Abstract
The kinetochore (centromeric DNA and associated proteins) mediates the attachment of chromosomes to the mitotic spindle apparatus and is required for faithful chromosome transmission. We established that evolutionarily conserved Saccharomyces cerevisiae SPT4, previously identified in genetic screens for defects in chromosome transmission fidelity (ctf), encodes a new structural component of specialized chromatin at kinetochores and heterochromatic loci, with roles in kinetochore function and gene silencing. Using chromatin immunoprecipitation assays (ChIP), we determined that kinetochore proteins Ndc10p, Cac1p, and Hir1p are required for the association of Spt4p to centromeric (CEN) loci. Absence of functional Spt4p leads to altered chromatin structure at the CEN DNA and mislocalization of the mammalian CENP-A homolog Cse4p to noncentromeric loci. Spt4p associates with telomeres (TEL) and HMRa loci in a Sir3p-dependent manner and is required for transcriptional gene silencing. We show that a human homolog of SPT4 (HsSPT4) complements Scspt4-silencing defects and associates with ScCEN DNA in an Ndc10p-dependent manner. Our results highlight the evolutionary conservation of pathways required for genome stability in yeast and humans.
Keywords: CSE4 , kinetochore, S. cerevisiae , silencing, SPT4
Introduction
In order to proliferate and develop, organisms depend on the accurate segregation of their chromosomes. Errors in this process lead to aneuploidy, birth defects, developmental disorders, and possibly cancer (Lengauer et al, 1998). CEN DNA is the cis-acting locus that specifies the binding of sequence-specific DNA-binding proteins and assembly of additional kinetochore proteins. Both the structural and regulatory components that define a functional kinetochore are essential for faithful chromosome transmission during mitosis and meiosis. The kinetochore maintains cohesion between sister chromatids, mediates the attachment of chromosomes to the mitotic spindle, and directs their subsequent movement to the spindle poles. The kinetochore is also the site through which completion of metaphase is sensed by the cell-cycle regulatory machinery, which coordinates the synchronous separation of chromosomes at the onset of anaphase (Kitagawa and Hieter, 2001).
In contrast to the complex centromeric structure of other eukaryotes, the CEN DNA sequence in Saccharomyces cerevisiae is relatively short (125 bp) with three conserved elements, CDEI, CDEII, and CDEIII (Fitzgerald-Hayes et al, 1982). The approximately 50 or more kinetochore proteins identified to date are classified based on whether they co-localize or interact with CEN DNA (inner kinetochore), spindle microtubules (outer kinetochore), or between the inner and outer kinetochore components (Kitagawa and Hieter, 2001; Cheeseman et al, 2002). Molecular analysis of the kinetochore complexes will aid in understanding of their functional roles. Binding of CBF3, a multiprotein complex containing Ctf13p, Ndc10p, Cep3p, and Skp1p to CDEIII, is critical for kinetochore assembly (Cheeseman et al, 2002).
In S. cerevisiae, the kinetochore chromatin domain is delimited on each side by strong nuclease-hypersensitive sites and is flanked by arrays of phased nucleosomes (Bloom and Carbon, 1982; Funk et al, 1989; Schulman and Bloom, 1991; Glowczewski et al, 2000). Modified and specialized histones are crucial for the assembly and function of the kinetochores and centromeric chromatin. The core centromeric chromatin contains the histone H3 variant Cse4p, the homolog of mammalian CENP-A (Stoler et al, 1995; Meluh et al, 1998). Genetic analyses have shown that increased histone levels impair mitotic chromosome segregation (Meeks-Wagner and Hartwell, 1986). Furthermore, mutations in genes encoding histones H2A, H2B, H4 and chromatin remodeling complex RSC proteins, (Sth1p, Snf5p), lead to chromosome missegregation, increased nuclease sensitivity in CEN DNA, and adjacent chromatin (Han et al, 1987; Saunders et al, 1990; Smith et al, 1996; Tsuchiya et al, 1998; Keith et al, 1999; Pinto and Winston, 2000; Hsu et al, 2003). These studies suggest a direct role for histones in centromere function and establish that nucleosome organization and function at the centromere is distinct from its function elsewhere in the genome.
In addition to kinetochore function, specialized chromatin is required for epigenetic functions such as the establishment and maintenance of gene silencing (Krude, 2002). Silent chromatin is present at the mating type loci (HMLα and HMRa) and at telomeric (TEL) DNA in S. cerevisiae (Grunstein, 1997). Using genetic analyses, we have previously shown that SPT4 is required for faithful chromosome transmission and kinetochore function. We determined that spt4 mutants show a 100-fold increase in the loss of a non-essential reporter chromosome, test positive in an in vivo assay for defects in kinetochore integrity, genetically interact with kinetochore mutants, and extracts exhibit defects in the binding of minichromosomes to microtubules (Basrai et al, 1996). The spt (suppressor of Ty) mutants were also identified in a genetic screen for suppression of transcriptional defects associated with Ty delta insertions in the 5′ regions of HIS4 and LYS2 (Winston et al, 1984). SPT4 interacts genetically and biochemically with SPT5 and SPT6 (Swanson and Winston, 1992; Krogan et al, 2003; Lindstrom et al, 2003), and the Spt4p/Spt5p/Spt6p complex is required for assembly or stabilization of nucleosomes (Swanson and Winston, 1992; Bortvin and Winston, 1996), and transcription elongation (Hartzog et al, 2002; Rondon et al, 2003).
In this paper, we establish that Spt4p is a new structural and functional component of the centromeric and heterochromatic loci linking chromatin structure with kinetochore function and gene silencing. We used ChIP experiments and mutants to analyze the specific association of Spt4p with different chromosomal loci. Consistent with a role in kinetochore structure and function, we determined that Spt4p is required for the integrity of centromeric chromatin and restriction of the localization of Cse4p to kinetochores. Also, we established a novel role of Spt4p in gene silencing and complementation of the yeast mutant phenotypes by human SPT4. Interestingly, we provide the first example of an in vivo association of a human protein, namely HsSpt4p, with CEN DNA in budding yeast. Our results further demonstrate how yeast can be used as a model system to study the fundamental process of chromosome segregation in humans.
Results
Spt4-GFP co-localizes with kinetochores in S. cerevisiae
Previously, we used genetic analyses to show that Spt4p is required for chromosome segregation and kinetochore integrity (Basrai et al, 1996; Kerscher et al, 2001). The spt4-138 and spt4Δ strains exhibit temperature-sensitive growth at 37°C and defects in chromosome transmission fidelity (ctf); so we decided to determine whether Spt4p co-localized with a kinetochore marker (Ndc10p). We tagged the C-terminus of Spt4p with green fluorescent protein (GFP) in a strain expressing Ndc10p tagged at the C-terminus with the hemagglutin epitope (HA). The Spt4-GFP fusion protein was functional, as the strain containing the tagged protein did not exhibit temperature-sensitive growth at 37°C (Figure 1A) or a ctf phenotype (data not shown). Subcellular localization showed that Ndc10p-HA was localized to the kinetochores as two or three distinct foci per nucleus (Figure 1B) (Sharp et al, 2002). In a majority of the cells (>90%), Spt4-GFP was localized to three to seven nuclear foci. One or two of these Spt4-GFP foci overlapped with kinetochore-containing Ndc10p-HA foci in greater than 90% of the cells (Figure 1B, merged). These results indicate that a subset of Spt4-GFP foci co-localize with the kinetochores. Spt4-GFP foci that do not overlap with Ndc10p-HA may represent an association with other chromosomal loci.
Figure 1.
