Abstract
BACKGROUND
The clinical utility of genotype-guided (pharmacogenetically based) dosing of warfarin has been tested only in small clinical trials or observational studies, with equivocal results.
METHODS
We randomly assigned 1015 patients to receive doses of warfarin during the first 5 days of therapy that were determined according to a dosing algorithm that included both clinical variables and genotype data or to one that included clinical variables only. All patients and clinicians were unaware of the dose of warfarin during the first 4 weeks of therapy. The primary outcome was the percentage of time that the international normalized ratio (INR) was in the therapeutic range from day 4 or 5 through day 28 of therapy.
RESULTS
At 4 weeks, the mean percentage of time in the therapeutic range was 45.2% in the genotype-guided group and 45.4% in the clinically guided group (adjusted mean difference, [genotype-guided group minus clinically guided group], −0.2; 95% confidence interval, −3.4 to 3.1; P=0.91). There also was no significant between-group difference among patients with a predicted dose difference between the two algorithms of 1 mg per day or more. There was, however, a significant interaction between dosing strategy and race (P=0.003). Among black patients, the mean percentage of time in the therapeutic range was less in the genotype-guided group than in the clinically guided group. The rates of the combined outcome of any INR of 4 or more, major bleeding, or thromboembolism did not differ significantly according to dosing strategy.
CONCLUSIONS
Genotype-guided dosing of warfarin did not improve anticoagulation control during the first 4 weeks of therapy. (Funded by the National Heart, Lung, and Blood Institute and others; COAG ClinicalTrials.gov number, NCT00839657.)
The need for clinical trials before widespread adoption of genotype-guided drug dosing and selection remains widely debated.1–4 Warfarin therapy has served as a model for the potential for pharmacogenetics to improve patient care.1 Observational studies have identified two genes, CYP2C9 and VKORC1, that are associated with variation in warfarin maintenance doses. However, the clinical utility of starting warfarin at the maintenance dose predicted by genotype-guided algorithms has been tested only in small trials, none of which were definitive.5–8 In contrast, observational studies have suggested potential benefits from genotype-guided dosing.9,10 In addition, previous clinical trials could not determine the usefulness of current dosing algorithms among black patients, for whom genotype-guided algorithms perform less well than for other populations.11–13
On the basis of available data, the Food and Drug Administration (FDA) has updated the label for warfarin twice, suggesting that variants in CYP2C9 and VKORC1 may be taken into consideration when choosing the initial warfarin dose. However, the Centers for Medicare and Medicaid Services did not find sufficient evidence to cover the cost of genotyping for warfarin dosing.14 Our study, called the Clarification of Optimal Anticoagulation through Genetics (COAG) trial, was designed to test the effect of genotype-guided dosing on anticoagulation control.
METHODS
STUDY DESIGN AND OVERSIGHT
The COAG trial was a multicenter, double-blind, randomized, controlled trial that compared a genotype-guided warfarin-dosing strategy with a clinically based dosing strategy during the first 5 days of therapy among patients initiating warfarin treatment.15–17 The study was designed by the authors and approved by the institutional review board at the University of Pennsylvania and at each participating clinical center. The data were collected, analyzed, and interpreted by the authors. A steering committee provided oversight of the trial (for details, see the Supplementary Appendix, available with the full text of this article at NEJM.org). An independent data and safety monitoring board monitored the trial and made recommendations to the National Heart, Lung, and Blood Institute. The first two authors wrote the first draft of the manuscript, which was edited and approved by all the authors.
The National Heart, Lung, and Blood Institute supported this study. Bristol-Myers Squibb donated Coumadin (warfarin). GenMark Diagnostics and AutoGenomics loaned genotyping platforms to the clinical centers. None of the companies supporting the trial had any role in the design of the protocol or in the collection, analysis, or interpretation of the data. The authors vouch for the data and the analyses, and for the fidelity of this report to the trial protocol, which is available at NEJM.org.
STUDY PATIENTS AND RANDOMIZATION
From September 2009 through April 2013, we enrolled both inpatients and outpatients at 18 clinical centers in the United States. All the patients were adults initiating warfarin therapy with a target international normalized ratio (INR) of 2 to 3. Detailed inclusion and exclusion criteria are provided in the Supplementary Appendix. All patients provided written informed consent.
Patients were randomly assigned, in a 1:1 ratio, to the use of a dosing algorithm that included both clinical variables and genotype data or to a clinically guided dosing strategy. Randomization was stratified according to clinical center and self-reported race (black vs. nonblack).
Genotyping for CYP2C9 and VKORC1 at each clinical center was performed with the use of one of two FDA-approved platforms, the Gen-Mark Dx eSensor XT-8 or the AutoGenomics INFINITI Analyzer. Per protocol, genotyping was performed in all patients immediately after blood-sample collection to maintain blinding to the treatment assignment. Genotyping was repeated at the central laboratory with the use of either pyrosequencing or real-time polymerase-chain-reaction assay to measure the accuracy at clinical centers.
