Abstract
The rapid increase in heat shock proteins upon exposure to damaging stresses and during plant development related to desiccation events reveal their dual importance in plant development and stress tolerance. Genome-wide sequence survey identified 20 non-redundant small heat shock proteins (sHsp) and 22 heat shock factor (Hsf) genes in barley. While all three major classes (A, B, C) of Hsfs are localized in nucleus, the 20 sHsp gene family members are localized in different cell organelles like cytoplasm, mitochondria, plastid and peroxisomes. Hsf and sHsp members are differentially regulated during drought and at different seed developmental stages suggesting the importance of chaperone role under drought as well as seed development. In silico cis-regulatory motif analysis of Hsf promoters showed an enrichment with abscisic acid responsive cis-elements (ABRE), implying regulatory role of ABA in mediating transcriptional response of HvsHsf genes. Gene regulatory network analysis identified HvHsfB2c as potential central regulator of the seed-specific expression of several HvsHsps including 17.5CI sHsp. These results indicate that HvHsfB2c is co-expressed in the central hub of small Hsps and therefore it may be regulating the expression of several HvsHsp subclasses HvHsp16.88-CI, HvHsp17.5-CI and HvHsp17.7-CI. The in vivo relevance of binding specificity of HvHsfB2C transcription factor to HSE-element present in the promoter of HvSHP17.5-CI under heat stress exposure is confirmed by gel shift and LUC-reporter assays. Further, we isolated 477 bp cDNA from barley encoding a 17.5 sHsp polypeptide, which was predominantly upregulated under drought stress treatments and also preferentially expressed in developing seeds. Recombinant HvsHsp17.5-CI protein was expressed in E. coli and purified to homogeneity, which displayed in vitro chaperone activity. The predicted structural model of HvsHsp-17.5-CI protein suggests that the α-crystallin domain is evolutionarily highly conserved.
Introduction
When plants are challenged by drought and temperature stresses, a wide array of interconnected cellular stress response systems is triggered. These cellular responses helps in readjustment of the growth of plants and its survival under abiotic stress exposure. An understanding of the molecular basis of these responses to stress adaptation is essential to make use of them in breeding programs. Heat shock responses (HSR) are temperature-related defense activities and include the induction of evolutionarily conserved chaperone proteins known as heat shock proteins (Hsps). Based on their molecular size, they are classified into different classes i.e Hsp100, Hsp90, Hsp70/DnaK, Hsp60/GroE and small heat shock proteins (sHsps) [1]. Generally, sHsps form large oligomeric complexes [2], ranging in size from 200–800 kDa, and are targeted to different cellular compartments. Though sHsps have been studied in different plant systems [3]; it is not completely known how different sHsps interact with their target proteins and why so many paralogues evolved. Synthesis of sHsps is not specific to heat stress response, but also expressed as part of the developmental program. sHsps are highly expressed in developmental stages like zygotic embryonic tissues, pollen maturation, embryogenesis and during seed maturation [4]–[7]. How these large gene family members of sHsps are finely regulated by a defined set of potential heat shock transcription factors (Hsfs) to control many vital processes important during plant development and stress response is largely obscure.
Plant Hsfs have a major role to play in the modulation of transcription during long-term heat shock response [8]. A typical Hsf protein contains a modular structure with an N-terminal DNA-binding domain (DBD), a nuclear localization signal (NLS), a nuclear export signal (NES), and in many cases a less conserved C-terminal activation domain rich in aromatic, hydrophobic and acidic amino acids (AHA) that have been reported to be crucial for activation function [8], [9]. Based on sequence homology and domain architecture, plant Hsfs have been divided into three conserved classes. Several heat shock factor (Hsf) complexes could be responsible for the developmental and stress inducible transcription of Hsp genes [10]. Rojas et al. [11] demonstrated transcriptional activation of a heat shock protein promoter by ABI3 and Hsf complex. These analyses indicated that genes are controlled by complex regulatory networks [12], [13]. The expression of Hsps during different stages of plant ontogeny and its stress-induction depend on the cis-motifs present in the respective genes; bound by different transcription factors especially Hsfs as demonstrated by transient reporter assays in Arabidopsis and sunflower embryos [14], [15]. The expression of particular isoforms of Hsp genes during seed development suggests that they may have distinct tissue and cell-specific functions during seed maturation and are regulated by a set of defined developmental programs. Though the importance of Hsfs as regulators of the heat shock response is known, the Hsfs-sHsp interconnections in plant development with special reference to developing seeds and stress responses remain unknown in cereals.
The present study was taken up to find out the expression profiles of sHsps and Hsfs in barley since that can ultimately help to identify key regulators promoting developmental events under stress. Here, we report a genome-wide survey of all non-redundant sets of HvsHsp and Hsf genes in the complex genome of barley, a model crop of tribe Triticeae. This survey provided first holistic insights into the interconnected responses of 20 sHsps and 22 Hsfs gene family members in drought stress response in vegetative tissues and also emphasized its role in seed development of barley. In silico motif analysis in the 5′ upstream regions of sHsp and Hsf genes revealed the presence of a distinct set of transcription factor binding sites (cis-elements) interlinking the role of ABA in mediating Hsf genes. This is the first comprehensive transcriptomic study that identified the differentially expressed sHsp and Hsf genes and the coexpressed gene networks involved in seed development and drought stress adaptation in barley. Our gene regulatory network analysis identified HvHsfB2c as central regulatory hub of sHsps. Our in vivo binding assays confirm that HvHsfB2c binds to HSE cis element in the HvHsp17.5CI promoter, its transcript is preferentially regulated under desiccation response. Further, we purified the recombinant HvsHsp17.5-CI protein (expressed preferentially in developing seeds and responsive to stress) to homogeneity and validated its chaperone activity.
Materials and Methods
Identification and Annotation of sHsp and Hsf Family Genes
The HarvEST Barley database (http://harvest.ucr.edu/) with 50,000 unigenes was searched for genes that encode proteins of sHsp and Hsf genes, sequence similarity searches were performed using Blastn and Blastx based on known rice and Arabidopsis sequence annotations retrieved from TIGR database (http://rice.plantbiology.msu.edu/) and TAIR database (http://www.arabidopsis.org/Blast/). The corresponding cDNA sequences were extracted from the database and subjected to a comparison with recently available 24K full length barley cDNA database to identify full-length clones [16]. cDNA sequences were translated and searched for conserved domains known from the corresponding rice proteins using NCBI database. Multiple sequence alignment and phylogenetic trees comprising barley and rice sHsp and Hsf full length protein sequences were generated by using ClustalW (DNAstar) program. Accession numbers of all identified genes are indexed in tables 1 and 2. Information about the number of amino acids (AA), molecular weights (M.Wt) and theoretical isoelectric point (pI) of all barley sHsps and Hsfs were predicted by using DNAstar software. Organellar targeting of these proteins were predicted by using pSORT (http://psort.nibb.ac.jp/) and TargetP (http://www.cbs.dtu.dk/services/TargetP/) programs. Information regarding ORF length and intron numbers was confirmed by comparing the respective cDNA and genomic clones. Conserved domains of the sHsp and Hsf proteins in barley were determined by Pfam program and from the existing literature.
Table 1. List of Hsf genes involved in different abiotic stress conditions and development of barley.
| Name | HarvESTID | Fl_cDNA ID | Affymetrix ID | Full/partial | ORF (bp) | Protein (AA) | M.wt (kDa) | pI | Intron | Chromosome Localization | Morex/Bac contig_ID | 5′ upstream region (bp) | Predicted localization |
| HvHsfA1a | 35_17839 | AK354917 | Contig8225_at | Full | 1533 | 510 | 56.58 | 5 | HVVMRX83KhA 0005M16_v61_c1 | 2007 | Nucleus | ||
| HvHsfA2a | 35_7894 | AK377082 | Contig18295_at | Full | 1116 | 371 | 41.08 | 5.61 | 1(77) | 4 | Contig_2161641 | 854 | Nucleus |
| HvHsfA2b | 35_9896 | AK359122 | Full | 1257 | 418 | 46.06 | 4.82 | 1(2446) | 2 | Contig_40049 | Nucleus | ||
| HvHsfA2c | 35_28323 | AK363263 | Full | 1119 | 372 | 42.11 | 4.9 | 1(891) | 1 | HVVMRX83KhA 0052F09_v62_c1 | 2094 | Nucleus | |
| HvHsfA2d | 35_48033 | AK364288 | rbaal35o24_at | Partial | 4 | Contig_5993 | Nucleus | ||||||
| HvHsfA2e | 35_23711 | AK358148 | Full | 1083 | 360 | 40.24 | 5.65 | 2 | Contig_2162221 | Nucleus | |||
| HvHsfA2f | AK372701 | Full | 1053 | 350 | 37.98 | 4.64 | 4 | Nucleus | |||||
| HvHsfA4b | 35_8623 | AK371121 | Contig23893_at | Full | 1299 | 432 | 48.54 | 5.41 | 1(543) | 3 | Contig_41104 | Nucleus | |
| HvHsfA4d | 35_9620 | AK362315 | Contig18961_at | Full | 1296 | 431 | 48.72 | 5.41 | 1(174) | 1 | Contig_1009401 | 2030 | Nucleus |
| HvHsfA3 | 35_41834 | HM446028 | Full | 1488 | 495 | 54.18 | 6.01 | 1(1050) | 2 | Contig_53615 | 1120 | Nucleus | |
| HvHsfA5 | 35_9627 | AK355564 | Contig18870_at | Full | 1374 | 457 | 50.17 | 5.51 | 1(1064) | 6 | HVVMRXALLh A0687I07_v22_c1 | 1986 | Nucleus |
| HvHsfA9 | 35_18881 | AK362997 | Contig10108_s_at | Partial | 7 | Nucleus | |||||||
| HHsfC1b | 35_36864 | AK353849 | HB03A08_T3_at | Full | 708 | 235 | 26.07 | 7.4 | 1(97) | 4 | Contig_1017877 | 798 | Nucleus |
| HvHsfB1 | AK366231 | Partial | 4 | Nucleus | |||||||||
| HvHsfB2a | 35_38788 | AK357873 | Full | 900 | 299 | 32.48 | 5.15 | 2 | Nucleus | ||||
| HvHsfB2b | 35_6843 | AK373036 | Contig18148_at | Full | 1173 | 390 | 41.88 | 5.53 | 7 | Contig_49614 | Nucleus | ||
| HvHsfB2c | 35_18576 | AK354133 | HV_CEa0014 A18r2_at | Full | 1209 | 402 | 41.74 | 4.78 | 7 | Nucleus | |||
| HvHsfB4b | AK376881 | Full | 951 | 316 | 34.69 | 7.11 | 2 | Nucleus | |||||
| HvHsfB4c | 35_31994 | AK356002 | Full | 1167 | 388 | 41.42 | 7.96 | 2 | Contig_221980 | Nucleus | |||
| HvHsfC1a | 35_26831 | CB860849 | Partial | 1(202) | 4 | Contig_61089 | 2030 | Nucleus | |||||
| HvHsfC2a | AK369002 | Full | 801 | 266 | 29.48 | 5.86 | 4 | HVVMRXALLhA 0053M06_v10_c1 | 2055 | Nucleus | |||
| HvHsfC2b | 35_16479 | AK354435 | Contig6968_at | Full | 801 | 266 | 28.27 | 6.35 | 7 | Contig_57251 | Nucleus |
The table shows the following details: Harvest unigene ID, full-length cDNA ID, Affymetrix ID, full-length/partial, open reading frame (ORF) size, predicted molecular mass for the deduced proteins, isoelectricpoint (pI), intron number with size, genomic sequence information (Assembly1 Morex ID), derived 5′ upstream of the translational start site and predicted subcellular localization.