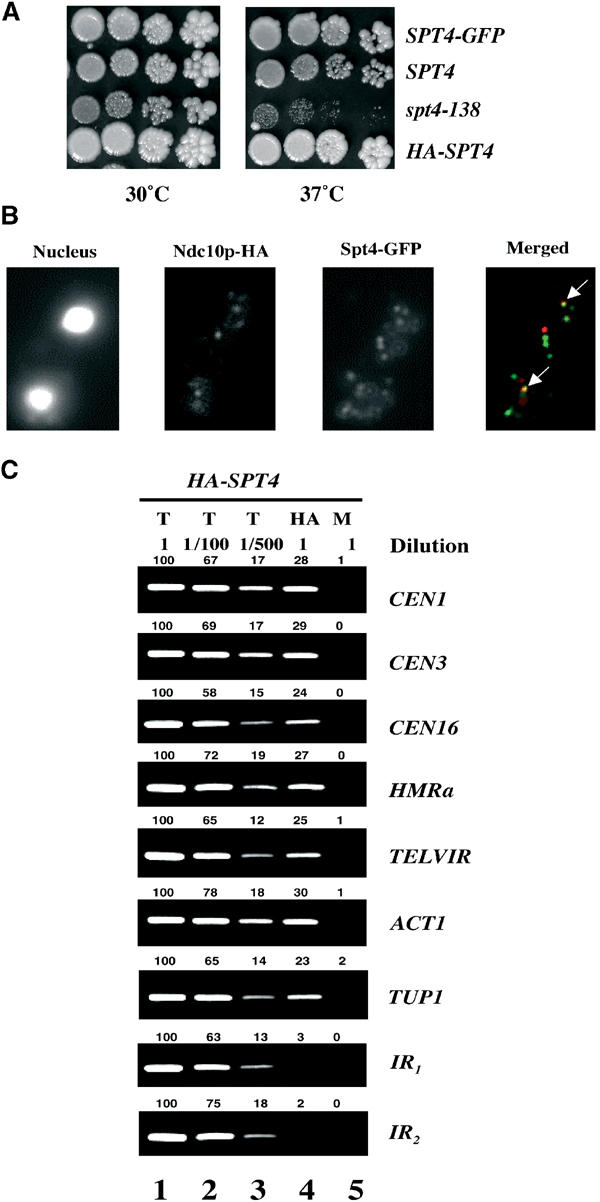
Spt4p co-localizes with kinetochores and associates with chromosomal loci. (A) Epitope-tagged SPT4 is functional. Growth of five-fold dilutions of wild-type (YPH499) and spt4-138 (YMB54) strains was compared to SPT4-GFP (YMB1859) and HA-SPT4 (GHY262) strains on YPD plates incubated at 30°C and 37°C for 3–4 days. The spt4-138 strain exhibits a slower growth compared to the wild-type strain even at the permissive temperature of 30°C. (B) Subset of Spt4-GFP foci co-localizes with kinetochore. Chromosome spreads from strain (YMB1859) expressing Ndc10p-HA, and Spt4-GFP probed with anti-GFP or anti-HA antibodies. The arrows in the merged panel indicate co-localization of Ndc10p-HA (red), and Spt4-GFP (green) is shown in yellow. (C) HA-Spt4p associates with chromosomal loci. ChIP experiments were carried out using wild-type strain (GHY262) expressing chromosomally tagged HASPT4 grown to logarithmic phase at 30°C. Different dilutions of chromatin samples from total (T) (undiluted, 1/100, 1/500), immunoprecipitated with anti-HA (HA), and mock (M) were analyzed using primers for core CEN1, core CEN3, core CEN16, HMRa, TELVIR, and ACT1 or intergenic regions (IR1 and IR2) devoid of ORFs (chromosome IV, co-ordinates 1 157 000–1 157 200 and 1 523 000–1 523 180, respectively). Values for the quantitation of the data are shown above each of the lanes, with the value for undiluted total set to 100 for each row (lane 1). These data show that the PCR yield is proportional to the amount of starting material.
HA-Spt4p associates with chromosomal DNA
We used a biochemical approach to assess the association of Spt4p with chromosomal loci using the ChIP technique. Strains containing an N-terminally tagged Spt4p with the HA epitope exhibit wild-type growth (Figure 1A). Chromatin extracted from wild-type cells expressing HASPT4 was crosslinked with formaldehyde and HA-Spt4p/DNA complexes were immunoprecipitated using anti-HA antibody. PCR analyses of total (T), immunoprecipitated (HA), and mock (M) samples were carried out using different primer pairs. Different dilutions of the T DNA (undiluted, 1/100, 1/500) used in PCR reactions verified that the PCR yield was proportional to the amount of starting DNA (Figure 1C). No PCR product was obtained from mock samples, samples precipitated with anti-GFP antibody, samples from a control strain lacking epitope tagged SPT4, or samples without addition of crosslinker (data not shown). Our results showed that HA-Spt4p associated with CEN DNA, heterochromatic loci HMRa, TELVIR and actively transcribing genes ACT1 and TUP1, but not with intergenic DNA regions (IR1 and IR2) devoid of ORFs. The enrichment of HA-Spt4p at various chromosomal loci was independent of the cell cycle stage (data not shown).
Kinetochore protein Ndc10p is required for the association of HA-Spt4p with core CEN loci
The association of HA-Spt4p with CEN DNA and other chromosomal loci led us to hypothesize that the association of HA-Spt4p with a locus may be dependent on the presence of specific protein(s) at these loci. Hence, we evaluated the association of HA-Spt4p with different chromosomal loci in ChIP experiments using kinetochore, RNAPII, or silencing mutants. In the first of such experiments, we used a temperature-sensitive ndc10-1 strain containing a mutation in the essential kinetochore gene NDC10, which fails to assemble a functional kinetochore at the nonpermissive temperature of 37°C (Goh and Kilmartin, 1993). The ndc10-1 strain has been used to establish the specificity of the interaction of kinetochore proteins with CEN DNA (Goshima and Yanagida, 2000; He et al, 2001). ChIP experiments were performed using wild-type and ndc10-1 strains expressing HASPT4 grown at the permissive temperature (30 or 25°C) or after a shift to 37°C. Our data showed that HA-Spt4p associated with CEN16, TELVIR, and ACT1 in the wild-type and ndc10-1 strains grown at 30 or 25°C, respectively (Figure 2A and B). However, after the shift of the ndc10-1 strain to 37°C, HA-Spt4p specifically failed to associate with the core CEN16 DNA; the chromosomal association of HA-Spt4p to non-CEN loci such as TELVIR and ACT1 was unaffected (Figure 2B). Similar results were obtained for association of HA-Spt4p with CEN1 and CEN3 (data not shown). Control experiments showed that the association of HA-Spt4p with CEN or non-CEN loci was unaffected when chromatin was prepared from wild-type cells shifted to 37°C (Figure 2A). These results show that Spt4p is a kinetochore protein and Ndc10p is required for the association of HA-Spt4p to CEN DNA.
Figure 2.
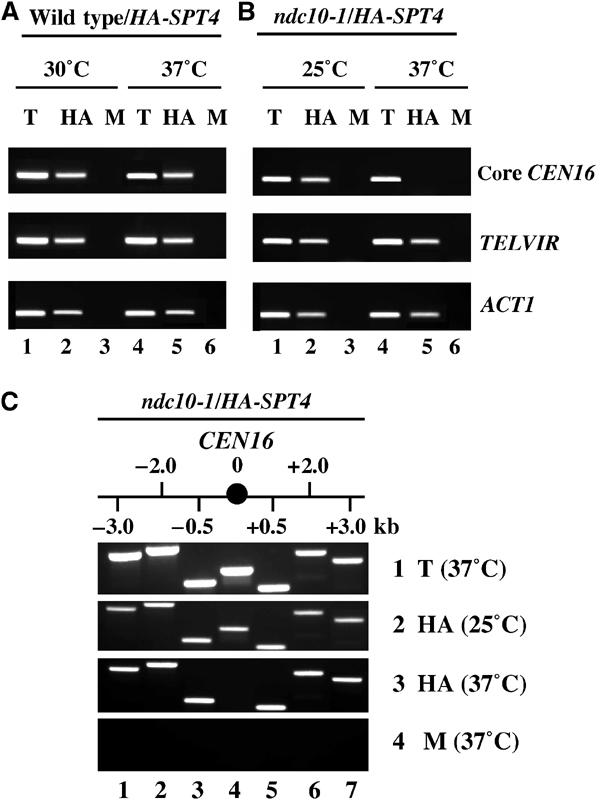
Ndc10p is required for the association of HA-Spt4p to the core CEN loci. ChIP experiments were carried out using wild-type strain (GHY262) or ndc10-1 strain expressing HASPT4 (YMB1659) grown at 30 or 25°C and shifted to 37°C for 4 h. (A, B) Chromatin samples from T, HA, and M were analyzed using primers for core CEN16, TELVIR, or ACT1. (C) Chromatin samples from T (row 1), HA (rows 2 and 3) and M (row 3) were analyzed using primers for core CEN16 (lane 4) and those that flank on either side (lanes 1–3 and 5–6).