STUDY INTERVENTION AND FOLLOW-UP
The study intervention period was the first 5 days of warfarin therapy. During this period, the pre-specified algorithms were used to determine the warfarin dose. For each dosing strategy, a dose-initiation algorithm was used during the first 3 days of therapy, and a dose-revision algorithm was used on day 4, 5, or both. The algorithms for the genotype-guided dosing strategy12,18 included clinical variables and genotype data for CYP2C9*2, CYP2C9*3, and VKORC1. The algorithms for the clinically based dosing strategy included clinical variables only. The dosing algorithms are provided in the Supplementary Appendix. If genotype information was not available for a patient in the genotype-guided dosing group before the administration of warfarin on any given day in the first 5 days, the clinical algorithm was used on that day.
During the first 4 weeks of therapy, patients and clinicians were unaware of the actual dose of warfarin that was administered, because the pills were encapsulated to prevent identification of the dose (see the Supplementary Appendix). After the 5-day initiation period, we adjusted the dose during the first 4 weeks using standardized dose-adjustment techniques,5,10 starting with the doses predicted by the algorithms and making the same relative adjustments on the basis of the INR in the two study groups. Clinicians were informed of the relative dose change (e.g., a 10% dose increase) at each INR measurement but not the actual dose of warfarin. Clinicians could contact the medical monitor (who was aware of the study-group assignments) to request an override of these relative dose changes without being informed of the actual dose. All patients were to be followed for a total of 6 months.
STUDY OUTCOMES
The primary outcome was the percentage of time in the therapeutic range (INR, 2 to 3) from the completion of the intervention period (day 4 or 5) through day 28 of therapy. We calculated the percentage of time in the therapeutic range using a standard linear interpolation method between successive INR values,19 as detailed in the Supplementary Appendix. Each clinical center measured INRs with the use of instruments certified by the Clinical Laboratory Improvement Amendments and following strict quality assurance.
Secondary outcomes included a composite outcome of any INR of 4 or more, major bleeding, or thromboembolism in the first 4 weeks (principal secondary outcome); the time to the first therapeutic INR; the time to the determination of a maintenance dose (which was defined as the time to the first of two consecutive INR measurements, measured at least 1 week apart, that were in the therapeutic range without a dose change); and the time to an adverse event (death from any cause, major bleeding, thromboembolism, or any clinically relevant nonmajor bleeding event20,21) in the first 4 weeks. Two physicians who were unaware of the study-group assignments adjudicated major bleeding and thromboembolic serious adverse events. The definitions of major bleeding,22 clinically relevant nonmajor bleeding, and thromboembolism are provided in the Supplementary Appendix.
STATISTICAL ANALYSIS
We analyzed the primary outcome in the modified intention-to-treat population, which included all patients who underwent randomization with the exception of patients for whom INR data were not available (Fig. S1 in the Supplementary Appendix). Safety outcomes were analyzed in the entire cohort, regardless of whether patients received the study drug. We used regression models to analyze the primary and secondary outcomes, using linear regression for the percentage of time in the therapeutic range and Cox regression for time-to-event outcomes. The protocol specified that we conduct coprimary analyses in which we evaluated the primary outcome in all patients and in a primary subgroup, which comprised patients who had an absolute difference of 1.0 mg or more in the predicted initial daily dose between the genotype-guided dosing algorithm and the clinical dosing algorithm. We used an alpha allocation approach, which formally allows for the evaluation of the treatment benefit in an enriched subgroup as a coprimary end point. In this approach, the overall type I error rate of 0.05 for the primary outcome was split between the analyses performed among all patients and among those in the primary subgroup.17 All models were adjusted for the stratification variables (center and race). Additional subgroups, which were prespecified, were race (black vs. nonblack), sex, and the total number of allelic variants (1 variant vs. 0 or >1 variant in either CYP2C9 or VKORC15). All statistical tests were two-sided. All analyses were performed with the use of the R statistical package, version 3.0.1 (R Development Core Team).
We specified a minimum detectable difference of 5.5% in the mean percentage of time in the therapeutic range between the genotype-guided group and the clinically guided group in the entire study population.16 We assumed a standard deviation for the percentage of time in therapeutic range of 25% and a potential dropout rate of 10%. On the basis of recruitment rates,15 the initial sample size of 1238 patients was revised to 1022 patients on September 16, 2012 (with the approval of the data and safety monitoring board). The revised sample size provided a power of at least 80% to detect a between-group difference of 5.5% at a type I error rate of 0.04 among all patients and a 9.0% difference at a type I error rate of 0.01 among patients in the coprimary analysis.
RESULTS
PATIENTS, GENOTYPING, AND FOLLOW-UP
A total of 1015 patients were enrolled and randomly assigned to either the genotype-guided dosing algorithm or the clinically guided dosing algorithm (Fig. S1 in the Supplementary Appendix). There were no significant between-group differences at baseline (Table 1). The characteristics of the patients according to self-reported race are provided in Table S1 in the Supplementary Appendix. A total of 60 participants (30 in each group) withdrew before completing the intervention period and did not have an available percentage of time in the therapeutic range, resulting in an analytic sample size of 955. A median of six INRs were measured during the first 4 weeks in each of the two study groups. Dispensed doses during the intervention period are summarized in Table S2 in the Supplementary Appendix.
Table 1.