Table 2. List of sHsp genes involved in different abiotic stress conditions and development of barley.
| Name | HarvEST ID | Fl cDNA ID | Affymetrix ID | Full/partial | ORF (bp) | Protein (AA) | M.wt (kDa) | pI | Intron | Chromosome localization | 5′ upstream region (bp) | Localization |
| HvHsp16.9-CI | 35_14860 | AK362925 | Contig2004_s_at | Full | 456 | 151 | 16.95 | 5.92 | 3 | Cytoplasm | ||
| HvHsp16.88-CI | 35_14863 | AK355146 | Contig2008_s_at | Full | 453 | 150 | 16.88 | 5.92 | 3 | 1367 | Cytoplasm | |
| HvHsp16.86-CI | AK376179 | Full | 456 | 151 | 16.86 | 5.79 | 3 | 2030 | Cytoplasm | |||
| HvHsp17.5-CI | 35_14859 | AK250749 | HB18H23r_s_at | Full | 477 | 158 | 17.57 | 5.9 | 4 | 1785 | Cytoplasm | |
| HvHsp16.7-CI | 35_14864 | AK252765 | Contig2010_at | Full | 453 | 150 | 16.76 | 5.46 | 4 | Cytoplasm | ||
| HvHsp17.7-CI | 35_1486635_30931 | AK368988 | Contig2012_s_at | Full | 486 | 161 | 17.79 | 5.52 | 4 | 2068 | Cytoplasm | |
| HvHsp17.3-CII | 35_15451 | AK375970 | Contig3284_x_at | Full | 480 | 159 | 17.33 | 6.05 | 3 | Cytoplasm | ||
| HvHsp17.7-CII | 35_15454 | BF263847 | Contig3287_x_at | Full | 489 | 162 | 17.7 | 6.07 | 3 | 2446 | Cytoplasm | |
| HvHsp-17.77CII | 35_15456 | AK355146 | Contig3289_at | Full | 489 | 162 | 17.77 | 6.41 | 3 | 1791 | Cytoplasm | |
| HvHsp19.0-CIII | 35_18565 | AK360636 | Contig10029_at | Full | 537 | 178 | 19.05 | 6.92 | 3 | Cytoplasm | ||
| HvHsp17.1-CV | 35_5795,35_27788 | AK373216 | Contig13073_at | Full | 468 | 155 | 17.14 | 6.69 | 1(92) | 2 | 2078 | Cytoplasm |
| HvHsp17.76-CIX | 35_5001 | AK370196 | Contig11961_at | Full | 498 | 165 | 17.65 | 4.68 | 3 | Cytoplasm | ||
| HvHsp19.2-CX | 35_5077 | AK370937 | Contig15445_at | Full | 534 | 177 | 19.26 | 7.53 | 6 | 2285 | Cytoplasm | |
| HvHsp15.1-Px | AK365443 | Full | 417 | 138 | 15.14 | 8.48 | 4 | 2000 | peroxisome | |||
| HvHsp21.9-ER | AK374148,AK374905 | Full | 618 | 205 | 21.97 | 6.64 | 6 | 2100 | endoplasmic reticulum | |||
| HvHsp-ER | 35_31108 | BQ766746 | EBro08_SQ007_C01_at | Partial | 3 | |||||||
| HvHsp21.3-MI | 35_16753,35_16752 | AK370932 | Contig6559_at | Full | 582 | 193 | 21.32 | 6.6 | 7 | mitochondria | ||
| HvHsp26.8-P | 35_21394 | AK376976 | EBem05_SQ003_L06_at | Full | 735 | 244 | 26.8 | 6.1 | 1(102) | 4 | 763 | chloroplast |
| HvHsp21.2 | 35_23844 | BF623486 | Contig21040_at | Full | 585 | 194 | 21.25 | 9.31 | 4 | |||
| HvHsp | 35_42702 | CA005065 | HU11O14u_at | Partial | 1 |
The table shows the following details: Harvest unigene ID, full-length cDNA ID, Affymetrix ID, full-length/partial, open reading frame (ORF) size, predicted molecular mass for the deduced proteins, isoelectricpoint (pI), intron number with size, genomic sequence information (Assembly1 Morex ID), derived 5′ upstream of the translational start site and predicted subcellular localization.
Expression Analysis Using the Barley Genechip and Gene Network Analysis
RNA isolation of flag leaf and developing seed tissue from control and drought stress treatments was performed as described previously [17]. Probe synthesis, labelling and hybridization were performed according to manufacturer's instructions (Affymetrix). The purified labelled cRNA samples prepared from various vegetative tissues, as well as flag leaf and developing grains collected under control and terminal drought were hybridized to Barley1 GeneChips as described by Close et al. [18]. Arrays were scanned on a GeneChip Scanner 3000. The raw gene expression data of flag leaf and developing grain under control and terminal drought collected from this study were normalized together with publicly available Affymetrix gene expression data obtained from drought-challenged seedlings (series GSE3170), drought stressed 21-day-old plants (series GSE6990) and awn, lemma and palea tissues collected during terminal drought (series GSE17669). The fold change calculations (control versus drought stress) derived from log transformed normalized expression data from every individual stage of plant development were extracted for sHsp and Hsf gene family members and shown in a heat map. Further, to create gene expression atlas from plant ontogeny all the publicly available gene expression covering various tissues and developmental stages from seed germination, seedling establishment, plant maturity, reproductive tissues and developing endosperm and embryo tissue during seed development were normalized using the CEL files in an R package. Log transformed quantile normalized expression values of sHsp and Hsf gene family were shown in a heat map. Using high throughput coexpression data covering the entire plant ontogeny of barley, the gene regulatory networks have been derived. Gene coexpression network of HsfB2c and HvsHsp17.5 is derived from Plant Network database using Heuristic Cluster Chiseling Algorithm [19]. The gene network vicinity of HsfB2c and HvsHsp17.5 are enriched with various sHsp genes as well other primary metabolism functional categories of coexpressed genes. Further, we used cornet database to predict the protein-protein interactions and coexpression network of AT3G46230 (orthologue of HvsHsp17.5) [20].
In Silico Promoter Analysis
To analyze putative cis-elements in the promoter region of sHsp and Hsf family genes, 1,500 bp DNA sequence up-stream of the 5′ end of the cDNAs was extracted from Whole Genome Shotgun sequencing of barley [21] using the viroblast database (http://webblast.ipk-gatersleben.de/barley/index.php). The sequences were further analysed by different web based softwares like PLACE [22] and PlantCARE [23] as well as motifs extracted from the literature. To find out the regulatory cis elements, whole promoter sequence was searched in both forward and reverse strands.
Electrophoretic Mobility Shift Assay (EMSA)
The coding sequence of HvHsfB2c was amplified from barley cv. Golden Promise leaf cDNA and transferred to pTOPO-cloning vector (Primer used for amplification: Forward TACCATGGGCAGCAGCCATCATCATCATCATCACAGCAGCGGCCTGGTGCCGCGCGGCAGCCA, Reverse: TACCATGGGCCTCACCTCGAGTTGGACCTGTCCTG). After sequencing of the transferred amplicon translation, HsfB2c protein was synthesized using the PURExpress in vitro system from NEB according to manufacturer's protocol. EMSA has been performed as described previously [24] in the presence of 100 ng pdIdC/rn.
The following Oligonucleotides, containing the HSE-binding box were used for the binding reactions: HSE-1: TCGAACAACCCAAAAT CCAAAAAATTCCACAACCCCAAAAAGGC, HSE-2: TCGAGCCTTTTTGGG GTTGTGGAATTTTTTGGATTTTGGGTTGT.