Ndc10p, a component of the inner kinetochore, associates with the core CEN DNA of about 250 bp (Meluh and Koshland, 1997). Hence, we carried out ChIP experiments to determine the extent to which perturbations in kinetochore structure due to the ndc10-1 mutation affected the association of HA-Spt4p with CEN and flanking sequences. We determined that HA-Spt4p associates with core CEN16 and the flanking DNA up to 3 kb on either side of CEN16 in the ndc10-1 mutant grown at 25°C (Figure 2C). However, HA-Spt4p specifically failed to associate with core CEN16 DNA, with no effect on chromosomal association to flanking CEN DNA in the ndc10-1 strain shifted to 37°C (Figure 2C). Based on these and earlier results (Figure 2B), we conclude that Ndc10p is required for the specific association of HA-Spt4p with core CEN DNA.
Centromeric chromatin structure is altered in spt4 mutants
The specific association of HA-Spt4p with CEN DNA led us to examine as to whether Spt4p was required for the proper configuration of centromeric chromatin, by analyzing the endonuclease accessibility of chromatin structure at CEN3 (Saunders et al, 1988, 1990). In wild-type cells, the kinetochore protects the three naturally occurring DraI sites in CDEII from digestion (Figure 3A). Mutations in CEN DNA, kinetochore proteins, or depletion of histones H2B or H4 increase the accessibility of CDEII to DraI (Saunders et al, 1988, 1990; Meluh et al, 1998). These experiments were carried out using Southern blot analysis of nuclear DNA digested with DraI (different concentrations) and then with EcoRI, using a 0.9 kb HindIII–BamHI fragment as a probe (Figure 3A). As can be seen in Figure 3B in a wild-type strain shifted to 37°C, the DraI sites within CDEII of CEN3 were minimally accessible (2.9 kb fragment). However, in the spt4-138 mutant, we observed a marked increase in the accessibility of centromeric DraI sites, with a five-fold increase in digestion as compared to the wild-type strain. We did not detect a significant difference in DraI accessibility at the permissive temperature of growth (data not shown). These results showed that Spt4p is required for the integrity of centromeric chromatin.
Figure 3.
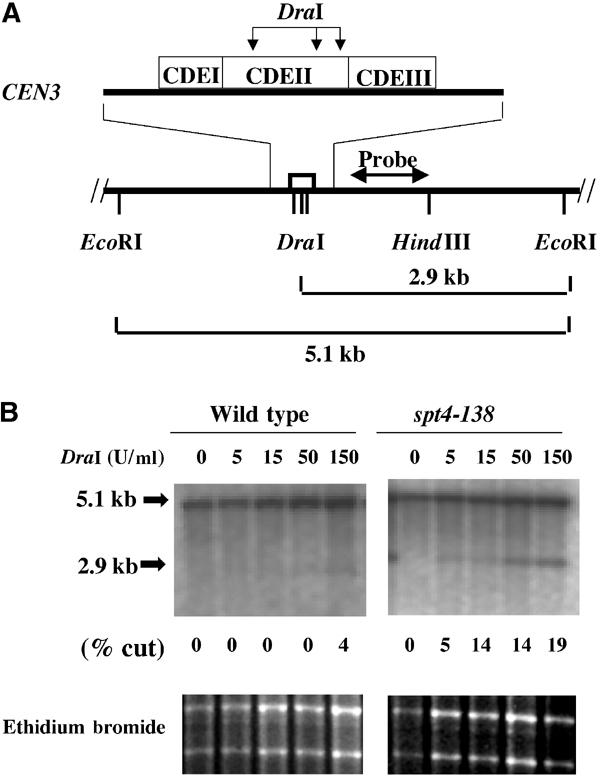
Centromeric chromatin structure is altered in the spt4-138 strain. (A) Representation of three DraI sites within CDEII of the CEN3, the resulting DraI–EcoRI or EcoRI–EcoRI fragments, and the probe used for Southern blot analysis. (B) CEN3 chromatin structure of wild-type and spt4-138 mutant. Nuclei prepared from logarithmic growing cultures of wild-type (PKY090) and spt4-138 (YMB2192) strains grown at 37°C for 6 h were incubated with 0, 5, 15, 50, and 150 U/ml of DraI at 37°C for 30 min. Southern blot analysis of the EcoRI digested DNA was performed and probed with a 32P-dCTP-labeled 0.9 kb HindIII–BamHI fragment as shown in (A). The fraction of CEN3 accessible to DraI (% cut) was determined by quantification of the cut band (2.9 kb) divided by the sum of the uncut (5.1 kb) and cut bands (2.9 kb) using a Fuji Phosphorimager. The results were reproducible in multiple experiments, with values not differing by more than 0.01%.
Spt4p is required for restricting the association of Cse4p-HA with kinetochores
The alteration of centromeric chromatin in the spt4-138 strain led us to examine as to whether the compromised kinetochore structure was accompanied by changes in the localization of a kinetochore protein Cse4p. Cse4p, a homolog of mammalian CENP-A, is present at the kinetochores in almost all systems and is critically required for chromosome segregation (Smith, 2002). We compared the localization pattern of chromosomally tagged Cse4p-HA from wild-type and spt4-138 strains using chromosome spreads and indirect immunofluorescence. In the majority of wild-type cells (>98%), Cse4p-HA was ‘clustered' and localized to one or two foci (Figure 4A), consistent with previous observations (Meluh et al, 1998). In contrast, in the spt4-138 strain, Cse4p-HA was localized to multiple foci within the nucleus (Figure 4A). Quantitative analysis showed that greater than 75% of the cells with spt4-138 showed multiple Cse4p-HA foci compared to only 2% of the cells containing wild-type SPT4 (Figure 4C). Similar results were obtained for the mislocalization of Cse4p-HA in the spt4Δ strain (data not shown). We next questioned whether Spt4p is required for localization of kinetochore protein Mtw1-GFP. Mtw1-GFP was localized to one or two kinetochore foci in greater than 95% of the wild-type cells (Figure 4B), as previously reported (Goshima and Yanagida, 2000; Iouk et al, 2002). An almost identical localization of Mtw1-GFP to one to two foci was also observed in the spt4-138 strain (Figure 4B and C). We also determined that Spt4p was not required for the localization of Ndc10p (data not shown). These results suggest that Spt4p is required for the optimal localization of a subset of kinetochore proteins such as Cse4p.
Figure 4.
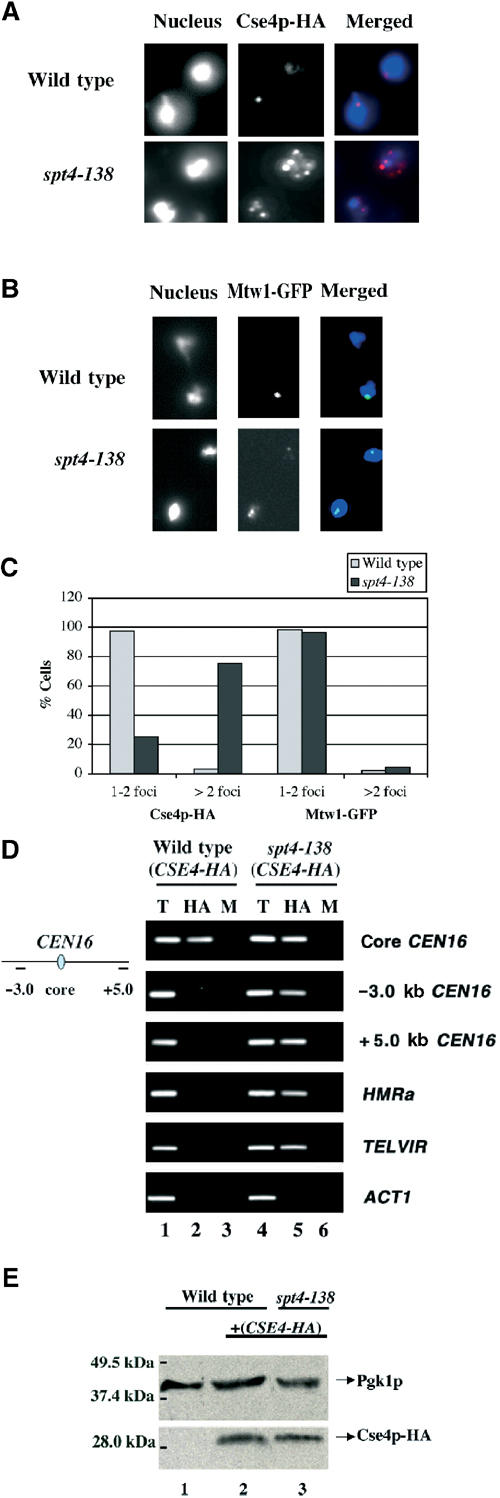
Spt4p is required for restricting the association of Cse4p-HA with kinetochores. (A, B) Cse4p-HA, but not Mtw1-GFP, shows an altered localization pattern in the spt4-138 strain. Chromosome spreads from wild-type (YPH98) and spt4-138 (YMB54) strains expressing CSE4-HA (YMB2142 and YMB2140, respectively) or MTW1 (YMB2231 and YMB2230, respectively) grown at 30°C were probed with anti-HA and anti-GFP antibodies. (C) Graphic representation of the percentage of wild-type and spt4-138 cells showing Cse4p-HA and Mtw1-GFP foci corresponding to data in (A) and (B), respectively. At least 100 nuclei were counted in two independent experiments. Similar results were obtained with the spt4Δ strain (data not shown). (D) Chromosomal association of Cse4p-HA is altered in the spt4-138 strain. ChIP experiments were carried out using wild-type (YMB2142) and spt4-138 (YMB2140) strains expressing Cse4p-HA grown at 30°C. Chromatin samples from T, HA, and M were analyzed using primers to core CEN16 and flanking DNA on either side at −3.0 or +5.0 kb, HMRa, TELVIR, or ACT1. (E) Expression of Cse4p-HA is not affected in the spt4-138 strain. Western blot analysis was performed using the following strains: wild-type (lane 1), wild-type, and spt4-138 strains expressing CSE4-HA (lanes 2 and 3). Blots were probed with anti-HA or anti-PGK (loading control) antibodies.