Characteristics of the Patients at Baseline.*
| Characteristic | Genotyped-Guided Group (N = 514) | Clinically Guided Group (N = 501) |
|---|---|---|
| Median age (IQR) — yr† | 59 (48–70) | 57 (46–68) |
| Male sex — no. (%) | 272 (53) | 246 (49) |
| Race or ethnic group — no. (%)‡ | ||
| Black† | 141 (27) | 134 (27) |
| Hispanic | 32 (6) | 33 (7) |
| Education — no. (%) | ||
| Did not complete high school | 52 (10) | 44 (9) |
| High-school diploma only | 131 (25) | 133 (27) |
| Post-secondary education | 308 (60) | 291 (58) |
| Did not respond | 23 (4) | 33 (7) |
| Current smoker — no. (%)† | 77 (15) | 68 (14) |
| Median body-surface area (IQR) — m2† | 2.01 (1.83–2.19) | 2.03 (1.85–2.23) |
| Warfarin and other therapies — no. (%) | ||
| Inpatient warfarin initiation | 348 (68) | 332 (66) |
| Indication for warfarin therapy | ||
| Deep-vein thrombosis or pulmonary embolism only | 289 (56) | 300 (60) |
| Atrial fibrillation or flutter only | 116 (23) | 105 (21) |
| Other indication only | 56 (11) | 53 (11) |
| Multiple indications | 49 (10) | 39 (8) |
| No indication given | 4 (1) | 4 (1) |
| Deep-vein thrombosis or pulmonary embolism as primary indication† | 305 (59) | 317 (63) |
| Expected duration of warfarin therapy | ||
| <1 mo | 33 (6) | 33 (7) |
| 1–3 mo | 35 (7) | 30 (6) |
| >3 mo | 446 (87) | 438 (87) |
| Previous warfarin use | 38 (7) | 48 (10) |
| Current amiodarone use† | 13 (3) | 10 (2) |
| Current fluvastatin use† | 2 (<1) | 1 (<1) |
| Current heparin use | 278 (54) | 281 (56) |
| Medical history — no. (%) | ||
| Congestive heart failure | 63 (12) | 64 (13) |
| Deep-vein thrombosis | 149 (29) | 146 (29) |
| Diabetes† | 118 (23) | 121 (24) |
| Hypertension | 280 (54) | 260 (52) |
| Myocardial infarction | 47 (9) | 48 (10) |
| Pulmonary embolism | 109 (21) | 105 (21) |
| Stroke† | 37 (7) | 31 (6) |
| Genetic variants — no. (%) | ||
| CYP2C9*2† | ||
| No variants | 414 (81) | 423 (84) |
| Heterozygous | 92 (18) | 70 (14) |
| Homozygous | 4 (1) | 7 (1) |
| Withdrew before genotyping | 4 (1) | 1 (<1) |
| CYP2C9*3† | ||
| No variants | 471 (92) | 451 (90) |
| Heterozygous | 38 (7) | 49 (10) |
| Homozygous | 1 (<1) | 0 |
| Withdrew before genotyping | 4 (1) | 1 (<1) |
| VKORC1 (VKORC1 3673G A)† | ||
| No variants (GG) | 250 (49) | 237 (47) |
| Heterozygous (AG or GA) | 201 (39) | 202 (40) |
| Homozygous (AA) | 59 (11) | 61 (12) |
| Withdrew before genotyping | 4 (1) | 1 (<1) |
| Total no. of variants§ | ||
| 0 | 204 (40) | 189 (38) |
| 1 | 178 (35) | 186 (37) |
| >1 | 128 (25) | 125 (25) |
| Withdrew before genotyping | 4 (1) | 1 (<1) |
There were no significant between-group differences for any characteristic. IQR denotes interquartile range.
This variable was used in the algorithms for dose initiation and dose revision in the two study groups. Dosing algorithms are provided in the Supplementary Appendix.
Race or ethnic group was self-reported.
The total number of variants was defined as the number of measured variants in CYP2C9*2, CYP2C9*3, and VKORC1.
Genotype data were available in the genotype-guided group for 45% of the patients before the first warfarin dose, for 94% before the second warfarin dose, and for 99% before the application of the dose-revision algorithm on day 4 or 5. The mean (±SD) difference between the dose calculated for patients without genotype data on day 1, as compared with the dose they would have received if genotype data had been available, was −0.1±0.4 mg per day during the first 3 days. The central laboratory confirmed 99.8% of all genotyping results from the clinical centers. All genotype distributions were in Hardy–Weinberg equilibrium (P>0.20 for all comparisons).
