shows excellent linear relationship between the concentration of the fluorescent marker and brightness of the fluorescence images within the range of concentrations tested. Based on the calibration curve, the patterned region was estimated to contain 0.97 μmol/mL of RGDS. Hydrogels used in the following cell migration studies were patterned with FITC-RGDSK-PEG-acrylate using the same procedure described above.
Figure 4.
Quantification of RGDS concentration conjugated in the hydrogels based on a standard calibration curve generated with known concentrations of FITC-RGDSK-PEG-acrylate. The patterned regions in the hydrogels used for cell migration studies were calculated to contain 0.97 μmol/mL of RGDS.
3D cell migration in RGDSK-patterned regions of degradable PEG hydrogel
Proteolytically degradable PEG hydrogels homogenously conjugated with RGDSK-PEG-acrylate were fabricated and used in this study as positive controls to demonstrate that these hydrogels are suitable materials to support cell migration. HDFs were encapsulated in fibrin gels, which in turn were encapsulated in collagenase-sensitive PEG hydrogels with RGDSK-PEG-acrylate via photopolymerization. HDFs migrated out radially from the cell clusters into the degradable PEG hydrogel networks (Figure 5A-C). On the other hand, cells entrapped in degradable PEG hydrogels without RGDSK or in non-degradable PEG hydrogels with RGDSK failed to migrate out from the cell clusters as expected (Figure 5D).
Figure 5.
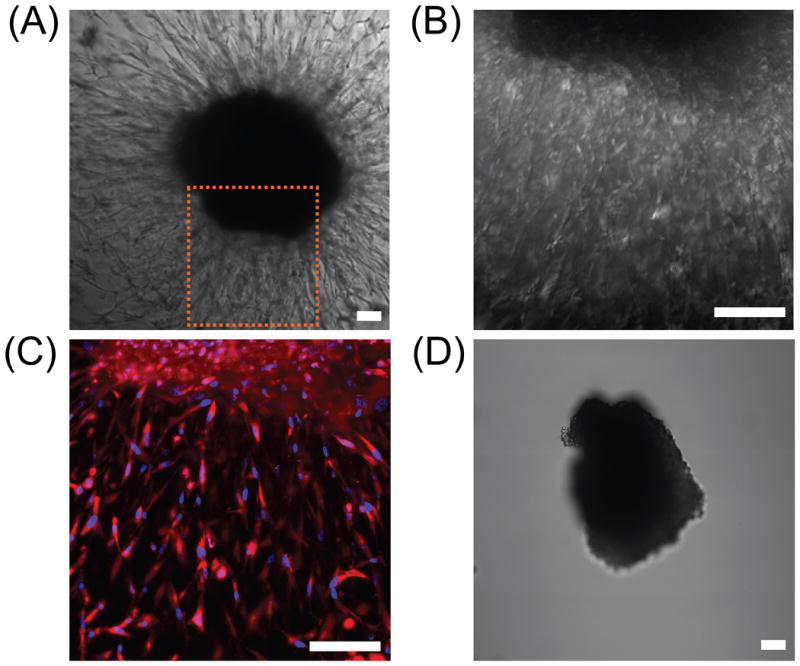
(A-C) DIC and confocal fluorescence images of HDFs undergoing 3D migration within degradable PEG hydrogel with homogenously distributed RGDS; (A) DIC image after 10 days of cell culture, (B) higher magnification DIC image of panel (A), and (C) DAPI and rhodamine-phallodin staining of cells shown in panel (B). (D) A cluster of HDFs encapsulated in a non-degradable hydrogel with RGDS did not show any signs of cell migration after 5 days of cell culture as cells could not penetrate the hydrogel network lacking collagenase-sensitive peptide linker. Scale bars = 100 μm.
To create collagenase-sensitive PEG hydrogels with patterned RGDS, HDF clusters encapsulated in collagenase-sensitive PEG hydrogels were submerged in 30 μmol/mL solution of FITC-RGDSK-PEG-acrylate for 30 min. Subsequently, using TPLS photolithography, conjugation of FITC-RGDSK-PEG-acrylate to the base PEGDA hydrogels was spatially controlled. Specifically, channels of RGDS were conjugated adjacent to HDF clusters to allow guided cell migration out from the cell clusters.
In these hydrogels spatially patterned with RGDSK, the HDFs were observed to extend numerous processes and migrate out from the fibrin gels into the regions patterned with RGDSK (Figure 6). On day 10, cells in the hydrogels were fixed and stained with rhodamine-phalloidin and DAPI to reveal F-actin and nuclei. Multiple nuclei in the RGDSK patterned regions indicate that the cells did not simply extend lamellipodia from the fibrin cell cluster, but actively migrated out en masse into the collagenase-sensitive hydrogels. This migration is not simply due to proliferation within the fibrin cluster since cell migration was selectively restricted to the RGDSK-patterned regions. Interestingly, when the migrating cells encountered an intersection of the RGDSK-patterned areas, they branched and migrated into two separate paths.