Cse4p has been shown to associate with core CEN DNA using the ChIP technique (Meluh et al, 1998). Our data suggested that the absence of functional Spt4p may result in mistargeting of Cse4p to non-CEN DNA. We used the ChIP technique to compare the association of Cse4p-HA to chromosomal loci in wild-type and spt4-138 strains. Consistent with previously published reports (Meluh et al, 1998), Cse4p-HA associated only with the core CEN16 DNA in wild-type cells (Figure 4D). In contrast, we observed a striking difference in the association of Cse4p-HA in the spt4-138 strain. In the spt4-138 strain, Cse4p-HA associated not only with the core CEN16 locus but also DNA flanking CEN16 (−3.0 and +5.0 kb) and heterochromatic loci such as HMRa and TELVIR. Cse4p-HA was not enriched at the ACT1 locus (Figure 4D). We determined that the mislocalization of Cse4p-HA in the spt4-138 strain was not due to its altered expression, as Western blot analyses showed similar levels of expression of Cse4p-HA in wild-type and spt4-138 strains (Figure 4E). Hence, based on subcellular localization and biochemical analyses, we conclude that in the absence of functional Spt4p, Cse4p-HA associates with additional centromeric and with noncentromeric loci such as TELVIR and HMRa.
Cac1Δ hir1Δ mutants show a defect in the association of HA-Spt4p with CEN loci
Recent data have shown that Cac1p, a chromatin assembly factor, and Hir1p, a histone regulatory protein, associate with kinetochores and that cac1Δ hir1Δ strains exhibit chromosome missegregation and declustering of Cse4p foci similar to that observed in spt4 mutants (Sharp et al, 2002). Hence, we determined whether Cac1p and Hir1p were required for the recruitment of HA-Spt4p to CEN DNA. ChIP experiments were carried out using wild-type, cac1Δ, hir1Δ, and cac1Δ hir1Δ strains expressing HASPT4. HA-Spt4p was enriched at core CEN3, HMRa, TELVIR, and ACT1 in wild-type, cac1Δ, and hir1Δ strains (Figure 5). However, in the cac1Δ hir1Δ strains we failed to detect an enrichment of HA-Spt4p specifically with core CEN3; the association of HA-Spt4p with non-CEN loci was unaffected. Similar results were obtained for lack of enrichment of HA-Spt4p to core CEN1 or CEN16 in the cac1Δ hir1Δ strain (data not shown). Western blot analysis showed similar levels of expression of HA-Spt4p in the single and double deletion strains (data not shown). Based on our results, we conclude that the absence of both Cac1p and Hir1p leads to a defect in the association of HA-Spt4p with core CEN DNA.
Figure 5.
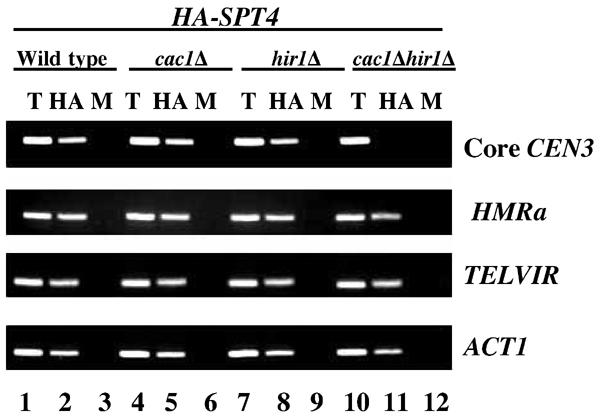
HA-Spt4p fails to associate with CEN DNA in a cac1Δ hir1Δ strain. ChIP experiments were carried out using wild-type (PKY090), cac1Δ (PKY638), hir1Δ (PKY117), or cac1Δ hir1Δ (PKY632) strains expressing HASPT4 (pMB237) grown at 30°C. Chromatin samples from T, HA, and M were analyzed using primers for core CEN3, HMRa, TELVIR, or ACT1.
RPB1 is not required for the association of HA-Spt4p with CEN DNA
A complex containing Spt4p/Spt5p has been shown to be required for transcription elongation (Hartzog et al, 2002; Rondon et al, 2003) and Spt5p associates with the open reading frames of actively transcribing genes (Andrulis et al, 2000; Kaplan et al, 2000; Pokholok et al, 2002; Simic et al, 2003). Hence, we determined whether the association of Spt4p to CEN DNA required a major component of the transcriptional machinery. We constructed an HASPT4 strain carrying a temperature-sensitive rpb1-1 mutation in the gene encoding the largest subunit of RNAPII. When rpb1-1 strains are shifted to the nonpermissive temperature, 37°C, transcription by RNAPII and growth of the strain are rapidly halted (Nonet et al 1987). ChIP experiments were carried out using rpb1-1 and a wild-type strain expressing HASPT4 grown at 25°C and after shift to 37°C for 1 h. Different dilutions of the T DNA (undiluted, 1/100, 1/500) used in PCR reactions verified that the PCR yield was proportional to the amount of starting DNA (Figure 6A). HA-Spt4p associated with core CEN1 and to actively transcribed genes ACT1 and TUP1 in both the wild-type and the rpb1-1 mutant grown at 25°C (Figure 6A, rows 1, 3, and 5). However, upon shift to 37°C, the association of HA-Spt4p with core CEN1 DNA (Figure 6A, row 2) was unaffected, even though there was lack of enrichment of HA-Spt4p with ACT1 or TUP1 in the rpb1-1 strain (Figure 6A, rows 4 and 6). HA-Spt4p was enriched at CEN DNA, ACT1, and TUP1 in the wild-type strain at 37°C. These results show that RPB1 is not required for the association of HA-Spt4p to core CEN DNA.
Figure 6.