PRIMARY OUTCOME
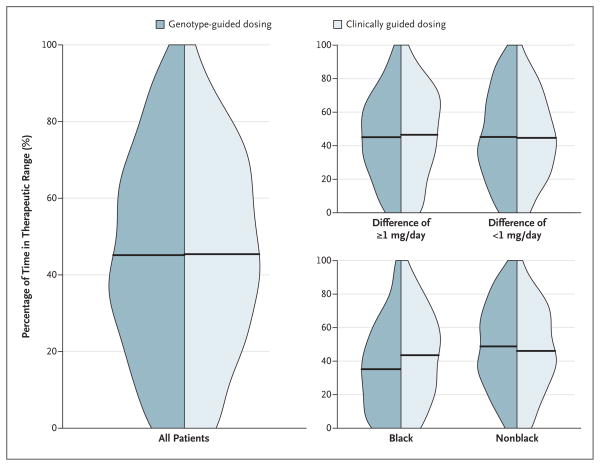
At 4 weeks, there was no significant between-group difference in the mean percentage of time in the therapeutic range: 45.2% in the genotype-guided group and 45.4% in the clinically guided group (adjusted mean difference [genotype-guided group minus clinically guided group], −0.2%; 95% confidence interval, −3.4 to 3.1; P=0.91) (Table 2 and Fig. 1). There was also no significant between-group difference in the percentage of time in the therapeutic range among patients in the coprimary analysis (Table 2). When the 4-week trial was divided into two 2-week intervals, there was also no significant difference between the groups in either interval (Table 2).
Table 2.
Percentage of Time in the Therapeutic INR Range through Week 4 of Therapy, According to Subgroup.*
| Variable | No. of Patients | Genotype-Guided Group | Clinically Guided Group | Mean Difference (95% CI) † | P Value |
|---|---|---|---|---|---|
| percent | percentage point | ||||
| Primary analyses | 955 | ||||
|
| |||||
| All patients | 45.2±26.6 | 45.4±25.8 | −0.2 (−3.4 to 3.1) | 0.91‡ | |
|
| |||||
| Patients stratified by absolute difference between algorithms in predicted dose | 0.63 ¶ | ||||
|
| |||||
| ≥1.0 mg/day§ | 392 | 45.1±25.5 | 46.5±27.1 | −1.1 (−6.2 to 4.0) | 0.67|| |
|
| |||||
| <1.0 mg/day | 563 | 45.2±27.4 | 44.7±24.8 | 0.5 (−3.7 to 4.8) | 0.81 |
|
| |||||
| Prespecified subgroup analyses | |||||
|
| |||||
| Race | 0.003¶ | ||||
|
| |||||
| Black | 255 | 35.2±26.0 | 43.5±26.5 | −8.3 (−15.0 to −2.0) | 0.01 |
|
| |||||
| Nonblack | 700 | 48.8±25.9 | 46.1±25.5 | 2.8 (−1.0 to 6.6) | 0.15 |
|
| |||||
| Sex | 0.71¶ | ||||
|
| |||||
| Male | 486 | 44.9±26.9 | 45.5±25.4 | 0.4 (−4.2 to 5.1) | 0.85 |
|
| |||||
| Female | 469 | 45.4±26.3 | 45.3±26.2 | −0.8 (−5.5 to 3.9) | 0.73 |
|
| |||||
| Total no. of genetic variants ** | 0.21¶ | ||||
|
| |||||
| 1 | 343 | 48.1±26.5 | 45.0±23.7 | 2.6 (−2.9 to 8.1) | 0.35 |
|
| |||||
| 0 or >1 variant | 612 | 43.6±26.5 | 45.7±27.0 | −1.7 (−5.8 to 2.4) | 0.41 |
|
| |||||
| Analysis according to 2-wk intervals | |||||
|
| |||||
| From day 4 or 5 to day 14 | 935 | 40.3±28.3 | 40.3±27.3 | 0.1 (−3.4 to 3.6) | 0.96 |
|
| |||||
| From day 15 to day 28 | 913 | 59.9±36.6 | 59.9±36.3 | 0.0 (−4.8 to 4.7) | 0.99 |
Plus–minus values are means ±SD. CI denotes confidence interval.
Values are the mean difference in the percentage of time in the therapeutic INR range in the genotype-guided group as compared with the clinically guided group, as estimated from multivariable linear regression models and adjusted for race and clinical center. A positive value indicates more time in the therapeutic range in the genotype-guided group.
The type I error rate was fixed at 0.04.
Patients who had an absolute difference of 1.0 mg or more in the predicted initial daily dose between the genotype-guided dose-initiation algorithm and the clinically guided dose-initiation algorithm were designated as the coprimary analysis group.
The P value for interaction was calculated to evaluate the equality of the mean difference between subgroups.
The type I error rate was calculated to be 0.016 on the basis of the alpha allocation approach.17
The total number of genetic variants was defined as the number of measured variants in CYP2C9*2, CYP2C9*3, and VKORC1.
Figure 1. Distribution of Time in the Therapeutic Range.
Side-by-side density plots show the distribution of the percentage of time in the therapeutic range of the international normalized ratio (INR) from the completion of the intervention period (day 4 or 5) to day 28 of therapy for the two study groups among all patients (at left), among patients stratified according to the absolute difference in the predicted initial daily dose of warfarin between the two algorithms (=1 mg [primary subgroup] vs. <1 mg) (at top right), and among patients stratified according to race (at bottom right). The horizontal lines indicate the mean percentage of time in the therapeutic range.