Figure 6.
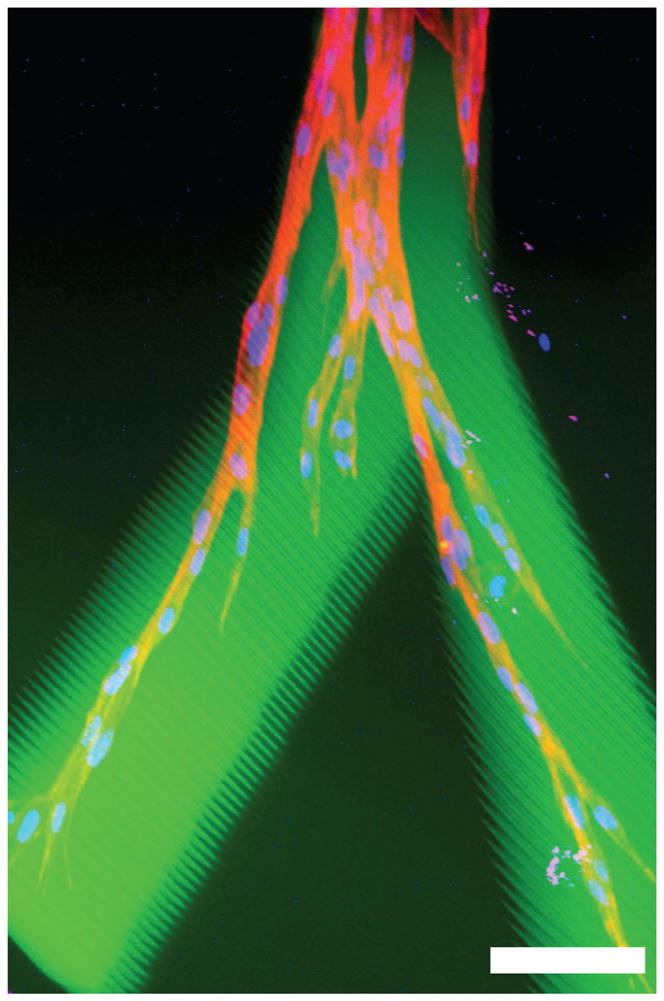
Confocal microscope image of HDFs undergoing 3D migration within a RGDS patterned region inside a collagenase-sensitive PEG hydrogel; an overlaid image shows FITC-RGDS-PEG-acrylate, rhodamine-phalloidin, and DAPI. Scale bar = 100 μm.
4. Discussion
The key for developing clinically relevant tissues in laboratories is in recreating the complex 3D distribution of biomolecules and cells in scaffold materials and maintaining their viability and functions. The work presented here represents a significant stride forward in development of functional tissues in vitro. We used the TPLS photolithographic technique to dictate the precise location of a cell adhesive ligand in hydrogels, and the cells encapsulated in these hydrogels were shown to be both viable and functional as they underwent guided 3D cell migration within the patterned regions of the hydrogels. These results demonstrate the prospect of guiding tissue regeneration at the microscale in 3D scaffolds by providing appropriate, spatially constrained bioactive cues. In these materials, cells proteolytically degrade the synthetic hydrogel locally and replace it with extracellular matrix proteins. The localization of cells to the patterned region is then maintained by the surrounding environment that is not permissive of cell invasion.
The use of laser photolithography to pattern biomolecules in 3D networks of hydrogels has been previously reported [9,23,24], but their applicability to create complex, functional tissues have been limited. For example, in the work by Luo et al. [9], ganglia cells seeded on the top surface of agarose gels were able to invade into the vertical GRGDS channels patterned with a beam of ultraviolet (UV) light; however, the precise control over the biomolecule distribution in z direction was inadequate due to the use of a conventional laser, requiring straight passes of light through the entirety of the material. Additionally, transmittance of the UV light decreased significantly through the hydrogel material and resulted in a decreasing concentration gradient of immobilized GRGDS from the surface, forming a barrier for chemotactic cell migration along the longitudinal direction of the channels. The same group recently addressed this issue by employing multi photon laser to immobilize biomolecules in micropatterns within agarose hydrogels; however, close guidance of cellular migration and tissue regeneration are yet to be demonstrated in these materials [23]. In another study [24], non-degradable PEGDA hydrogels were used as a patterning substrate, drastically limiting its application to simple cell encapsulation studies.