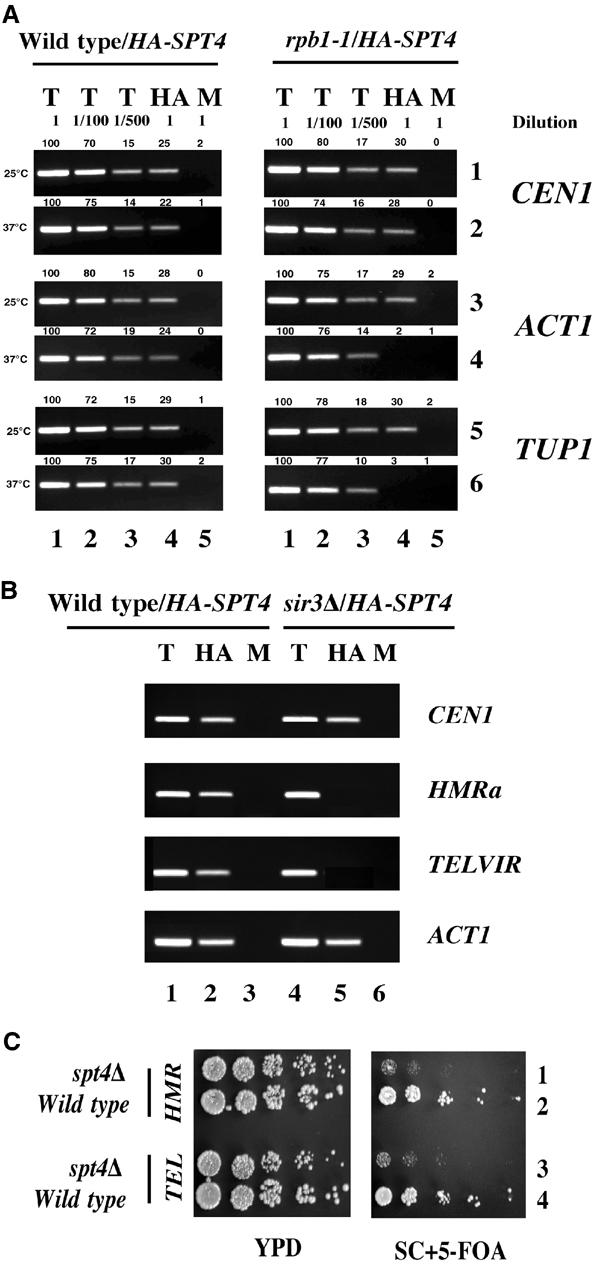
RNAPII and Sir3p are not required for the association of HA-Spt4p to core CEN and spt4 strains display a defect in silencing of reporter genes at heterochromatic loci. (A) HA-Spt4p associates with CEN DNA, but not with the actively transcribed genes ACT1 and TUP1 in a temperature-sensitive rpb1-1 strain. ChIP experiments were carried out using wild-type (GHY501) and rpb1-1 (GHY560) strains expressing HASPT4 (pMB237) grown at 30°C and shifted to 37°C for 1 h. Different dilutions of chromatin samples from total (T) (undiluted, 1/100, 1/500), immunoprecipitated with anti-HA (HA), and mock (M) were analyzed using primers for core CEN1, ACT1, or TUP1. Values for the quantitation of the data are shown above each of the lanes, with the value for undiluted total set to 100 for each row (lane 1). These data show that the PCR yield is proportional to the amount of starting material. (B) HA-Spt4p associates with core CEN DNA, but not with HMRa and TEL in an sir3Δ strain. ChIP experiments were carried out using wild-type (BUY668) and sir3Δ (BUY671) strains expressing HASPT4 (pMB237) grown at 30°C. Chromatin samples from T, HA, and M were analyzed using primers for core CEN1, HMRa, TELVIR, or ACT1. These results suggest that Spt4p may not be required for the transcription of HMRa. (C) Spt4p is required for silencing at HMRa and TEL. Silencing phenotypes were assayed by expression of URA3 integrated adjacent to either HMRa in wild-type (BUY545) and spt4Δ (YMB1871) or TELVIIL (TEL) in wild-type (BUY668) and spt4Δ (YMB1849) strains. Five-fold serial dilutions of cells were plated on YPD, and SC+5-FOA, and incubated for 3–4 days at 30°C. As a control, we determined that spt4Δ strains auxotrophic for URA3 (ura3) do not exhibit growth defects on media containing 5-FOA.
Sir3p is required for association of HA-Spt4p with TEL and HMRa
Our data showed that kinetochore proteins, Ndc10p, Cac1p, and Hir1p, are required for the association of HA-Spt4p with CEN DNA, but not TEL and HMRa. We reasoned that the Spt4p complex at the centromere may be different from the one present at the TEL or HMRa loci. Hence, we questioned whether Sir3p, a component of TEL and HMRa chromatin, is required for the association of HA-Spt4p at these loci. ChIP experiments were carried out using wild-type and sir3Δ strains expressing HASPT4. Consistent with our previous observations, we determined that HA-Spt4p associates with CEN1, HMRa, TELVIR, and ACT1 in the wild-type strain (Figure 6B). In contrast, the absence of SIR3 (sir3Δ) resulted in a loss of enrichment of HA-Spt4p, specifically at HMRa and TELVIR. Sir3p was not required for the association of HA-Spt4p to CEN1 or ACT1. Our results show that Sir3p is required for the enrichment of HA-Spt4p at HMRa and TELVIR. These results support our hypothesis for the presence of distinct HA-Spt4p complexes at CEN, TEL, and HMRa.
Spt4p is required for heterochromatin gene silencing
The association of HA-Spt4p with TEL and HMRa in an Sir3p-dependent manner suggested that Spt4p may have a role in heterochromatic gene silencing. In wild-type cells, silencing of a URA3 reporter gene placed adjacent to HMRa or TELVIIL leads to the transcriptional inhibition of URA3 (Kamakaka and Rine, 1998). The expression of URA3 is assayed by comparing the growth of strains on rich media (YPD), and synthetic complete medium containing the metabolic poison 5-fluoro-orotic acid (SC+5FOA), which inhibits the growth of URA3 prototrophs (Boeke et al, 1984). Thus, growth on 5-FOA media reflects silencing, whereas growth inhibition is indicative of a defect in silencing of URA3. Our results showed that unlike wild-type cells in which the expression of URA3 is silenced, the spt4Δ strain does not repress URA3 expression at either the HMRa or TEL (Figure 6C), as evidenced by reduced growth on 5-FOA medium. Together, these data demonstrate that spt4Δ cells are defective in gene silencing at HMRa and TEL, and provide the first evidence for a functional role of Spt4p at heterochromatic loci.
Human SPT4 (HsSPT4) complements the silencing defects of S. cerevisiae spt4 (Scspt4) mutants and shows an Ndc10p-dependent association with S. cerevisiae CEN DNA
We have previously shown that a human homolog of ScSPT4 is able to complement functionally the ctf, ts, and spt phenotypes of Scspt4 mutants (Hartzog et al, 1996). The results presented in this paper provide new evidence for a role for Spt4p in heterochromatic gene silencing. Hence, we determined whether HsSPT4 functionally complements the silencing defects of Scspt4 mutants. Consistent with our previous observations (Figure 6C), we determined that spt4 transformants with vector alone showed growth inhibition on media containing 5FOA. Both HsSPT4 and ScSPT4 were able to complement the silencing defect of the URA3 gene at TEL and HMRa in the spt4Δ strain (Figures 7A and B). Also, expression of HsSPT4 does not alter the expression of the URA3 gene at either TEL or HMRa in wild-type cells. These results show that HsSPT4 is able to functionally complement the silencing phenotypes of Scspt4 mutants.
Figure 7.
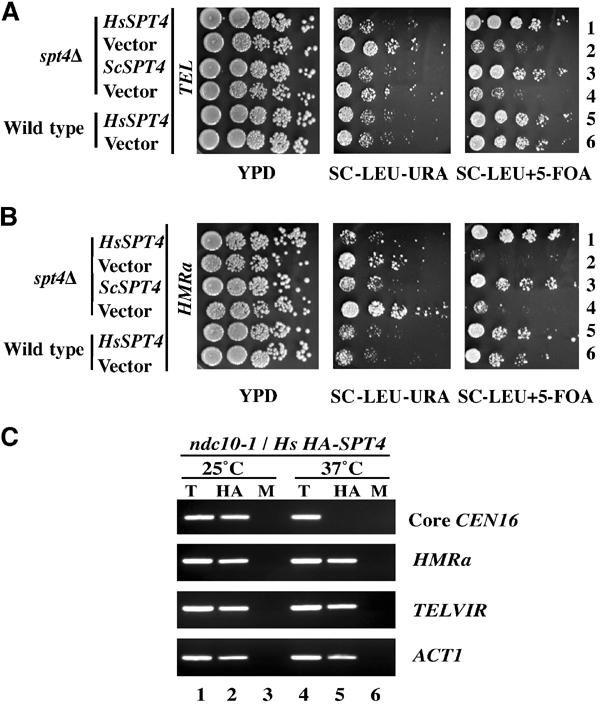
Human SPT4 complements the silencing phenotypes of an Scspt4 mutant and associates with S. cerevisiae core CEN DNA in an Ndc10p-dependent manner. (A) and (B) Silencing phenotypes were assayed by determining the expression of URA3 integrated adjacent to telomere VIIL (TEL) and HMRa in wild-type (BUY671 and BUY545, respectively) and spt4Δ strains (YMB1849 and YMB1871, respectively) expressing human SPT4 (HsSPT4) (pMB299), ScSPT4 (ScSPT4) (pMB237), or vector (vector). Serial dilutions of cells were plated on YPD, SC-LEU-URA, and SC-LEU+5-FOA and incubated for 3–4 days at 30°C. (C) ChIP experiments were carried out using ndc10-1 expressing pHsSPT4/LEU2 (YMB1827) strain grown at 25°C and shifted to 37°C for 4 h. Chromatin samples from T, HA, and M were analyzed using primers for core CEN16, HMRa, TELVIR, or ACT1.