However, there was a significant interaction between race and dosing strategy (P = 0.003) (Table 2). Among black patients, the mean percentage of time in the therapeutic range was less in the genotype-guided group than in the clinically guided group (35.2% vs. 43.5%; adjusted mean difference, −8.3%; P = 0.01). Among nonblack patients, the mean percentage of time in the therapeutic range was slightly higher in the genotype-guided group than in the clinically guided group (48.8% vs. 46.1%; adjusted mean difference, 2.8%; P = 0.15). There were no significant differences in the percentage of time in the therapeutic range according to sex or the total number of genetic variants (Table 2).
ANTICOAGULATION CONTROL AND DOSE PREDICTION
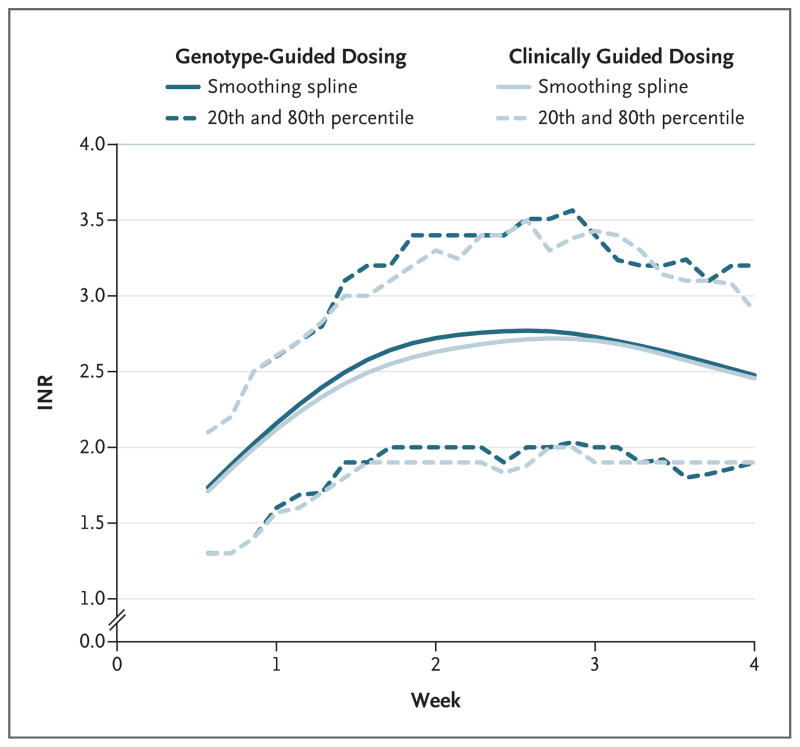
There were no significant between-group differences in the mean percentage of time above the therapeutic range (INR, >3) or below the therapeutic range (INR, <2) (Fig. 2, and Table S3 in the Supplementary Appendix). However, black patients in the genotype-guided group were more likely to have INRs above the therapeutic range than were those in the clinically guided group (Fig. S2 and Table S3 in the Supplementary Appendix).
Figure 2. Range of INRs during the 4-Week Study.
Shown are the INRs from the completion of the intervention period (day 4 or 5) to day 28 of therapy in the two study groups. Solid lines represent smoothing splines with 5 degrees of freedom. Dashed lines represent the 20th and 80th percentiles of INR values calculated over a 3-day window.
There was no overall between-group difference in the time to the first INR in the therapeutic range (Table S4 in the Supplementary Appendix). However, black patients in the genotype-guided group took longer on average to reach the first therapeutic INR than did those in the clinically guided group (Table S4 and Fig. S3 in the Supplementary Appendix). The time to the determination of the maintenance dose did not differ significantly between the two groups overall or according to the primary subgroup, race, or total number of genetic variants (Table S5 in the Supplementary Appendix).
The performance characteristics of the dosing algorithms with respect to the maintenance dose that was determined are shown in Table S6 (which includes the accuracy of a hypothetical, empirical dosing strategy of 5 mg per day) and in Figure S4, both in the Supplementary Appendix. The genotype-guided algorithms performed better at predicting the maintenance dose among nonblack patients than among black patients. Dose overrides during the first 4 weeks were rare, occurring in only 3.9% of doses in the genotype-guided group and 3.6% of those in the clinically guided group; rates of overrides did not differ according to race.
ADVERSE EVENTS
At 4 weeks, there were no significant between-group differences in the principal secondary outcome (the time to any INR of =4, major bleeding, or thromboembolism) or any other adverse events (Table 3, and Table S7 in the Supplementary Appendix). Safety data for the entire duration of follow-up (i.e., past the primary outcome duration) are provided in Table S8 in the Supplementary Appendix.
Table 3.