Transient expression of HvHsfB2c-derivatives
Arabidopsis Col-0 mesophyll protoplast isolation and transformation was carried out with plants grown in soil under controlled conditions in a phytochamber for 4 weeks (8 h light/16 h dark at 20°C and 18°C, respectively) [25]. For microscopic localization studies, HvHsfB2c was cloned into pENSG-/pEXSG vectors (N-/C-terminal fusion with CFP, respectively) and 10 µg of DNA per 100 µl protoplasts was transformed. After 16 h incubation in the dark, CFP fluorescence was evaluated using the LSM 710 Laser Scanning System (Zeiss, Oberkochen, Germany). In case of the luciferase reporter assay, a HvsHsp17.5-CI promoter-luciferase construct, pUBQ10-GUS for normalization [26] and either pEXSG-HvHsfB2c or pUGW15-CFP [27] as control were transformed (10 µg total DNA per 100 µl protoplasts; ratio 1∶1∶1). Heat stress mimicking condition was applied by temporal increase of the incubation temperature for 10 min to 35°C. The used HvsHsp17.5-CI promoter fragment contained the region 700 bp upstream of the start ATG. Primer used for amplification are Forward: pHsp17.5_BamHI CAGGATCCTGTTGAGGACTGACA, Reverse: pHsp17.5_NcoI CACCATGGCGATCGGGTACTCGG. The luciferase-assay was performed as described in [28].
Homology Modeling of HvsHsp17.5-CI
HvsHsp17.5-CI molecular model was generated using the homology modeling server SWISS-MODEL [29] utilizing Triticum aestivum sHsp16.9 protein crystal structure as a template (PDB No:1gmeA). Following PROCHECK analysis, the model with the best Z-score −0.92 showed an RMSD of 2.70 Å and 70% sequence identity with respect to the template. The modeled residue range was taken from amino acids 2–158 by I-TASSER. Dimeric structure was generated by aligning the monomeric structure with the α-crystallin domain of TasHsp16.9 and HvsHsp17.5-CI using the program I-TASSER server [30], [31] with the best C- score 0.897, an RMSD of 2.40 Å and 0.699 TM-score with respect to the template.
Expression and Purification of HvsHsp17.5-CI Recombinant Protein
HvsHsp17.5-CI specific oligonucleotide primers were designed one for the N-terminus region (5′-ATACTACATATGTCGCTGATCCGTCGCAGCAACGT-3′) and the other for C-terminus region (5′-TAATGCGGCCGCCTAGCCGGAGATCTGGATGGAC-3′). The 5′ and 3′ untranslated regions in the cDNA were removed and an NdeI site at the translation initiation and a NotI site just downstream of the translation termination codon were introduced. HvsHsp17.5-CI PCR amplified cDNA product was digested and cloned into NdeI and NotI sites of pET28a (+) expression vector. The sequences adjoining the 5′ and 3′ ends of the cloned segment were confirmed by sequencing. This construct resulted in the expression of HvsHsp17.5-CI polypeptide with additional extra 20 amino acids including hexa histidine tag at the N-terminus. The recombinant pET28a-HvsHsp17.5-CI plasmid was transformed into BL21 (DE3) cells and grown in LB-medium supplemented with 50 µg/ml kanamycin at 37°C. As absorbance at 600 nm (A600) reached a value of about 0.5–0.6, the expression of recombinant HvsHsp17.5-CI was induced by adding IPTG (isopropyl β-D-1-thiogalactopyranoside, 1 mM) and the cells were allowed to grow for an additional period of 3 h at 37°C. After induction of recombinant protein, E. coli cells were lysed by sonication. Native recombinant HvsHsp17.5-CI protein was purified from clarified E. coli lysate through Ni-NTA column chromatography, following the manufacturer's instructions (Qiagen, Germany) and protein samples were analyzed by SDS-PAGE.
Stress Tolerance of E. coli Overexpressing Recombinant HvsHsp17.5-CI and its Chaperone Activity
E. coli BL21 (DE3) cells transformed with pET28a (+) (vector control) or with pET28a-HvsHsp17.5-CI plasmids. Transformed E. Coli were grown overnight in fresh LB medium containing 50 µg/ml kanamycin. When the absorbance at 600 nm reached a value of 0.25, varying concentrations of NaCl (0–750 mM) for salinity stress, and 0–25% of polyethylene glycol (PEG, molecular weight of 3,350) were added to impose dehydration stress after the addition of IPTG. For temperature stress, cultures were grown at 37 to 55°C after IPTG treatment. After induction with the addition of IPTG (1 mM), cultures were kept at 37°C for 12 h in a shaking incubator. Cell growth was monitored by measuring the absorbance at 600 nm. Each experiment was repeated thrice and average readings were taken. Recombinant HvsHsp17.5-CI chaperone activity was assayed by using thermo labile restriction enzyme, SwaI (New England Biolabs, Beverly, MA) as previously described [32], [33]. The heat-labile SwaI enzyme was pre-incubated at a range of temperatures (25, 30, 35, 40, 45 and 50°C) for 60 min in the presence of either BSA (5 µg) or recombinant HvsHsp17.5-CI (5 µg). After pre-incubation, the reaction mixture was cooled to 25°C and plasmid DNA (500 ng) with a unique SwaI recognition site was added and further incubated at 25°C for 60 min for restricting digestion of plasmid DNA. The restriction digested plasmid DNA samples were separated by electrophoresis on 1% agarose gel and stained with ethidium bromide. Plasmid digestion profiles were compared with their respective controls.
Results
Identification of sHsp and Hsf Gene Families in Barley
The conserved amino acid sequence of α-crystallin domain (ACD) for sHsps and DNA binding domain (DBD) for Hsfs was adapted as a query to search possible homologs encoded in the barley genome using HarvEST (http://harvest.ucr.edu/) and NCBI databases. This resulted in the retrieval of 20 sHsp and 22 Hsf encoding gene sequences. Further, functional annotations of these sequences were verified by using BlastX, BlastN and BlastP programmes of NCBI. Details of all the genes encoding for barley sHsps and Hsfs are represented in the Tables 1 and 2 respectively. Comparative sequence alignment of the genomic and cDNA sequences of sHsp and Hsf genes revealed the predicted exons and introns. Only certain classes of sHsps possess introns. HvsHsp proteins showed variation in length (from 138 to 244 amino acids), isoelectric point (pI) values (4.68 to 9.31) and molecular weights (15.14 to 21.25 kDa). Prediction of subcellular localization of these proteins using pSORT and TargetP programmes [34]–[36] indicated that 13 sHsp proteins are located in the cytoplasm and one each in the mitochondria, ER, plastid and peroxisome. In case of Hsf family proteins, 21 are distributed in the nucleus and one in the cytoplasm.
Phylogenetic Analysis of Hsf Gene Family Members
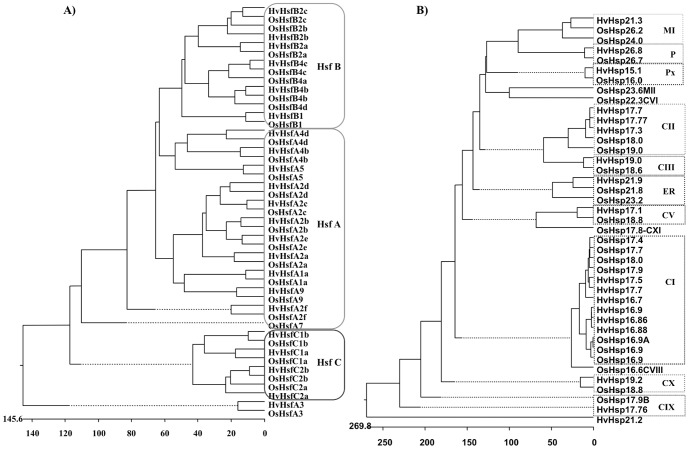
A phylogenetic tree was constructed for 22 barley and 25 rice Hsf genes. All Hsfs clustered broadly into three major cluster groups A, B and C, which included representative genes of barley and rice (Figure 1A). The major cluster class A is further divided into several subclasses based on its phylogenetic relationship and designated as A1, A2, A3, A4, A5 and A9. HvHsfA3 and its rice ortholog OsHsfA3 did not cluster with class A but grouped separately (Figure 1A). This however appears to be closer to HsfC. All class B Hsfs showed divergence from a common point but are closer to HsfA class genes compared to class C (Figure 1A). The motif distribution also followed the same scenario with the phylogenetic analysis. Therefore, it looks from the phylogenetic tree that one subgroup of class A Hsf gave rise to class B and C (Figure 1A).
Figure 1. Phylogenetic relationships between barley and rice sHsp and Hsf proteins.
The phylogenetic tree was drawn from the deduced amino acid sequences of sHsp (A) and Hsf (B) proteins from the barley and rice genome using the ClustalW (MegAlign, DNAStar). Subfamilies are shaded in different colours.
The detailed knowledge of tomato, rice, maize and Arabidopsis Hsf functional domains and motifs enabled us to analyze similar kind of domains for the 22 Hsfs identified in the barley genome (Table 1 and Table S1). Motif analysis and sequence alignment of HvHsfs showed a highly structurally conserved DBD domain which contained 3 α-helix bundles and 4 β-strands in the N-terminal region (Figure S1) as described earlier in other plant species [37]–[39]. Hsfs function as transcriptional activators because of AHA motif characterized by aromatic (W, F, Y), large hydrophobic (L, I, V) and acidic (E, D) amino acid residues in their C-terminus (Figure S1A). However, class B and C putative AHA motifs could not be predicted in barley. Due to the absence of AHA motif in class B and class C subfamilies, they probably lack activator function. The C-terminal activation domain was found only in HsfA, suggesting that only class A Hsfs can activate autonomously.