The functional complementation of the ctf phenotype of Scspt4 mutants by HsSPT4 (Hartzog et al 1996) led us to examine whether HsSpt4p associates specifically with CEN DNA. ChIP experiments were carried out using kinetochore mutant ndc10-1 expressing HsHA-SPT4 grown at 25°C and after shift to 37°C for 4 h. Similar to the results for ScSpt4p-HA, we were able to observe an enrichment of HsHA-Spt4p with core CEN16, HMRa, TELVIR, and ACT1 at 25°C (Figure 7C). However, HsHA-Spt4p failed to associate specifically with only core CEN16 upon shift to 37°C. Based on these results, we conclude that HsHA-Spt4p associates with core CEN DNA of S. cerevisiae in an Ndc10p-dependent manner.
Discussion
The results presented in this paper provide direct evidence that evolutionarily conserved Spt4p is a new component of specialized chromatin at the centromere and heterochromatic regions in S. cerevisiae, with key roles in kinetochore structure and function as well as gene silencing. Our results show that Spt4p is required for: (a) integrity of centromeric chromatin and kinetochore structure and function, potentially mediated by restricting the localization of Cse4p, the homolog of mammalian CENP-A, to centromeric chromatin and (b) heterochromatic gene silencing. We determined that kinetochore proteins Ndc10p, Cac1p, Hir1p, the silencing protein, Sir3p, and RNA polymerase II subunit Rpb1p are required for the in vivo association of Spt4p to core CEN DNA, heterochromatic loci TEL and HMRa, and actively transcribing genes, respectively. These results show the presence of distinct HA-Spt4p complexes at CEN, TEL, HMRa, and actively transcribed loci. Furthermore, the human homolog of SPT4 (HsSPT4) functionally complements the yeast mutant phenotypes and associates with S. cerevisiae core CEN DNA in an Ndc10p-dependent manner. These results underscore the evolutionary conservation of pathways required for genome stability and illustrate how the yeast model system can be used to understand the complex molecular architecture of the kinetochore and its role in the fundamental process of chromosome segregation in humans.
SPT4 is a component of centromeric chromatin
Our previous analysis of SPT4 demonstrated that spt4 mutants show a 100-fold increase in the loss of a non-essential reporter chromosome, test positive in in vivo assays for defects in kinetochore integrity, and genetically interact with kinetochore mutants. Also, extracts from spt4 mutants exhibit defects in the binding of minichromosomes to microtubules (Basrai et al, 1996). Combined, these data support the hypothesis that kinetochore structure and/or function are altered in the spt4 mutants. The co-localization of a subset of Spt4-GFP foci with the kinetochore protein Ndc10p-HA and the in vivo association of HA-Spt4p with centromeric DNA provided further evidence for the role for Spt4p in kinetochore structure/function. We determined that components that constitute the inner kinetochore (Ndc10p) and chromatin (Cac1p, Hir1p) are required for the specific association of HA-Spt4p to core CEN DNA, but not to other loci. Ndc10p has been shown to be required for the association of inner, central, and outer kinetochore components to CEN DNA (Goshima and Yanagida, 2000; Cheeseman et al, 2002).
To gain direct evidence for the role of Spt4p in kinetochore structure, we determined whether the chromosome segregation and kinetochore integrity defects observed in the spt4 mutants (Basrai et al, 1996) may be due to an alteration of centromeric chromatin structure. We determined that in an spt4 mutant the CEN3 chromatin is at least five times more accessible to DraI digestion than CEN3 chromatin from a wild-type strain. Similar increases in DraI accessibility have been previously used to establish a structural role for histones (H2A, H2B, and H4), Cse4p, Cac1p, Hir1p, and chromatin remodeling proteins, Sth1p and Snf5p, in centromere structure (Han et al, 1987; Saunders et al, 1990; Smith et al, 1996; Tsuchiya et al, 1998; Pinto and Winston, 2000; Hsu et al, 2003).
Having established that Spt4p is a component of kinetochores, we addressed the impact of loss of functional Spt4p on kinetochore structure by examining whether Spt4p was required for the optimal localization of the kinetochore protein Cse4p. It has been suggested that the CEN DNA is wrapped around a Cse4p variant nucleosome (Basrai and Hieter, 1995; Stoler et al, 1995; Meluh et al, 1998; Keith and Fitzgerald-Hayes, 2000; Bjerling and Ekwall, 2002; Smith, 2002). We determined that Spt4p restricts the localization of Cse4p-HA exclusively to the kinetochores. ChIP experiments supported this conclusion, as the absence of functional Spt4p led to the association of Cse4p-HA to centromeric and noncentromeric loci such as TELVIR and HMRa. Overexpression of CSE4 from a GAL1 inducible promoter does not result in increased chromosome loss in wild-type strains (Sakelaris and Basrai, unpublished data). We have previously shown that there is a significant increase in chromosome loss in double mutants that combined mutations in spt4 with those of either CDEI or CDEII, but not with those of CDEIII (Basrai et al, 1996). These results suggest that Spt4p interacts with CDEI and CDEII, but not CDEIII. A similar model has been proposed for the interaction of Cse4p based on the increased chromosome loss observed for cse4 mutants when combined with mutations in CDEI and CDEII, but not CDEIII (Keith and Fitzgerald-Hayes, 2000; Smith, 2002). So far, we have failed to detect a genetic interaction between SPT4 and CSE4 alleles (data not shown). We speculate that the association of Cse4p-HA with noncentromeric DNA in an spt4 mutant may: (a) lead to the recruitment of one or more kinetochore proteins to the noncentromeric sites affecting the steady-state levels of these proteins including Cse4p at the kinetochore or (b) affect the chromatin structure at the noncentromeric loci.
Cac1p and Hir1p have been identified as proteins that associate with centromeric chromatin. Strains lacking both CAC1 and HIR1 exhibit chromosome missegregation phenotypes and show a declustering of Cse4p foci similar to that observed in the spt4 mutants (Sharp et al, 2002). spt4 and hir1 mutants are synthetically lethal and Spt4p and Hir1p interact biochemically (DeSilva et al, 1998; Formosa et al, 2002; G Hartzog and J Speer, personal communication). Furthermore, mutations in SPT4, SPT5, and SPT6 confer phenotypes similar to those due to mutations in the HIR genes (Compagnone-Post and Osley, 1996). Our data on lack of association of Spt4p is the first example of a kinetochore protein that requires both Cac1p and Hir1p for centromeric association. It is possible that mislocalization of Cse4p in cac1Δ hir1Δ strains may be due to lack of enrichment of Spt4p at the kinetochores. Based on genetic and biochemical interactions as well as phenotypes for single- and double-mutant combinations between cac1Δ, hir1Δ, and spt4Δ, we propose that SPT4 may be downstream of CAC1 and HIR1 in chromatin assembly at CEN loci.
Hence, based on genetic analyses, subcellular localization, in vivo association with CEN DNA, and centromeric chromatin studies, we propose that Spt4p is a key component of centromeric chromatin that is required for kinetochore function. Further studies with proteins such as Spt4p and associated proteins such as Spt5p and Spt6p may help us understand the molecular complexity of centromeric chromatin and understand the role of these and other proteins in kinetochore function.
Novel role of Spt4p in heterochromatic gene silencing
Several proteins including the Sir proteins (Sir1p, Sir2p, Sir3p, and Sir4p) are required for the formation of repressive chromatin structure at heterochromatic loci such as TEL and HMRa (Hoppe et al, 2002; Rusche et al, 2002). Absence of Sir3p or other silencing protein(s) leads to defects in transcriptional silencing of reporter genes at these sites. Our results suggest that Sir3p is required for the association of HA-Spt4p with TEL and HMRa. Coupled with the fact that spt4 mutants show defects in silencing at TEL and HMRa, these data show that Spt4p has a role in heterochromatic gene silencing. Studies with S. pombe have shown that several kinetochore components co-localize to other silenced regions (Allshire and Pidoux, 2001). Cac1p, Hir1p, and our data on Spt4p represent examples of S. cerevisiae proteins with roles in gene silencing and kinetochore function in addition to other functions (Enomoto et al, 1997; Game and Kaufman, 1999; Sharp et al, 2002).