Adverse Events through Day 28 of Warfarin Therapy.
| Outcome | Genotype-Guided Group (N = 514) | Clinically Guided Group (N = 501) | Hazard Ratio (95% CI)* | P Value |
|---|---|---|---|---|
| no. (%) | ||||
| Any INR ≥4, major bleeding, or thromboembolism† | 105 (20) | 103 (21) | 1.01 (0.77–1.33) | 0.93 |
|
| ||||
| Any INR ≥4 | 100 (19) | 92 (18) | 1.08 (0.81–1.44) | 0.59 |
|
| ||||
| Major bleeding‡ | 4 (1) | 10 (2) | 0.41 (0.13–1.31) | 0.13 |
|
| ||||
| Thromboembolism | 5 (1) | 4 (1) | 1.27 (0.34–4.73) | 0.72 |
|
| ||||
| Clinically relevant nonmajor bleeding§ | 13 (3) | 20 (4) | 0.62 (0.30–1.27)§ | 0.18 |
|
| ||||
| Death from any cause | 2 (<1) | 1 (<1) | 2.09 (0.19–23.22) | 0.55 |
Hazard ratios are for the comparison between the genotype-guided dosing group and the clinically guided dosing group, as estimated from multivariable Cox regression models and adjusted for race and clinical center. A hazard ratio of more than 1 indicates that patients in the genotype-guided had, on average, a shorter time to an adverse event than did those in the clinically guided dosing group. Follow-up time began at randomization. Censoring events for major bleeding and thromboembolic events were death and administrative censoring at day 28. The censoring event for death was administrative censoring at day 28.
This composite was the principal secondary outcome.
The INR at the time of the bleeding event was available for all but one patient (in the clinically guided dosing group). The INR was elevated (>3) in three patients in the genotype-guided dosing group and in one patient in the clinically guided dosing group.
The binary outcome of any clinically relevant nonmajor bleeding event was analyzed with the use of a multivariable logistic-regression model, adjusted for race and clinical enter. The point estimate and confidence interval are estimated odds ratios for a clinically relevant nonmajor bleeding event in the genotype-guided group as compared with the clinically guided group.
DISCUSSION
In our study, we found no benefit of genotype-guided dosing of warfarin with respect to the primary outcome of the percentage of time in the therapeutic INR range, either overall or among patients with a predicted dose difference between the genotype-guided algorithm and the clinically guided algorithm of at least 1 mg per day. Our findings exclude a meaningful effect of genotype-guided dosing on the percentage of time in the therapeutic range during the first month of warfarin treatment. However, there was a significant difference in the effects of the algorithms in the prespecified subgroup of black patients, as compared with nonblack patients. Although the interaction between race and dosing strategy with respect to the primary outcome could be due to chance, the analysis was prespecified and was consistent with our a priori hypothesis that there would be race-based differences.
The dosing algorithms that we used in the trial have been validated and account for race (specifically black vs. nonblack).11–13,18 The genotype-guided algorithm performed as well as anticipated on the basis of previous studies,5,8,10–12,18,23 with an R2 of 0.48 and a mean absolute error of 1.3 mg per day for the dose-initiation algorithm and an R2 of 0.69 and a mean absolute error of 1.0 mg per day for the dose-revision algorithm. Despite this accuracy in predicting maintenance doses, there was no benefit of genotype-guided dosing with respect to anticoagulation control.
Observational studies have shown an association between the use of genetic algorithms and improved outcomes, but because of limitations in the study design, they were unable to assess whether the observed associations were causal.1,9,10 Previous clinical trials have produced equivocal results,5–8 but these trials were limited by a small size and lack of blinding to the warfarin dose. The two trials that suggested possible benefit also were limited by large numbers of dropouts6 and a comparison with nonalgorithm-based dosing.8 Previous studies also enrolled either no black patients6–8 or a minimal number of black patients5 (a total of 3) (Anderson J: personal communication).
The average percentage of time in the therapeutic range of 45% in our study is similar to that in other trials, taking into account the range of INRs used for the calculation and the timing and duration of therapy (Tables S9A and S9B in the Supplementary Appendix).5,10,24,25 Unlike previous trials that used only a baseline genotype-guided algorithm, our study used both a dose-initiation and a dose-revision algorithm. A recent study comparing a similar initiation algorithm with a combined initiation and revision algorithm showed no effect on the percentage of time in the therapeutic range with the addition of the revision algorithm.10
There are several questions that our study was not designed to answer. First, the trial did not compare genotype-based dosing with usual care or a fixed initial dose (e.g., 5 mg per day). However, such a comparison could not have discerned whether differences in outcomes were due to the marginal benefit of genetic information or to the use of the clinical information that is included in all genotype-guided dosing algorithms. Second, our study does not address the question of whether a longer duration of genotype-guided dosing would have improved INR control,26 an issue that is being addressed in another trial.27 Third, the dosing algorithms that we used included the three single-nucleotide polymorphisms among the two genes that are most likely to influence warfarin dosing. Although other genes may contribute to warfarin dosing, it is unlikely that they have a substantial effect, particularly in white populations.28 Fourth, although there were no significant between-group differences in the rates of bleeding or thromboembolic events during the primary follow-up period of 4 weeks, the trial was not powered for these outcomes. Fifth, the first dose of warfarin was not informed by genotyping in 55% of the patients; whether this influenced the results is unknown. However, the effect of missing genetics data on day 1 on the dose administered during the first 3 days of therapy was trivial.
In conclusion, our findings do not support the hypothesis that initiating warfarin therapy at a genotype-guided maintenance dose for the first 5 days, as compared with initiating warfarin at a clinically predicted maintenance dose, improves anticoagulation control during the first 4 weeks of therapy. Our results emphasize the importance of performing randomized trials for pharmacogenetics, particularly for complex regimens such as warfarin.