Phylogenetic Analysis of HvsHsp Gene Family Members
We performed phylogenetic analysis using the deduced amino acid sequences of different isoforms of barley and rice sHsps. The sHsp sequences were clustered into different groups based on their subcellular localization (Figure 1B). Most of the genes from barley and rice fell into the same sub-clusters, which indicated that they are highly conserved. Using rice sHsp family classification, we identified sHsp subfamilies in barley and annotated 20 sequences. These are distributed into 10 different classes, including cytoplasmic (CI, CII, CIII, CV, CIX and CX), mitochondrial (MI), ER, plastidial (P) and peroxisomal (Px) proteins. While 6 proteins have fallen into CI, 3 into CII and one each into the remaining classes (Figure 1B). Furthermore, one sHsp gene named HvsHsp21.2 stood apart and was not clustered with any of the sHsp genes from rice. Its subcellular location is also not known. The CI gene family is generally the largest sHsp subfamily in rice and A. thaliana, it has 7 and 6 gene members respectively. The phylogenetic relationships within the cytosolic I subfamily deserves particular attention. In barley, there are two sub-clusters of cytosolic I genes (Figure 1B). One cluster comprising of HvsHsp16.9, HvsHsp16.86 and HvsHsp16.88 members, closely related to O. sativa 16.9A, 16.9B and 16.9C and the other cluster members HvsHsp17.7, HvsHsp17.5 and HvsHsp16.7 clustered to O. sativa 17.4, 17.7, 17.9A and 18.0. While all the Os16.9 sHsps are located on chromosome 1, Os17.4 is located on chromosome 3. The six sHsps of barley have been analyzed for their chromosomal localization. While sHsps 17.5, 17.7, 16.7 are localized on chromosome 4 in barley, other sHsps 16.9, 16.86 and 16.88 are located on chromosome number 3 (Table 2). The number of sHsp gene family members in the cytosol is larger compared to other cell organelles, indicating that cytosol might be the primary site of action for the function of sHsps. The pattern distribution among different classes also suggested that this small Hsp gene family is highly conserved among cereals. Sequence homology among these different classes ranged from 62 to 80%, but the functional relationships of these individual subfamilies to each other is not clear.
Sequence alignment of sHsp subfamilies revealed some interesting patterns of sequence conservation. The main characteristic feature of all the sHsps is the presence of an evolutionarily conserved central domain of 80–90 amino acids named α- crystallin domain (ACD) but have divergent N- and C-terminal extensions. N-terminal preceding the ACD region displayed variability in length and amino acid composition that contributed to a large extent for the structural diversity among subfamilies of sHsps [40]. The ACD region further can be divided into consensus I and II domains separated by a hydrophilic domain of variable length. All HvsHsps shared a consensus region I, which is highly conserved throughout the eukaryotes but the second consensus region II is unique and conserved only in plants [40] (Figure S2). The residues Pro-X (14)-Gly-Val-Leu in consensus region I are a conserved signature motif present in almost all sHsps. A similar motif Pro-X (14)-X-Val/Leu/Ile-Val/Leu/Ile also appeared in the consensus region II [41]. Outside the α-crystallin domain, a typical “I/V-X-I/V” motif in the C-terminal extension can be recognized in most sHsps except in class V (Figure S2D). Arginine residue present in ß7 strand among sHsps represented the most conserved site in the eukaryotes. This arginine in barley is located at the same position as in the case of α-crystallin structure of wheat (Figure S2). The class I cytosolic proteins have a consensus region of 15 amino acids at the N-terminus (Figure S2A), class II have 11 amino acids (Figure S2B) and class P proteins comprises 24 amino acids which do not exist in other classes (Figure S2J). The amino acid similarity between individual sHsps belonging to different groups range from 62% to 80%, whereas the similarity between individual sHsps within the groups ranged from 85% to 99%. However, there are a number of secondary structural features that are conserved across sub families irrespective of the species.
Differential Expression of sHsp and Hsf Genes During Plant Development and Drought Stress Response
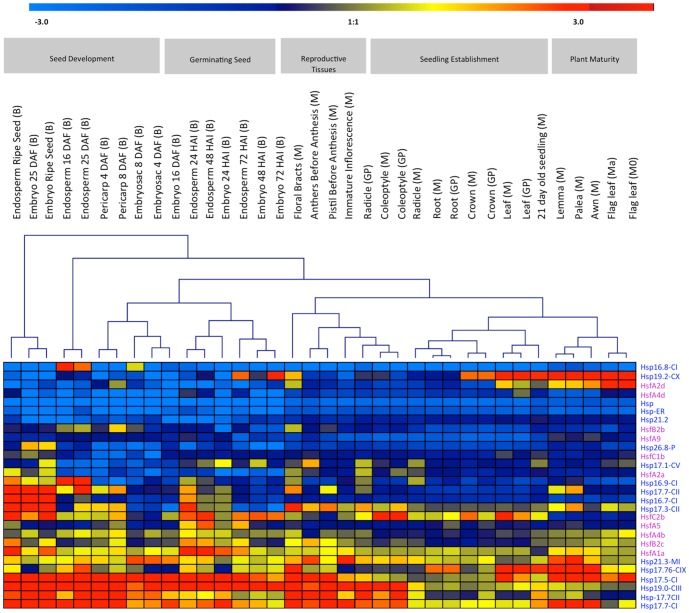
Unraveling the co-expression patterns can render important clues regarding the gene function. To understand the potential interlinking role of sHsp and Hsf genes, we monitored the expression profile (a) during plant ontogeny and (b) in response to drought stress. Microarray data revealed that sHsp17.7 (CI and CII), 17.5 (CI), and 19 (CIII) are mostly expressed in developing seed tissues like endosperm (25 DAF and ripe seed), embryo (25 DAF), pericarp (4 DAF) and in reproductive tissues such as mature floral bracts (Figure 2). The expression of these genes was however, less obvious during seedling establishment in tissues like coleoptile, root and crown. Among Hsfs, HsfA1a, HsfA2a, HsfB2c, HsfC2b and HsfA4b were more preferentially expressed in endosperm and embryo (25 DAF) than others (Figure 2), suggesting a tighter coexpression with sHsp17.7 (CI and CII), 17.5 (CI), and 19 (CIII). These specific gene family members of sHsp and Hsf genes are regulated by a defined developmental program such as embryogenesis and seed maturation events.
Figure 2. Expression profiles of sHsp and Hsf family genes during various stages of plant ontogeny analyzed by the Affymetrix 22K barley gene chip.
Horizontal rows represent expression patterns of individual gene. Trivial names of genes as well as the corresponding Affymetrix IDs are given. Vertical lines represent the developmental stages and investigated tissues. Signal intensities: red, high expression; yellow, moderate expression; blue, low expression. Represented cultivars are named as M, ‘Morex’; Mo, ‘Morocco’, Ma ‘Martin’, B, ‘Barke’; GP, ‘Golden Promise’. Quantile normalized expression values are given as log2.
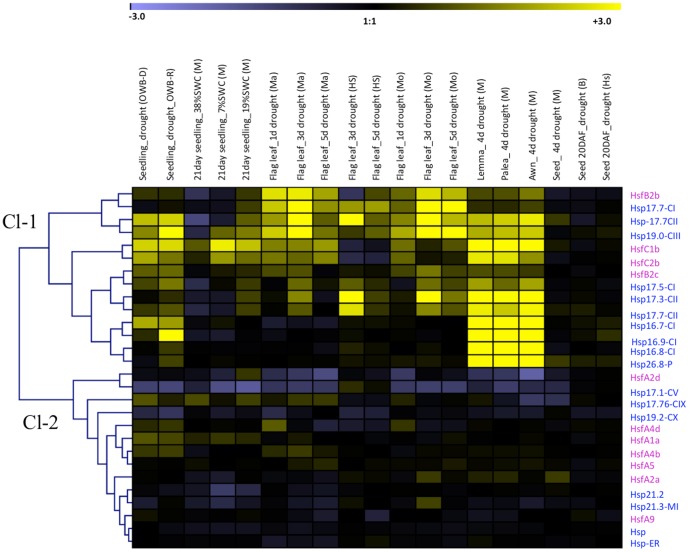
Interestingly, many sHsps in barley which are highly expressed in developing seed were also found to be preferentially upregulated under drought in vegetative tissues. The clustering process identified the genes that were highly up-regulated under drought stress in different stages of the plant development (cluster-1) and other gene sets were down regulated in majority of the stages with different time points (cluster-2) (Figure 3). Genes included in the cluster-1 are both sHsps (HvHsp17.5-CI, 17.7-CI, 17.7-CII, 19-CIII, 17.3-CII, 17.77-CII, 16.7-CI, 16.9-CI, 16.8-CI and 26.8-P) and Hsfs (HvHsfB2b, B2c, C1b and C2b). They are up-regulated under drought in all the developmental stages (early seedlings, 21-day-old seedlings, flag leaf, lemma and palea) and also abundantly expressed in a range of organs (floral bracts, pistils before anthesis, 5-day-old caryopsis and 22 DAP embryo) under normal conditions (Figs. 2 and 3). These genes perhaps have a protective chaperone role both during critical stages of plant development as well under drought. Thus, cluster 1 genes could be considered to represent as a core set of drought responsive genes. Within cluster 2, several sHsps (HvHsp17.1-CV, 17.76-CIX, 19.2-CX, 21.2, HvHsp-ER, 21.3 MI genes) and Hsf A1a, A2a, A2d, A4b, A4d, A5 and A9 members were either down-regulated or non-differential regulation in most of the developmental stages of plant during water stress or their expression was low.