Function of Spt4p is evolutionarily conserved
SPT4 is conserved evolutionarily with homologs in Schizosaccharomyces pombe, Mus musculus, and humans (Chiang et al, 1996; Hartzog et al, 1996; Wood et al, 2002). We have shown previously that HsSpt4p, which is 42% identical to ScSpt4p, functionally complements the ctf, ts, and spt phenotypes of S. cerevisiae mutants (Hartzog et al, 1996). Here we show that HsSPT4 complements the defects in the silencing of reporter genes at heterochromatic loci TEL and HMRa in Scspt4 mutants. Our data on the association of HsSpt4p with S. cerevisiae CEN DNA in an Ndc10p-dependent manner are consistent with the complementation of ctf phenotypes. These studies represent the first example of the in vivo association of a human protein to the kinetochores of budding yeast. Human homologs for key kinetochore proteins such as CENP-A, CENP-C, Skp1p, Sgt1p, and others have been described; however, only HsSGT1 has been shown to complement the chromosome loss phenotype of S. cerevisiae sgt1 mutants (Kitagawa and Hieter, 2001). Our results with Spt4p provide an exciting opportunity to study the association and order of assembly of human kinetochore proteins in budding yeast. The relatively simple S. cerevisiae centromere may provide clues to the fundamental order of assembly of the kinetochore of mammalian cells.
The significance of our studies with SPT4 is further strengthened by the observations that most human cancers are associated with abnormal karyotypes and increased rates of chromosome missegregation (Lengauer et al, 1998). Mutations that cause genome instability in model organisms such as S. cerevisiae may help us identify genes such as SPT4 that represent ‘cross species candidate genes' for cancer predisposition. It is possible that HsSPT4 is a prime target for mutation, leading to chromosome missegregation in human cancer. Future studies with SPT4 in yeast and humans will help us determine the role of this gene in genome stability.
Materials and methods
Strains, plasmids, and growth conditions
Yeast growth media and protocols are as described (Rose et al, 1990). The list of strains is given in Table I. The SPT4 ORF was tagged in frame with GFP or HA as described (Longtine et al, 1998). The plasmids used were pMB237 (HA-tagged SPT4) (Basrai et al, 1996), pMB299 (HA-tagged human SPT4) (Hartzog et al, 1996), and BMB 1058 (MTW1-GFP) (Iouk et al, 2002). CSE4 was tagged at its chromosomal locus using pCSE4HA/URA3 (pPM204)) (a gift from Pam Meluh, Memorial Sloan-Kettering Cancer CTR, NY, USA). Tagged strains were confirmed for in-frame fusions by PCR and Western blot analysis.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YMB54 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 spt4-138 | Basrai et al (1996) |
| YMB1849 | MATα ade2-1 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 ppr1Δ::HIS3 URA3-TELVIIL spt4Δ::KAN | This study |
| YMB1859 | MATa ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 bar1-1 LYS2 NDC10:HA3:URA3:HA3 SPT4GFP/HIS | This study |
| YMB1871 | MATa ade2-1 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 ppr1Δ::HIS3 HMRI::URA3 spt4Δ::KAN | This study |
| YMB2140 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 spt4-138+ [pCSE4-HA/URA3] | This study |
| YMB2142 | MATa ura3-52 lys2-801 ade2-101 trp1Δ1 leu2Δ1+[pCSE4-HA/URA3] | This study |
| YMB2230 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 spt4-138 CFVII (RAD2d.YPH277) URA3 SUP11+[pMTW1-GFP/LEU2] | This study |
| YMB2231 | MATa ura3-52 lys2-801 ade2-101 trp1Δ1 leu2Δ1+[pMTW1-GFP/LEU2] | This study |
| YPH98 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 | Spencer et al (1990) |
| YPH499 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1trp1Δ63 | Sikorski and Hieter (1989) |
| BUY545 | MATa ade2-1 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 ppr1Δ::HIS3 HMRΔI::URA3 | R Kamakaka |
| BUY668 | MATα ade2-1 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 ppr1Δ::HIS3 URA3-TELVIIL | Dhillon and Kamakaka (2000) |
| BUY671 | MATa/α ade2-1 his3-11 leu2-3,112 LYS2 trp1-1 ura3-1 ppr1Δ::HIS3 URA3-TELVIIL sir3Δ::TRP1 | R Kamakaka |
| GHY262 | MATα his4-912δ lys2-128δ ura3-52Δ1 pep4::LEU2 HA-SPT4 SPT5-MYC | G Hartzog |
| GHY501 | MATα his3-200δ his4-912δ lys2-128Δ ura3-52Δ1 leu2Δ1 rpb1Δ187::HIS3+[pRP112=RPB1CEN/URA3] | G Hartzog |
| GHY560 | MATα his3-200Δ his4-912δ lys2-128δ ura3-52Δ1 leu2Δ1 rpb1Δ187::HIS3+[pRP1-1= rpb1-1CEN/URA3] | G Hartzog |
| JK421 | MATa ade2-1 ura3-1 his3-11,1 trp1-1 leu2-3,112 can1-100 ndc10-1 | Goh and Kilmartin (1993) |
| PKY090 | MATa leu2-3,112 ura3-1 his3-11,15 trp1-1 ade2-1 can1-100 adh4::URA3-TEL VIIL | P Kaufman |
| PKY117 | MATa leu2-3,112 ura3-1 his3-11,15 trp1-1 ade2-1 can1-100 hir1::HIS3 adh4::URA3-TEL VIIL | P Kaufman |
| PKY632 | MATa leu2-3,112 ura3-1 his3-11,15 trp1-1 ade2-1 can1-100 cac1Δ::hisG hir1Δ::HIS3 URA3- ΔVIIL | P Kaufman |
| PKY638 | MATa leu2-3,112 ura3-1 his3-11,15 trp1-1 ade2-1 can1-100 cac1Δ::hisG URA3- VIIL | P Kaufman |
Chromatin immunoprecipitation
‘In vivo' crosslinking and chromatin immunoprecipitation (ChiP) were performed as described previously (Meluh and Broach, 1999), with the following modifications. Yeast strains were grown in YPD or selective media to an OD600 of 1.0–1.2, then treated ‘in situ' with 1% paraformaldehyde for 30 min. Immunoprecipitation was performed on the diluted chromatin solution with anti-HA (Roche) or anti-GFP (Roche) or no antibody addition (mock). PCR analyses of total (T), immunoprecipitated (HA), and mock (M) samples were performed using primer pairs (sequences are available upon request). All ChIP experiments were carried out at least three times and often four times. Different dilutions of the template DNA were used in PCR reactions to verify that the PCR yield was proportional to the amount of starting DNA. Quantitation of the data in Figures 1C and 6A was performed using ImageQuant software.
Chromosome spreads and fluorescence microscopy
Chromosome spreads were performed as described (Loidl et al, 1991). HA-tagged proteins were detected using antibody 16B12 (Covance) with secondary antibody Cy3-conjugated Affinipure Goat Anti-mouse IgG (Jackson ImmunoResearch). GFP-tagged proteins were detected using antibody Rabbit Alexa Fluor 488 (Molecular Probes). Nuclear morphology was examined by DAPI (4′,6-diamidino-2-phenylindole) staining. Fluorescence microscopy utilized a Zeiss Axioscope 2 microscope (Carl Zeiss Inc.) with a Cooke Sensicam (Cooke), a Chroma GFP filter set (CZ909, Chroma Technology Corp.), and a Uniblitz Shutter assembly (Uniblitz).
Western blot analysis
Western blot analysis was as described previously (Iouk et al, 2002). The primary antibody used was anti-HA (clone 12CA5—Roche) or anti-Pgk1p (A-6457—Molecular Probes). The secondary antibody was HRP-conjugated sheep anti-mouse IgG (NA931V—Amersham).
Centromeric chromatin structure accessibility assay
The DraI assay was performed as described previously (Saunders et al, 1990; Meluh et al, 1998). Logarithmically growing cultures at 30°C were shifted to 37°C for 6 h. Nuclei were isolated and incubated with different concentrations of DraI at 30°C for 30 min, followed by digestion with EcoRI and Southern blotting using a 32P-dCTP-labeled 0.9 kb HindIII–BamHI fragment as a probe.