Supplementary Material
Acknowledgments
Supported under a contract (HHSN-268200800003C) with the National Heart, Lung, and Blood Institute. Bristol-Myers Squibb donated Coumadin (warfarin). GenMark Diagnostics and AutoGenomics loaned genotyping platforms to the clinical centers.
Dr. Kimmel reports receiving consulting fees from Pfizer and Janssen; Dr. Kasner, receiving personal fees from Novartis, Pfizer, Merck, Medtronic, Photothera, AstraZeneca, and Cardionet and grant support from W.L. Gore, Acorda, and Daiichi Asubio; Dr. Gage, receiving consulting fees from Boehringer Ingelheim; Dr. Eby, receiving grant support from Siemens and serving on an advisory board for Instrumentation Laboratory; Dr. Stevens, receiving grant support from Iverson Genetics; Dr. Muldowney, receiving consulting fees from MCG (formerly Milliman Care Guidelines), lecture fees from Bristol-Myers Squibb, Pfizer, and Boehringer Ingelheim, and grant support from Novartis and Theravasc; Dr. Ortel, receiving consulting fees from Boehringer Ingelheim, Bayer, and Instrumentation Laboratory and grant support from Eisai, GlaxoSmithKline, Pfizer, Daiichi Sankyo, and Instrumentation Laboratory; Dr. Goldhaber, receiving consulting fees from Bristol-Myers Squibb, Daiichi Sankyo, Baxter, Boehringer Ingelheim, Eisai, Merck, Pfizer, Portola, and Sanofi-Aventis and grant support from Bristol-Myers Squibb, Daiichi Sankyo, EKOS, and Johnson & Johnson; Dr. Caldwell, holding a patent, issued to the Marshfield Clinic, on the use of CYP4F2 for warfarin dosing; Dr. Califf, serving as a board member at Portola, receiving consulting fees from Novartis, Johnson & Johnson, Bayer, Roche, Pfizer, Bristol-Myers Squibb Foundation, Regeneron, CV Sight, Daiichi Sankyo, Eli Lilly, Gambro, Kowa, Genentech, GlaxoSmithKline, Laboratoires Servier, and Portola, receiving grant support from Amylin, Johnson & Johnson, Merck/Schering Plough, Novartis, Bristol-Myers Squibb Foundation, and Eli Lilly, and having an equity interest in N30 Pharma; and Dr. Ellenberg, receiving consulting fees from Bristol-Myers Squibb.
We thank the study patients, the technologists and research pharmacists at the clinical center genotype laboratory, and Ms. Sandra Barile for her assistance in the preparation of the manuscript.
APPENDIX
The authors’ affiliations are as follows: the University of Pennsylvania, Perelman School of Medicine, Philadelphia (S.E. Kimmel, B.F., S.E. Kasner, R.A.M., E.R.M., J.H.E.); University of Florida, Gainesville (J.J.A.); Intermountain Medical Center, Murray, UT (J.L.A., S.M.S.); Washington University School of Medicine, St. Louis (B.F.G., C.S.E.); National Institutes of Health, National Heart, Lung, and Blood Institute, Bethesda, MD (Y.D.R., N.L.G.); Mayo Clinic College of Medicine, Rochester, MN (R.B.M.); University of Texas Medical Branch at Galveston, Galveston (S.Z.A.-R.); Marshfield Clinic Research Foundation, Marshfield, WI (S.Y., M.D.C.); University of California, San Francisco, San Francisco (M.C.F.); Henry Ford Hospital, Detroit (V.S.); University of Maryland School of Medicine, Baltimore (R.B.H.); University of Alabama at Birmingham, Birmingham (N.A.L.); Vanderbilt University, Nashville (J.A.S.M.); Georgia Regents Medical Center, Augusta (J.G.); Tulane University, New Orleans (P.D.); Mount Sinai School of Medicine (R.J.D., J.L.H.) and Montefiore Medical Center (H.H.B.) — both in New York; Duke University Medical Center, Durham, NC (T.L.O., R.M.C.); University of Utah Health Care, Provo (R.C.P.); and Brigham and Women’s Hospital, Boston (S.Z.G.).
Footnotes
The authors’ affiliations are listed in the Appendix.
The opinions expressed in this article are those of the authors and do not necessarily reflect the views of the National Institutes of Health.