Figure 3. Expression profiles of barley genes responsive to drought.
Expression ratios (drought vs control) are calculated based 3 replications. Fold change values are colour-coded: dark yellow >6 fold up-regulated, black no change, violet >6 fold down-regulated. Horizontal rows represent gene expression patterns. Vertical lines represent different stress treatments. Gene expression data refers to cvs. Brenda (B), Morex (M), Morocco (Mo), Martin (Ma), Oregon Wolf Barley-Dominant (OWB-D), Oregon Wolf Barley-Recessive (OWB-R), Hs (H. spontaneum HS584).
In Silico Analysis of sHsp and Hsf Family Promoter Regions
The regulatory cis-acting transcription factor binding sites in the sHsp and Hsf promoters were identified using PlantCARE and PLACE databases and details are depicted in Figure S3 and Figure S4. The cis-motif ABRE [42] required for ABA response is present in the promoters of all the Hsf genes except in HsfA2c, suggesting that these Hsf genes are involved in ABA mediated signal transduction. The HSE motifs responsible for the expression of Hsp genes during high-temperature stress are present in the promoter regions of many sHsps (HvHsp16.86-CI, 17.5-CI, 17.7-CI, 17.77-CI, 17.1-CV, 19.2-CX, 26.8-P), as well in HsfA class members (HsfA1a, A2a and A2c genes). Besides, within barley promoter sequences of sHsp and Hsf, many motifs which are associated with abiotic stress responses (heat shock element, HSE; drought-inducibility, MBS; low temperature responsive, LTR; anaerobic induction, ARE), hormonal responses (MeJA-responsiveness, TGACG-motif) and seed development (Skn-1 motif and GCN4 motif) are enriched (Figure S3 and Figure S4).
Among all, Hsp17.5-CI, 17.77-CII and HsfA2a have large number of different cis-motifs, related to seed development and drought stress. Thus, it is expected that many of these sHsp genes are found to be regulated both under stress as well as during seed development, perhaps due to common physiological cause of desiccation related events. Notably, many HvsHsp and Hsf genes containing Myb binding site (MBS) element in the upstream region (Figure S3 and Figure S4), which has a role in desiccation stress response, were found to be inducible by drought stress treatment in a tissue specific manner (Figure 2, 3). Seed development specific motifs such as Skn-1 and GCN4 conferring endosperm specific expression were also found in many sHsp and Hsfs. Seven Skn-1 motifs were noticed in HvsHsp17.1-CV, 4 in 17.7-CII, 5 in HsfC2a and 4 in A5 (Figure S3 and Figure S4) and displayed the highest expression in endosperm and embryo compared to other plant parts (Figure 2). In addition, cis-motifs like CCGTCC, TGA and ARE/GC related to meristem expression, salicylic acid and anaerobic inductions were observed in most of the HvsHsp and Hsf genes. Such a tissue specific expression of heat shock genes reveals important developmental role in the reproductive tissues during development.
Gene Network of HvHsfb2c Based on Genome-Wide Coexpression Data
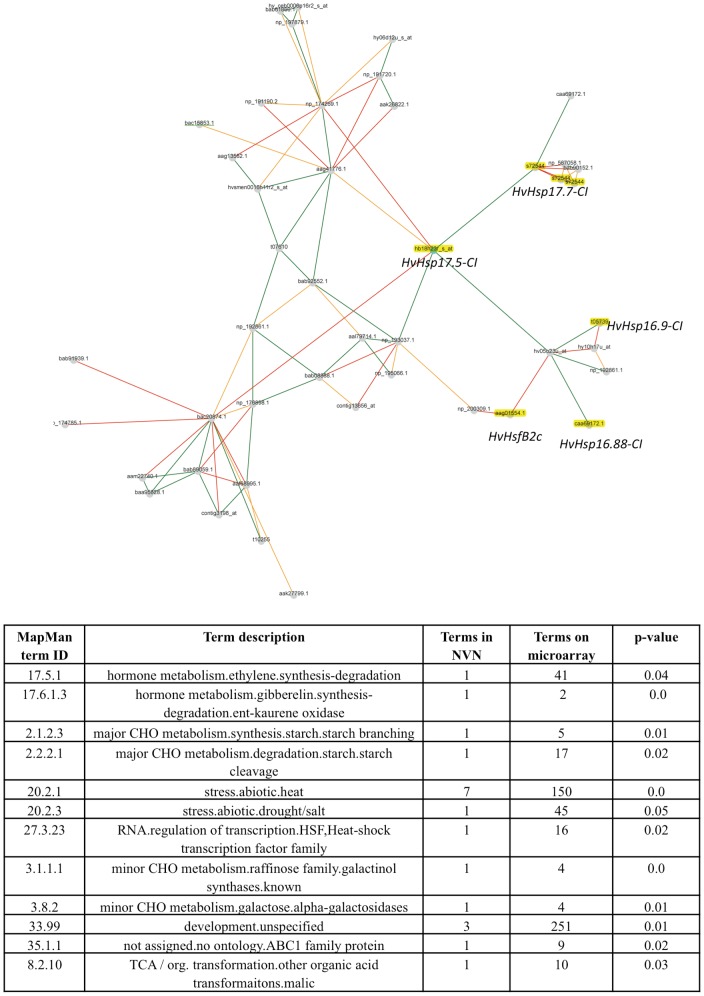
In contrary to Arabidopsis, HvHsfB2c possess a nuclear signal and was preferentially expressed in developing seeds as well prominently upregulated under drought. Our genome-wide gene expression data covering plant ontogeny suggest that HvHsfB2c is the central hub in the derived gene network (Figure 4). Network data also emphasize that HvHsfB2c is coexpressed in the central hub of several small Hsp members (HvHsp16.9-CI, HvHsp16.88-CI, HvHsp16.87-CI, HvHsp17.5-CI, and HvHsp17.7-CI), which are preferentially expressed in developing and imbibed seeds (Figure 4 and Table S2). Also, HsfB2c is coexpressed under drought stress together with several HvHsp CI, CII and CIII family members. Thus, HsfB2c transcription factor is likely to play an important role in both events (a) desiccation tolerance during seed maturation and (b) drought tolerance in possibly regulating the expression of several subclasses of HvHsp. Further, we performed cornet analysis to unravel the potential protein interaction partners and coexpressed gene regulatory networks of AT3G46230 (orthologous gene of sHsp17.5 in barley) in Arabidopsis. The predicted protein-protein interaction analysis suggests that AT3G46230 is likely to form protein complexes with several small heat shock proteins Hsp17.4, Hsp17.6 class I, Hsp17.6 class II, Hsp 17.6A and Hsp70 family (Figure 5).
Figure 4. Gene network analysis.
Gene co-expression network of HvHsfB2c/HvHsp17.5-CI, is derived from Plant Network using Heuristic Cluster Chiseling Algorithm based on genome-wide plant ontology high throughput gene expression data. Meta-network containing genes of HvHsfB2c cluster are enriched for several sHsps (highlighted in yellow colour) and also enriched Hsp class in the MapMan functional categories are represented (see table). For further details refer Table S1.
Figure 5. Integrative view of protein-protein interactions and coexpression networks of AT3G46230 (orthologous gene of sHsp17.5 in barley) derived in Arabidopsis based on CORNET correlation networks [20].
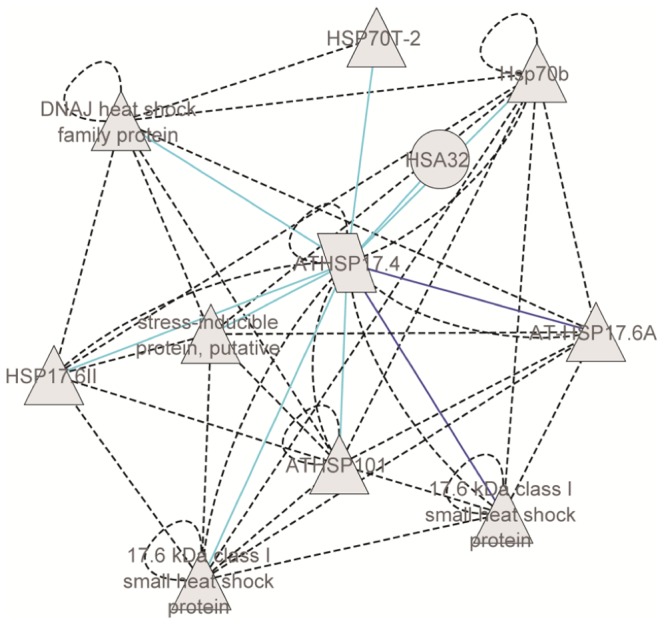
Predicted protein interactions are highlighted in dotted black colour and also autoregulatory loops are shown for several Hsp proteins. The embedded coexpression network of AT3G46230 includes several direct and indirect targets identified through microarray experiments from 256 experimental data sets generated from abiotic stress treatments. Significance of coexpression is measured by the Pearson correlation coefficient (dark blue lines represent positive correlation of 0.9; light blue lines represent positive correlation of 0.8).