Acknowledgments
We thank G Hartzog and O Kerscher for support and advice throughout the course of this work. We are grateful to W Au, S Biggins, K Bloom, N Dhillon, L Espinoza, G Hartzog, A Hong, P Hieter, Y-J Kim, R Kamakaka, M Karthikeyan, P Kaufman, P Meluh, and V Measday for strains, reagents, and advice. We acknowledge M Nau, T Rice, J Smith, and members of the Basrai laboratory for discussions and comments on this manuscript. We owe special gratitude to J Kastenmayer and C Carter for critical evaluation of the data and insightful comments on the manuscript and to D Horowitz for his support during the completion of this work.
References
- Allshire R, Pidoux A (2001) Centromeres. Curr Biol 11: R454. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Guzman E, Doring P, Werner J, Lis JT (2000) High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev 14: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai MA, Hieter P (1995) Is there a unique form of chromatin at the Saccharomyces cerevisiae centromeres? BioEssays 17: 669–672 [DOI] [PubMed] [Google Scholar]
- Basrai MA, Kingsbury J, Koshland D, Spencer F, Hieter P (1996) Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol Cell Biol 16: 2838–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerling P, Ekwall K (2002) Centromere domain organization and histone modifications. Braz J Med Biol Res 35: 499–50700 [DOI] [PubMed] [Google Scholar]
- Bloom KS, Carbon J (1982) Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29: 305–317 [DOI] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197: 345–346 [DOI] [PubMed] [Google Scholar]
- Bortvin A, Winston F (1996) Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272: 1473–1476 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Drubin DG, Barnes G (2002) Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J Cell Biol 157: 199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang PW, Wang SQ, Smithivas P, Song WJ, Crombez E, Akhtar A, Im R, Greenfield J, Ramamoorthy S, Van Keuren M, Blackburn CC, Tsai CH, Kurnit DM (1996) Isolation and characterization of the human and mouse homologues (SUPT4H and Supt4h) of the yeast SPT4 gene. Genomics 34: 368–375 [DOI] [PubMed] [Google Scholar]
- Compagnone-Post PA, Osley MA (1996) Mutations in the SPT4, SPT5, and SPT6 genes alter transcription of a subset of histone genes in Saccharomyces cerevisiae. Genetics 143: 1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSilva H, Lee K, Osley MA (1998) Functional dissection of yeast Hir1p a WD repeat-containing transcriptional corepressor. Genetics 148: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N, Kamakaka RT (2000) A histone variant, Htz1p and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol Cell 6: 769–780 [DOI] [PubMed] [Google Scholar]
- Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders MA, Berman J (1997) RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev 11: 358–370 [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M, Clarke L, Carbon J (1982) Nucleotide sequence comparison and functional analysis of yeast centromere DNAs. Cell 29: 235–244 [DOI] [PubMed] [Google Scholar]
- Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ (2002) Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway. Polymerase passage may degrade chromatin structure. Genetics 162: 1557–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk M, Hegemann JH, Philippsen P (1989) Chromatin digestion with restriction endonucleases reveals 150–160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae. Mol Gen Genet 219: 153–160 [DOI] [PubMed] [Google Scholar]
- Game JC, Kaufman PD (1999) Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics 151: 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowczewski L, Yang P, Kalashnikova T, Santisteban MS, Smith MM (2000) Histone-histone interactions and centromere function. Mol Cell Biol 20: 5700–5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh PY, Kilmartin JV (1993) NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol 121: 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Yanagida M (2000) Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100: 619–633 [DOI] [PubMed] [Google Scholar]
- Grunstein M (1997) Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol 9: 383–387 [DOI] [PubMed] [Google Scholar]
- Han M, Chang M, Kim UJ, Grunstein M (1987) Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 48: 589–597 [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Basrai MA, Ricupero-Hovasse SL, Hieter P, Winston F (1996) Identification and analysis of a functional human homolog of the SPT4 gene of Saccharomyces cerevisiae. Mol Cell Biol 16: 2848–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Speer JL, Lindstrom DL (2002) Transcript elongation on a nucleoprotein template. Biochim Biophys Acta 1577: 276–286 [DOI] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK (2001) Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106: 195–206 [DOI] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D (2002) Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22: 4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JM, Huang J, Meluh PB, Laurent BC (2003) The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol Cell Biol 23: 3202–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW (2002) The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol 159: 807–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka RT, Rine J (1998) Sir- and silencer-independent disruption of silencing in Saccharomyces by Sas10p. Genetics 149: 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Morris JR, Wu C, Winston F (2000) Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev 14: 2623–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith KC, Baker RE, Chen Y, Harris K, Stoler S, Fitzgerald-Hayes M (1999) Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol Cell Biol 19: 6130–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith KC, Fitzgerald-Hayes M (2000) CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere DNA around a cse4p variant nucleosome. Genetics 156: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Hieter P, Winey M, Basrai MA (2001) Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics 157: 1543–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Hieter P (2001) Evolutionary conservation between budding yeast and human kinetochores. Nat Rev Mol Cell Biol 2: 678–687 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J (2003) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T (2002) Chromatin assembly: the kinetochore connection. Curr Biol 12: R256–R258 [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B (1998) Genetic instabilities in human cancers. Nature 396: 643–649 [DOI] [PubMed] [Google Scholar]
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR III, Hartzog GA (2003) Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol 23: 1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Nairz K, Klein F (1991) Meiotic chromosome synapsis in a haploid yeast. Chromosoma 100: 221–228 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Meeks-Wagner D, Hartwell LH (1986) Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44: 43–52 [DOI] [PubMed] [Google Scholar]
- Meluh PB, Broach JR (1999) Immunological analysis of yeast chromatin. Methods Enzymol 304: 414–430 [DOI] [PubMed] [Google Scholar]
- Meluh PB, Koshland D (1997) Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev 11: 3401–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM (1998) Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613 [DOI] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R (1987) Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol 7: 1602–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto I, Winston F (2000) Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J 19: 1598–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell 9: 799–809 [DOI] [PubMed] [Google Scholar]
- Rondon AG, Garcia-Rubio M, Gonzalez-Barrera S, Aguilera A (2003) Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J 22: 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P (1990) Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J (2002) Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell 13: 2207–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M, Fitzgerald-Hayes M, Bloom K (1988) Chromatin structure of altered yeast centromeres. Proc Natl Acad Sci USA 85: 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders MJ, Yeh E, Grunstein M, Bloom K (1990) Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol Cell Biol 10: 5721–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman I, Bloom KS (1991) Centromeres: an integrated protein/DNA complex required for chromosome movement. Annu Rev Cell Biol 7: 311–336 [DOI] [PubMed] [Google Scholar]
- Sharp JA, Franco AA, Osley MA, Kaufman PD (2002) Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev 16: 85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM (2003) Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 22: 1846–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM (2002) Centromeres and variant histones: what, where, when and why? Curr Opin Cell Biol 14: 279–285 [DOI] [PubMed] [Google Scholar]
- Smith MM, Yang P, Santisteban MS, Boone PW, Goldstein AT, Megee PC (1996) A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol Cell Biol 16: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Gerring S, Connelly C, Hieter P (1990) Mitotic chromosome segregation fidelity mutants in Saccharomyces cerevisiae. Genetics 124: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M (1995) A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev 9: 573–586 [DOI] [PubMed] [Google Scholar]
- Swanson M, Winston F (1992) SPT4, SPT5, and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132: 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya E, Hosotani T, Miyakawa T (1998) A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res 26: 3286–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Chaleff DT, Valent B, Fink GR (1984) Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107: 179–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O'Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schafer M, Muller-Auer S, Gabel C, Fuchs M, Dusterhoft A, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, Dreano S, Gloux S, Lelaure V, Mottier S, Galibert F, Aves SJ, Xiang Z, Hunt C, Moore K, Hurst SM, Lucas M, Rochet M, Gaillardin C, Tallada VA, Garzon A, Thode G, Daga RR, Cruzado L, Jimenez J, Sanchez M, del Rey F, Benito J, Dominguez A, Revuelta JL, Moreno S, Armstrong J, Forsburg SL, Cerutti L, Lowe T, McCombie WR, Paulsen I, Potashkin J, Shpakovski GV, Ussery D, Barrell BG, Nurse P, Cerrutti L (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 [DOI] [PubMed] [Google Scholar]