Departing from these previous studies, we took advantage of high resolution TPLS photolithographic technique developed in our laboratory [10] and sought to spatially control the movements of migrating cells in 3D network of biodegradable hydrogels. Specifically, the hydrogel precursor solution was photopolymerized around fibroblast clusters in fibrin gels, and the hydrogels were patterned with RGDS in selective regions with the TPLS photolithographic technique. Two-photon laser delivers highly focused laser beam to achieve two-photon excitation within minuscule focal volume, confining RGDSK conjugation reactions within the micro-environment. As shown in Fig. 6, fibroblasts migrated into the local regions decorated with RGDSK, demonstrating applications of TPLS photolithography to create hydrogels that can guide and restrict cell migrations in 3D.
In these hydrogels, fibroblasts extended long sprouts from the fibrin cellular clusters into the hydrogels by day 10. The cellular sprouts overlapped with FITC label in RGDSK in the horizontal optical sections indicating that the cellular migration occurred successfully within the 3D paths patterned with RGDSK. Multiple nuclei staining along the patterned regions confirmed that the cells actively migrated out into the hydrogels.
In this study, relatively high PEG-RGDS concentration of 30 μmol/mL was used to allow its diffusion into hydrogels and subsequent conjugation to hydrogels by the TPLS technique. These hydrogels were found to have RGDS concentration of 0.97 μmol/mL as measured by its fluorescence intensity, indicating a low (ca. 3%) conjugation efficiency of the TPLS technique. The TPLS technique was used in this study to allow conjugation of acrylate-containing moieties into pre-formed hydrogels. Since the majority of acrylate groups in the PEG polymer was crosslinked to form the bulk of hydrogels, only a small number of free acrylate groups was available for subsequent conjugation with soluble acrylate-containing moieties, hence its low conjugation efficiency. This contrasts to the high conjugation efficiency (ca. 100%) of PEG-RGDS added in precursor solution, in which PEG-RGDS was thoroughly mixed with PEGDA and photopolymerized, grafting the majority of PEG-RGDS into the crosslinked PEGDA networks [22]. As a result of the low conjugation efficiency, the RGDS concentration of 0.97 μmol/mL in the hydrogels used in this study was less than one third of the optimal RGDS concentration of 3.5 μmol/mL for migration of HDFs and smooth muscle cells in the hydrogels [13]. Albeit less than the optimal RGDS concentration, HDFs were still able to migrate out from the cell clusters into the hydrogels. Future modifications to the patterning condition should yield hydrogels with an optimal RGDS concentration for cell migration. In addition, by varying the exposure settings during the patterning procedure, it will be possible to create gradients of RGDS inside the 3D network of hydrogels to stimulate chemotactic cell migration.
5. Conclusions
We have demonstrated versatile applications of TPLS photolithographic technique to create PEG hydrogel scaffolds with complex 3D distribution of biomolecules seamlessly integrated with resident cells. This biomimetic hydrogel system can be readily modified with various biomolecules such as peptides, growth factors, and other signaling molecules, and this system can be applied to basic science to investigate various aspects of cell locomotion to advance our knowledge in wound healing, inflammation, embryogenesis, and tumor cell metastasis. In addition, applications of TPLS photolithographic technique in tissue engineering may lead to development of more native tissue-like constructs that can support and direct the complex processes of tissue regeneration.