Transcriptional Regulation of HvsHsp17.5-CI by HvHsfB2c under heat stress
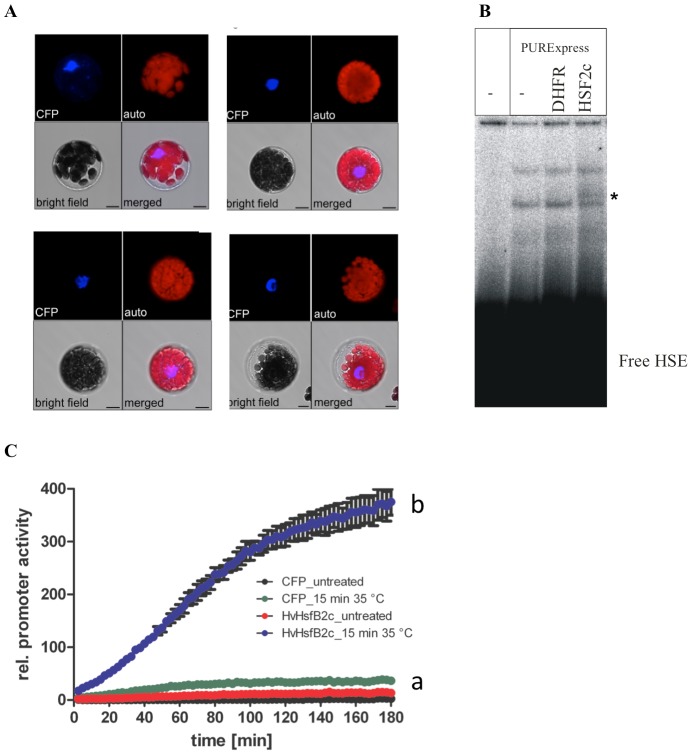
At first, the sub-cellular localisation of N- and C-terminal cyan fluorescing protein (CFP)-tagged HvHsfB2c was tested in a transient expression assay using Arabidopsis thaliana mesophyll protoplasts. The protein sequence of HvHsfB2c contains two signatures for nuclear localisation (pat4: HRRK at 131 amino acid and bipartite: RRGEKRLLCDIHRRKVV at 120 amino acid). In agreement with WoLFPSORT algorithm [43] the predominant nuclear localisation of HvHsfB2c could be confirmed for the C-terminal (Figure 6A) and the N-terminal CFP-fusion derivatives (Figure S5). As HvHsfB2c was found predominantly in the nucleus, regulation via heat stress induced translocation of the protein is very unlikely. In order to verify the in silico predicted binding of HvHsfB2c on HSE-box from the HvsHsp17.5-CI promoter, Electrophoretic Mobility Shift Assay (EMSA) was performed. For the analysis, a 44 bp DNA fragment was used containing the HSE-Box (AAATTCC) as core element. The coding region of HvHsfB2c was amplified from cDNA derived from barley cv. Golden Promise leaf RNA. As indicated in Figure 6B, the appearance of an additional band in the EMSA study refers to the DNA binding ability of in vitro translated HvHsfB2c. For analysis of the binding specificity, luciferase (LUC)-reporter assays were performed (Figure 6C). The LUC-gene was fused to a fragment of the HvsHsp17.5-CI promoter containing the region 700 bp upstream of the start ATG including the HSE-element. As indicated by the reporter gene assay no LUC-activity could be detected under control conditions. The presence of the co-transformed HvHsfB2c alone was not sufficient to increase the LUC-reporter gene activity. A strong increase of LUC-activity could be detected in the HvHsfB2c co-transformed mesophyll protoplasts under heat stress conditions (Figure 6C).
Figure 6. Transcriptional regulation of HvsHSP17.5-CI by HvHSF2c under heat stress.
A. Subcellular localisation of HvHsfB2c-CFP in transient transformed Arabidopsis thaliana mesophyll protoplasts, scale = 10 µM. B. EMSA with in vitro translated HvHsfB2c SFB2c on HSE-box from the ProHvHsp17.5-CI. Extract of PURExpress without template DNA and translated DHFR (E.coli dihydrofolate reductase) were included as negative controls. * indicates HsfB2c specific band. C. Luciferase reporter gene assay. ProHvHsp17.5::LUC was used as reporter gene in Arabidopsis thaliana Col-0 protoplast co-transformation experiments with HvHsfB2c expression vector at 35°C (blue line, a: significant difference to controls), (b) CFP-control vector at 21°C (black line), HvHsfB2c expression vector at 21°C (red line) and CFP-control vector at 35°C (green line). Results are depicted as LUC/GUS ratios. The experiment was repeated twice in triplicates with similar results. Error bars indicate the standard error of the mean of 3 replicates.
In Silico Structural Analysis and Homology Modeling of the HvsHsp17.5-CI Protein
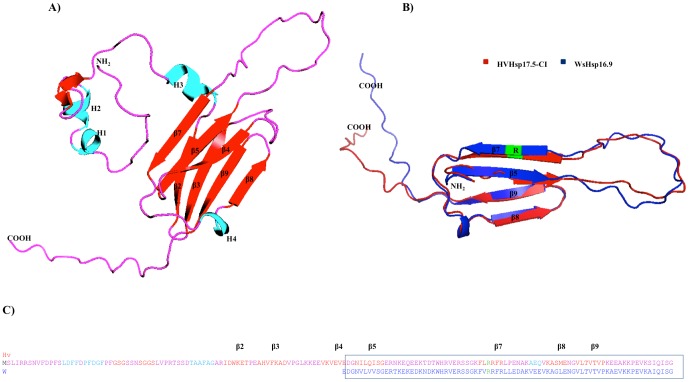
The crystal structure of TaHsp16.9 protein (PDB No: 1gmeA) was chosen in the present study as a template for homology modeling of HvsHsp17.5-CI (Figure 7A) using the server SWISS-MODEL [44]. It has been found that the two proteins have an identity of 70% at the amino acid level and a similarity of 81%. Our annotation revealed that the evolutionarily conserved α-crystallin domain contains a compact ß-strand that was responsible for dimer formation while the rest of the protein forms a conserved secondary structure despite large levels of sequence diversity [45]. There are a number of secondary structural features that are highly conserved across subfamilies, such as the ß3, ß4, ß5, ß8 and ß9 (Figure 7A). We also generated dimeric structure alignment by superimposing the monomeric structure with the α-crystallin of barley and crystallin structure of TaHsp16.9, using the program I-Tasser [30] and showed the dimer interface of both proteins is virtually identical. HvsHsp17.5 and TaHsp16.9 differed in the length of beta strands but they maintain same number in their conserved “α-crystallin domains” (Figure 7B). Considering the number of β-strands and their positions in the domain of α-crystallin, it is expected that the structure of this domain is closer to that of TaHsp16.9.
Figure 7. Structural organization of the HvsHsp17.5-CI protein.
A. The α-helix and ß-strands held between the two surface loops are shown in red and light blue colors. The N and C termini are indicated by NH2, COOH, letters respectively. B. Structural alignment of the crystallin domain of TasHsp16.9 and HvsHsp17.5-CI proteins are labeled as blue and red respectively. Highly conserved arginine (R) residue is shown in green color. C. Structural alignment of HvsHsp17.5-CI and TasHsp16.9. The conserved regions of HvsHsp17.5-CI and TasHsp16.9 are labeled with respective colors of figure A and B.
Prevention of High Temperature-Induced Thermal Inactivation of Swa I Restriction Enzyme by Recombinant HvsHsp17.5-CI
In order to obtain large quantity of highly purified HvsHsp17.5-CI protein, a heterologous system was used to overexpress recombinant HvsHsp17.5-CI protein in E. coli. The protein profiles of the E. coli BL21 (DE3) strain carrying the HvsHsp17.5-CI-pET28a construct revealed overexpression of an approximately 20-kDa recombinant protein (Figure 8A), and most of it was located in the soluble fraction of the E. coli lysate. The recombinant HvsHsp17.5-CI protein was purified to near-homogeneity (Figure 8A) from clarified E. coli lysate by Ni-NTA column chromatography. In contrast to Hsp60 and Hsp70 proteins, the chaperone activity of sHsps is ATP-independent [46]. The molecular chaperone activity of plant sHsps has been demonstrated both in vitro and in vivo [47]. Swa I restriction enzyme is thermo-labile and loses its enzymatic activity by pre-incubating at 37°C and above. We tested the protection of SwaI restriction enzyme (sensitive to high temperature) against thermal inactivation by pre-incubating 5 units of SwaI with 5 µg of either recombinant HvsHsp17.5-CI or acetylated BSA at 25–50°C for 60 min before assaying the residual activity of SwaI on plasmid DNA. The restriction endonuclease SwaI completely lost its activity after 60 min of incubation at temperatures above 35°C. The pre-incubated SwaI was tested at 25°C for 60 min using supercoiled plasmid DNA (500 ng) containing a unique SwaI recognition site. DNA digestion profiles (Figure 8B) suggested that the recombinant protein and BSA were both able to protect SwaI activity up to 30°C (Figure 8B, lanes 6, 7). At temperatures above 35°C, BSA failed to protect against thermal inactivation of SwaI (Figure 8B, lanes 7, 9, 11, 13), whereas HvsHsp17.5-CI provided significant protection (Figure 8B, lanes 6, 8, 10). Above 40°C, HvsHsp17.5-CI provided only marginal protection. However, it was ineffective against thermal inactivation of SwaI at or above 45°C (Figure 8B, lanes 12, 14). These results suggest that the recombinant HvsHsp17.5 keeps the Swa I restriction enzyme in folding competent state at higher temperature and the N-terminus hexahistidin tag is not interfering in this activity. The amount of the residual activity of Swa I restriction enzyme can be quantified by measuring the amount of DNA restricted.
Figure 8. Expression, purification and chaperone activity of recombinant HvsHsp17.5-CI.