No other potential conflict of interest relevant to this article was reported.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Ginsburg GS, Voora D. The long and winding road to warfarin pharmacogenetic testing. J Am Coll Cardiol. 2010;55:2813–5. doi: 10.1016/j.jacc.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Burke W, Laberge AM, Press N. Debating clinical utility. Public Health Genomics. 2010;13:215–23. doi: 10.1159/000279623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashley EA, Hershberger RE, Caleshu C, et al. Genetics and cardiovascular disease: a policy statement from the American Heart Association. Circulation. 2012;126:142–57. doi: 10.1161/CIR.0b013e31825b07f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodcock J, Lesko LJ. Pharmacogenetics — tailoring treatment for the outliers. N Engl J Med. 2009;360:811–3. doi: 10.1056/NEJMe0810630. [DOI] [PubMed] [Google Scholar]
- 5.Anderson JL, Horne BD, Stevens SM, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–70. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 6.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83:460–70. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 7.Burmester JK, Berg RL, Yale SH, et al. A randomized controlled trial of genotype-based Coumadin initiation. Genet Med. 2011;13:509–18. doi: 10.1097/GIM.0b013e31820ad77d. [DOI] [PubMed] [Google Scholar]
- 8.Wang M, Lang X, Cui S, et al. Clinical application of pharmacogenetic-based warfarin-dosing algorithm in patients of Han nationality after rheumatic valve replacement: a randomized and controlled trial. Int J Med Sci. 2012;9:472–9. doi: 10.7150/ijms.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epstein RS, Moyer TP, Aubert RE, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study) J Am Coll Cardiol. 2010;55:2804–12. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Anderson JL, Horne BD, Stevens SM, et al. A randomized and clinical effectiveness trial comparing two pharmacogenetic algorithms and standard care for individualizing warfarin dosing (Couma-Gen-II) Circulation. 2012;125:1997–2005. doi: 10.1161/CIRCULATIONAHA.111.070920. [DOI] [PubMed] [Google Scholar]
- 11.Schelleman H, Chen J, Chen Z, et al. Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin Pharmacol Ther. 2008;84:332–9. doi: 10.1038/clpt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–31. doi: 10.1038/clpt.2008.10. Erratum, Clin Pharmacol Ther 2008;84: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavallari LH, Perera MA. The future of warfarin pharmacogenetics in under-represented minority groups. Future Cardiol. 2012;8:563–76. doi: 10.2217/fca.12.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Medicare & Medicaid Services. Medicare coverage database — potential NCD topics. ( http://www.cms.hhs.gov/mcd/ncpcviewdocument.asp?id=19)
- 15.Kimmel SE, French B, Anderson JL, et al. Rationale and design of the Clarification of Optimal Anticoagulation through Genetics trial. Am Heart J. 2013;166:435–41. doi: 10.1016/j.ahj.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.French B, Joo J, Geller NL, et al. Statistical design of personalized medicine interventions: the Clarification of Optimal Anticoagulation through Genetics (COAG) trial. Trials. 2010;11:108. doi: 10.1186/1745-6215-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joo J, Geller NL, French B, Kimmel SE, Rosenberg Y, Ellenberg JH. Prospective alpha allocation in the Clarification of Optimal Anticoagulation through Genetics (COAG) trial. Clin Trials. 2010;7:597–604. doi: 10.1177/1740774510381285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenzini P, Wadelius M, Kimmel S, et al. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clin Pharmacol Ther. 2010;87:572–8. doi: 10.1038/clpt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosendaal FR, Cannegieter SC, van der Meer FJ, Briët E. A method to determine the optimal intensity of oral anticoagulant therapy. Thromb Haemost. 1993;69:236–9. [PubMed] [Google Scholar]
- 20.The van Gogh Investigators. Idraparinux versus standard therapy for venous thromboembolic disease. N Engl J Med. 2007;357:1094–104. doi: 10.1056/NEJMoa064247. [DOI] [PubMed] [Google Scholar]
- 21.ROCKET AF Study Investigators. Rivaroxaban-once daily, oral, direct factor Xa inhibition compared with vitamin K antagonism for prevention of stroke and Embolism Trial in Atrial Fibrillation: rationale and design of the ROCKET AF study. Am Heart J. 2010;59(3):340.e1–347.e1. doi: 10.1016/j.ahj.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 22.Palareti G, Leali N, Coccheri S, et al. Bleeding complications of oral anticoagulant treatment: an inception-cohort, prospective collaborative study (ISCOAT): Italian Study on Complications of Oral Anticoagulation Therapy. Lancet. 1996;348:423–8. doi: 10.1016/s0140-6736(96)01109-9. [DOI] [PubMed] [Google Scholar]
- 23.The International Warfarin Pharmacogenetics Consortium. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–64. doi: 10.1056/NEJMoa0809329. Erratum, N Engl J Med 2009;361:1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia DA, Wallentin L, Lopes RD, et al. Apixaban versus warfarin in patients with atrial fibrillation according to prior warfarin use: results from the Apixaban for Reduction in Stroke and Other Thromboembolic Events in Atrial Fibrillation trial. Am Heart J. 2013;166:549–58. doi: 10.1016/j.ahj.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Ezekowitz MD, Wallentin L, Connolly SJ, et al. Dabigatran and warfarin in vitamin K antagonist-naive and -experienced cohorts with atrial fibrillation. Circulation. 2010;122:2246–53. doi: 10.1161/CIRCULATIONAHA.110.973735. [DOI] [PubMed] [Google Scholar]
- 26.Horne BD, Lenzini PA, Wadelius M, et al. Pharmacogenetic warfarin dose refinements remain significantly influenced by genetic factors after one week of therapy. Thromb Haemost. 2012;107:232–40. doi: 10.1160/TH11-06-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Do EJ, Lenzini P, Eby CS, et al. Genetics informatics trial (GIFT) of warfarin to prevent deep vein thrombosis (DVT): rationale and study design. Pharmacogenomics J. 2012;12:417–24. doi: 10.1038/tpj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper GM, Johnson JA, Langaee TY, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–7. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.