Supplementary Material
Acknowledgments
This work was supported by NIH (R01 HL070537) and Korea Research Foundation Grant (M01-2003-000-20016-0).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu VA, Bhatia SN. Three-dimensional photopatterning of hydrogels containing living cells. Biomedical Microdevices. 2002;4:257–266. [Google Scholar]
- 2.Yu TY, Ober CK. Methods for the topographical patterning and patterned surface modification of hydrogels based on hydroxyethyl methacrylate. Biomacromolecules. 2003;4:1126–1131. doi: 10.1021/bm034079m. [DOI] [PubMed] [Google Scholar]
- 3.Ward JH, Bashir R, Peppas NA. Micropatterning of biomedical polymer surfaces by novel UV polymerization techniques. Journal of Biomedical Materials Research. 2001;56:351–360. doi: 10.1002/1097-4636(20010905)56:3<351::aid-jbm1103>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 4.Suh KY, Seong J, Khademhosseini A, Laibinis PE, Langer R. A simple soft lithographic route to fabrication of poly(ethylene glycol) microstructures for protein and cell patterning. Biomaterials. 2004;25:557–63. doi: 10.1016/s0142-9612(03)00543-x. [DOI] [PubMed] [Google Scholar]
- 5.Tien J, Nelson CM, Chen CS. Fabrication of aligned microstructures with a single elastomeric stamp. Proc Natl Acad Sci U S A. 2002;99:1758–62. doi: 10.1073/pnas.042493399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan W, Desai TA. Microfluidic patterning of cellular biopolymer matrices for biomimetic 3-D structures. Biomedical Microdevices. 2003;5:235–244. [Google Scholar]
- 7.Fernandes R, Wu LQ, Chen TH, Yi HM, Rubloff GW, Ghodssi R, et al. Electrochemically induced deposition of a polysaccharide hydrogel onto a patterned surface. Langmuir. 2003;19:4058–4062. [Google Scholar]
- 8.Mironov V, Boland T, Trusk T, Forgacs G, Markwald RR. Organ printing: computer-aided jet-based 3D tissue engineering. Trends in Biotechnology. 2003;21:157–161. doi: 10.1016/S0167-7799(03)00033-7. [DOI] [PubMed] [Google Scholar]
- 9.Luo Y, Shoichet MS. A photolabile hydrogel for guided three-dimensional cell growth and migration. Nat Mater. 2004;3:249–53. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 10.Hahn MS, Miller JS, West JL. Three-dimensional biochemical and biomechanical patterning of hydrogels for guiding cell behavior. Advanced Materials. 2006;18:2679–2684. [Google Scholar]
- 11.Denk W, Strickler JH, Webb WW. 2-Photon Laser Scanning Fluorescence Microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 12.Mann BK, Gobin AS, Tsai AT, Schmedlen RH, West JL. Smooth muscle cell growth in photopolymerized hydrogels with cell adhesive and proteolytically degradable domains: synthetic ECM analogs for tissue engineering. Biomaterials. 2001;22:3045–51. doi: 10.1016/s0142-9612(01)00051-5. [DOI] [PubMed] [Google Scholar]
- 13.Gobin AS, West JL. Cell migration through defined, synthetic ECM analogs. FASEB J. 2002;16:751–3. doi: 10.1096/fj.01-0759fje. [DOI] [PubMed] [Google Scholar]
- 14.West JL, Hubbell JA. Polymeric biomaterials with degradation sites for proteases involved in cell migration. Macromolecules. 1999;32:241–244. [Google Scholar]
- 15.DeLong SA, Moon JJ, West JL. Covalently immobilized gradients of bFGF on hydrogel scaffolds for directed cell migration. Biomaterials. 2005;26:3227–34. doi: 10.1016/j.biomaterials.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 16.Lee SH, Miller JS, Moon JJ, West JL. Proteolytically degradable hydrogels with a fluorogenic substrate for studies of cellular proteolytic activity and migration. Biotechnology Progress. 2005;21:1736–1741. doi: 10.1021/bp0502429. [DOI] [PubMed] [Google Scholar]
- 17.Lee SH, Moon JJ, Miller JS, West JL. Poly(ethylene glycol) hydrogels conjugated with a collagenase-sensitive fluorogenic substrate to visualize collagenase activity during three-dimensional cell migration. Biomaterials. 2007;28:3163–70. doi: 10.1016/j.biomaterials.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Moon JJ, Lee SH, West JL. Synthetic biomimetic hydrogels incorporated with ephrin-A1 for therapeutic angiogenesis. Biomacromolecules. 2007;8:42–9. doi: 10.1021/bm060452p. [DOI] [PubMed] [Google Scholar]
- 19.Mann BK, West JL. Cell adhesion peptides alter smooth muscle cell adhesion, proliferation, migration, and matrix protein synthesis on modified surfaces and in polymer scaffolds. J Biomed Mater Res. 2002;60:86–93. doi: 10.1002/jbm.10042. [DOI] [PubMed] [Google Scholar]
- 20.Steinbrink DR, Bond MD, Van Wart HE. Substrate specificity of beta-collagenase from Clostridium histolyticum. J Biol Chem. 1985;260:2771–6. [PubMed] [Google Scholar]
- 21.Barrett AJ, Rawlings ND, Woessner JJF. Handbook of Proteolytic Enzymes. San Diego: Academic Press; 1998. [Google Scholar]
- 22.Hern DL, Hubbell JA. Incorporation of adhesion peptides into nonadhesive hydrogels useful for tissue resurfacing. J Biomed Mater Res. 1998;39:266–76. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 23.Wosnick JH, Shoichet MS. Three-dimensional chemical Patterning of transparent hydrogels. Chemistry of Materials. 2008;20:55–60. [Google Scholar]
- 24.Cheung YK, Gillette BM, Zhong M, Ramcharan S, Sia SK. Direct patterning of composite biocompatible microstructures using microfluidics. Lab on a Chip. 2007;7:574–579. doi: 10.1039/b700869d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.