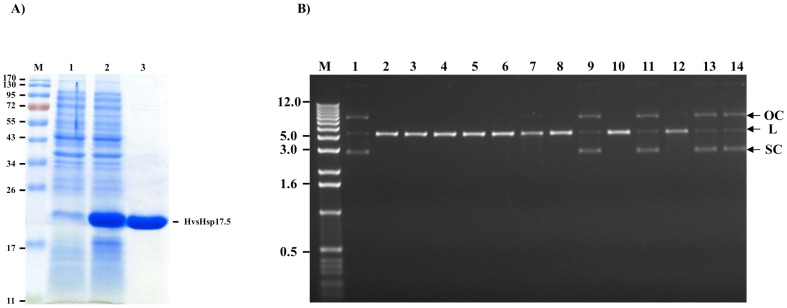
A) Expression of recombinant HvsHsp17.5-CI in E. coli. Lane M, molecular weight marker; lane 1, uninduced; lane 2, induced; lane 3, purified recombinant HvsHsp17.5-CI protein. Figures on the left indicate molecular weight in kDa. B) Prevention of thermal inactivation of SwaI restriction enzyme by recombinant HvsHsp17.5-CI. The SwaI restriction enzyme was preincubated at 25, 30, 35, 40, 45 or 50°C in the presence of either BSA or recombinant HvsHsp17.5-CI for 60 min. Residual activity of SwaI was determined by incubation with 300 ng plasmid at 25°C for 60 min, followed by electrophoresis on a 1% agarose gel. Lane M, 1-Kb DNA ladder; lane 1, plasmid DNA control (without SwaI digestion); lane 2, plasmid DNA digested with the SwaI restriction enzyme; lanes 3, 5, 7, 9, 11, and 13, plasmid DNA digested with SwaI after preincubation at 25, 30, 35, 40, 45 and 50°C, respectively, in the presence of BSA; lanes 4, 6, 8, 10, 12, and 14, plasmid DNA digested with SwaI after preincubation at 25, 30, 35, 40, 45 and 50°C, respectively, in the presence of recombinant HvsHsp17.5-CI. SC, supercoiled plasmid; OC, open circular plasmid; L, linear plasmid. The numbers on the left represent the DNA markers in kb.
Discussion
The structure and function of the sHsp and Hsf family genes have been widely addressed in model plants like Arabidopsis, maize and rice [8], [13], [37]–[39], [48]. Most of the sHsps respond to a wide range of environmental stresses like heat, cold, drought, high light, salt, UV, oxidative stress and plant-pathogen interaction [5] and their concentration may even go up to 1% of the total proteins under high temperature stress. Proteins that are destabilized during cellular stresses are reactivated in the presence of ATP-dependent chaperones. We applied various bioinformatics approaches to analyze the phylogenetic relationship, conserved domains, localization prediction of proteins, in silico promoter analysis, transcript profiling and coexpression gene network analysis in barley to unravel its expression divergence and functional relevance of sHsps and Hsfs gene family members. Specialized sHsp and Hsf members which are preferentially expressed in specific organs/developmental stages (developing and imbibed seeds) and also influenced by drought stress conditions seem to suggest a conserved function of desiccation tolerance both during seed maturation events as well as during drought prone response in vegetative tissues.
The presence of HSE cis element is often correlated with the expression of respective sHsp and Hsf genes under heat stress as shown in the microarray analysis of Arabidopsis, rice and maize [5], [38], [39], [49]. The cis-motif ABRE [42] required for ABA response is present in the promoters of all the Hsf genes except in HsfA2c, suggesting that these Hsf genes are involved in ABA mediated signal transduction. The role of ABA in seed development [50], [51] and drought responses [52] is well known. Thus, ABA may be another important molecule in the regulation HSFs during seed development or drought stress. Within HsfA class, developmental expression of HsfA9 in Arabidopsis is regulated by the seed specific transcription factor ABSCISIC ACID-INSENSITIVE3 (ABI3). ABI3 knock out lines lack HsfA9 transcript and also seed abundant heat stress proteins like Hsp 17.4-CI, Hsp 17.7-CII and Hsp101 [15]. They concluded that HsfA9 acts as a potent activator for the seed specific or developmental specific expression of Hsp genes in Arabidopsis. Recently, functional analysis of rice heat shock factor binding proteins OsHSBP1 and OsHSBP2 revealed their involvement in heat shock response [53]. Both these genes have been found important for seed development as their knockout lines are associated with significant seed abortion. Further research is required to clarify the expressions of sHsps and Hsf genes and their interplay during specific sexual processes.
Among Hsfs, HsfA1a, HsfA2a, HsfB2c, HsfC2b and HsfA4b were more preferentially expressed in endosperm and embryo (25 DAF) than others (Figure 2), suggesting a tighter coexpression with sHsp17.7 (CI and CII), 17.5 (CI), and 19 (CIII). These specific gene family members of sHsp and Hsf genes are regulated by a defined developmental program such as embryogenesis and seed maturation events, a situation resembling with Arabidopsis and sunflower [15], [54]. Within this category, HsfB2c is identified as a putative central regulator represented in the vicinity of gene network of HvHsp CI, CII and CIII family members, and also the respective promoters are enriched with HSE cis element. Thus, HsfB2c is likely to mediate expression of these sHsp members. The analysis also identified the importance of HsfB2c that has not been previously implicated in plant stress responses and development and therefore HsfB2c might be one of the primary regulators of the HSR in barley. To prove the in vivo relevance of the in silico identified gene network, the molecular interaction of HvHsfB2c transcription factor on HSE-box cis element within the HvsHsp17.5-CI promoter was analysed. These results suggest that HvHsfB2c is responsible for the heat inducible transcriptional activation of HvsHsp17.5-CI. This transcriptional activation is mostly achieved by binding of HvHsfB2c to the HSE-box under heat stress conditions. It is rather surprising to note that typical feature of class A Hsfs which are known to possess transcriptional activator domain [55] remains unregulated under drought, while HsfB (HvHsfB2b, HvHsfB2c) and HsfC class (HvHsfC1b, HvHsfC2b) transcription factors were upregulated under drought in barley (Figure 3). While class B-Hsfs differ with class A not only in the lack of transcriptional activator AHA-motif, but also differ within oligomerization domain [56]. In this context, it is interesting to note the first emerging evidence we have shown in barley that HvHsfB2c as a central regulator mediates transcriptional response of HvsHsp17.5-CI gene, which is preferentially regulated during desiccation responses. Moreover, out of 5 reported class B-Hsfs in Arabidopsis, B3 and B4 are expressed at low level and HsfB1, HsfB2a and HsfB2b are significantly increased upon heat stress treatment [57], [58]. Similarly, the orthologue of HsfB2C in Arabidopsis, AtHsf7 (AT4G11660) is expressed in developing seeds and found to be induced under stress. Also, the coexpressed genes depicted in the gene network of AtHsf7 show embryo arrest in mutants and defective in thermo tolerance. These results suggest that HvHsfB2c in barley and AtHsf7 are not only highly conserved in sequence and expression specificity between monocot and dicot lineages, but also seems to possess conserved function in seed development and stress tolerance. While none of the HsfB class members are characterized in monocots, studies from Ikeda et al. [59] indicate that HsfB factors suppress the general heat shock response under non-heat-stress conditions and in attenuating period they appear to be necessary for the expression of heat stress-inducible heat shock protein genes under heat stress conditions, which is necessary for acquired thermotolerance. The authors also mentioned that the heat stress response is finely regulated by activation and repression activities of Hsfs in Arabidopsis. Also, recent studies suggest that Arabidopsis HsfB4 possess regulatory function in root development [60].
Based on coexpression network, it appears that HvsHsp 17.5 plays a vital role in barley seed development (Figure 4). Also, a strong interaction of a putative serine/threonine protein kinase, a calcyclin binding protein, ATP dependent RNA helicase and a metallothionein protein were observed in the gene network generated with that of sHsp17.5 (Figure 4 and Table S2). Serine/theronine protein kinase and calcyclin as a calcium binding protein acts as second messenger associated with the signal transduction. Metallothionins are not only associated with drought stress, but also involved in regulating zinc ion mobilization and metal homeostasis in late embryo developmental stages [61]. Cornet analysis emphasized the potential protein interaction partners as well the coexpressed gene regulatory networks of AT3G46230 (orthologous gene of sHsp17.5 in barley) in Arabidopsis. These results suggest that AT3G46230 is likely to form protein complexes with several small heat shock proteins Hsp17.4, Hsp17.6 class I, Hsp17.6 class II, Hsp 17.6A and Hsp70 family (Figure 5). The presence of sHsps in different cell organelles indicate that potentially these genes might act as chaperones in protecting the various cellular compartments under stress as reported in case of mitochondrial Hsp22 of Drosophila [62].
Our results suggest that the 17.5CI sHsp that we isolated from barley has chaperone activity and is able to protect the growth of E. coli and the activity of a thermolabile chaperone enzyme under heat stress. Based on our results, it is reasonable to speculate that the recombinant HvsHsp17.5-CI protein might function as a molecular chaperone that conferred a moderate protective function against stress-induced protein damage in bacterial cells, as mostly these class of proteins might be involved in stabilization but not necessarily in refolding of denatured proteins under stress [3], [40]. This situation differs from high molecular weight Hsps which could refold denatured proteins under stress but requires ATP [33]. Cytosolic Hsp17.7 and Hsp17.3 of tomato have been shown to act as molecular chaperones in vivo [63] and also an overexpression of AtHSP17.6A lead to enhance osmotolerance [64]. Chauhan et al. [49] showed that a chloroplastic TasHSP26 is involved in seed maturation and germination and its heterologous expression results in tolerance to heat stress in Arabidopsis. Thus, it appears that some of these plant specific sHsps act not only as molecular chaperones under stress, but also likely to possess similar function in developmental programs especially in the seeds.
Supporting Information
Multiple sequence alignment of the Hsf protein family in barley with corresponding members of rice. Different classes of the HSF numbers correspond to the order of the alignment. The multiple alignment results clearly show the highly conserved DBD domains among all the Hsf genes which are marked with dotted boxes. The secondary structure elements of DBD (α1-β1- β2-a2-a3- β3-β4) are shown above the alignment. These were predicted based on the PSIPRED protein structure prediction server. The scheme at the top depicts the locations and boundaries of the HR-A core, insert and HR-B regions within the HR-A/B regions which are marked with thick boxes. Positions of the other identified motifs nuclear localization signals (NLS) are highlighted with yellow colour, nuclear export signal (NES) highlighted in yellow colour with underline and activator (AHA) motif sequences are shown in the red colour and each motif name is mentioned above the alignment. Alignment was performed by using ClustalW (DNAstar) program.
(PDF)
Multiple alignment of different sub-classes of Hv sHsps. The conserved α- crystallin domain was labeled with dotted box. The defined consensus regions I and II are marked with underline below the sequences. Highly conserved and semi-conserved regions are shown in “*” and “.”, respectively. Small Hsp region specific to respective subclasses was labeled with thick boxes. The chloroplast localized proteins have transit sequences that are specific for organelle and is labeled with red colour. The conserved Arg is displayed in red colour in the β7 strand. The secondary structure assignments for all classes of HvsHsps were labeled above the sequences. The predicted β-strands depicted by thick lines above the alignment are based on their position in known secondary structure of Hsp16.9 from T. aestivum (van Montfort et al. 2001). The IXI/V motif in the C-terminal extension is shown in green. The SXXFD motif and interacting residues in the conserved alpha crystallin domain are in pink colour. Alignment was performed by using ClustalW (DNAstar) program.
(PDF)
Position of putative cis -elements present in the promoter regions of barley Hsf genes. The analysis was performed using PlantCARE and PLACE databases. The “rectangle mark” shows the relative position of the different motifs.
(PDF)
Position of putative cis -elements present in the promoter regions of barley sHsp genes. The analysis was performed using PlantCARE and PLACE databases. The “rectangle mark” shows the relative position of the different motifs.
(PDF)
Subcellular localisation of HvHsfB2c in transient transformed Arabidopsis thaliana mesophyll protoplasts for the the N-terminal CFP-fusion derivatives. scale = 10 µM. A, N-terminal CFP-fusion; B, C-terminal CFP fusion.
(PDF)
Barley sHsp and Hsf Orthologous genes in Arabidopsis .
(PDF)
Additional file 5: Detailed list of functional annotations of genes represented in the network of HvHsfB2c/HvHsp17.5-CI .
(PDF)
Funding Statement
The authors work in the NS laboratory has been supported by grants from the IB-BMBF (IND 09/526) and from the Ministry of Education, SaxonyAnhalt. PSR acknowledges the Leibniz-DAAD post doctoral fellowship award (Number: A/11/94309) from Germany Academic Exchange programme (DAAD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9: 244–252. [DOI] [PubMed] [Google Scholar]
- 2. van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E (2001) Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol 8: 1025–1030. [DOI] [PubMed] [Google Scholar]
- 3. Basha E, Jones C, Wysocki V, Vierling E (2010) Mechanistic differences between two conserved classes of small heat shock proteins found in the plant cytosol. J Biol Chem 285: 11489–11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Puigderrajols P, Jofre A, Mir G, Pla M, Verdaguer D, et al. (2002) Developmentally and stress- induced small heat shock proteins in cork oak somatic embryos. J Exp Bot 53: 1445–1452. [PubMed] [Google Scholar]
- 5. Sarkar NK, Kim YK, Grover A (2009) Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wehmeyer N, Hernandez LD, Finkelstein RR, Vierling E (1996) Synthesis of small heat-shock proteins is part of the developmental program of late seed maturation. Plant Physiol 112: 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wehmeyer N, Vierling E (2000) The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol 122: 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scharf KD, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta 1819: 104–119. [DOI] [PubMed] [Google Scholar]
- 9. Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Doring P (2004) Characterization of C- terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39: 98–112. [DOI] [PubMed] [Google Scholar]
- 10. Prandl R, Schoffl F (1996) Heat shock elements are involved in heat shock promoter activation during tobacco seed maturation. Plant Mol Biol 31: 157–162. [DOI] [PubMed] [Google Scholar]
- 11. Rojas A, Almoguera C, Jordano J (1999) Transcriptional activation of a heat shock gene promoter in sunflower embryos: synergism between ABI3 and heat shock factors. Plant J 20: 601–610. [DOI] [PubMed] [Google Scholar]
- 12. Long TA, Brady SM, Benfey PN (2008) Systems approaches to identifying gene regulatory networks in plants. Annu Rev Cell Dev Biol 24: 81–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma S, Bohnert HJ (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hong SW, Vierling E (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27: 25–35. [DOI] [PubMed] [Google Scholar]
- 15. Kotak S, Vierling E, Baumlein H, von Koskull-Doring P (2007) A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19: 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsumoto T, Tanaka T, Sakai H, Amano N, Kanamori H, et al. (2011) Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol 156: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, et al. (2008) Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146: 1738–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, et al. (2004) A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol 134: 960–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, et al. (2011) PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23: 895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Bodt S, Hollunder J, Nelissen H, Meulemeester N, Inze D (2012) CORNET 2.0: integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol 195: 707–720. [DOI] [PubMed] [Google Scholar]
- 21. Mayer KF, Martis M, Hedley PE, Simkova H, Liu H, et al. (2011) Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23: 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, et al. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuhlmann M, Horvay K, Strathmann A, Heinekamp T, Fischer U, et al. (2003) The alpha-helical D1 domain of the tobacco bZIP transcription factor BZI-1 interacts with the ankyrin-repeat protein ANK1 and is important for BZI-1 function, both in auxin signaling and pathogen response. J Biol Chem 278: 8786–8794. [DOI] [PubMed] [Google Scholar]
- 25. Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- 26. Sun CW, Callis J (1997) Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. Plant J 11: 1017–1027. [DOI] [PubMed] [Google Scholar]
- 27. Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, et al. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41. [DOI] [PubMed] [Google Scholar]
- 28. Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D (2011) Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage- associated molecular patterns. Plant J 68: 100–113. [DOI] [PubMed] [Google Scholar]
- 29. Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- 30. Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reddy PS, Reddy GM, Pandey P, Chandrasekhar K, Reddy MK (2012) Cloning and molecular characterization of a gene encoding late embryogenesis abundant protein from Pennisetum glaucum: protection against abiotic stresses. Mol Biol Rep 39: 7163–7174. [DOI] [PubMed] [Google Scholar]
- 33. Reddy PS, Thirulogachandar V, Vaishnavi CS, Aakrati A, Sopory SK, et al. (2011) Molecular characterization and expression of a gene encoding cytosolic Hsp90 from Pennisetum glaucum and its role in abiotic stress adaptation. Gene 474: 29–38. [DOI] [PubMed] [Google Scholar]
- 34. Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- 35. Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–36. [DOI] [PubMed] [Google Scholar]
- 36. Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590. [DOI] [PubMed] [Google Scholar]
- 37. Chauhan H, Khurana N, Agarwal P, Khurana P (2011) Heat shock factors in rice (Oryza sativa L.): genome-wide expression analysis during reproductive development and abiotic stress. Mol Genet Genomics 286: 171–187. [DOI] [PubMed] [Google Scholar]
- 38. Lin YX, Jiang HY, Chu ZX, Tang XL, Zhu SW, et al. (2011) Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A (2009) Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem 47: 785–795. [DOI] [PubMed] [Google Scholar]
- 40. Waters ER (2013) The evolution, function, structure, and expression of the plant sHSPs. J Exp Bot 64: 391–403. [DOI] [PubMed] [Google Scholar]
- 41. Caspers GJ, Leunissen JA, de Jong WW (1995) The expanding small heat-shock protein family, and structure predictions of the conserved “alpha-crystallin domain”. J Mol Evol 40: 238–248. [DOI] [PubMed] [Google Scholar]
- 42. Straub PF, Shen Q, Ho TD (1994) Structure and promoter analysis of an ABA- and stress- regulated barley gene, HVA1. Plant Mol Biol 26: 617–630. [DOI] [PubMed] [Google Scholar]
- 43. Horton P, Park KJ, Obayashi T, Fujita N, Harada H, et al. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37: D387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stamler R, Kappe G, Boelens W, Slingsby C (2005) Wrapping the alpha-crystallin domain fold in a chaperone assembly. J Mol Biol 353: 68–79. [DOI] [PubMed] [Google Scholar]
- 46. Lee GJ, Pokala N, Vierling E (1995) Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem 270: 10432–10438. [DOI] [PubMed] [Google Scholar]
- 47. Siddique M, Port M, Tripp J, Weber C, Zielinski D, et al. (2003) Tomato heat stress protein Hsp16.1-CIII represents a member of a new class of nucleocytoplasmic small heat stress proteins in plants. Cell Stress Chaperones 8: 381–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Waters ER, Aevermann BD, Sanders-Reed Z (2008) Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperones 13: 127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chauhan H, Khurana N, Nijhavan A, Khurana JP, Khurana P (2012) The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ 35: 1912–1931. [DOI] [PubMed] [Google Scholar]
- 50. Sreenivasulu N, Radchuk V, Strickert M, Miersch O, Weschke W, et al. (2006) Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA- regulated maturation in developing barley seeds. Plant J 47: 310–327. [DOI] [PubMed] [Google Scholar]
- 51. Sreenivasulu N, Wobus U (2013) Seed-development programs: a systems biology-based comparison between dicots and monocots. Annual review of plant biology 64: 189–217. [DOI] [PubMed] [Google Scholar]
- 52. Sreenivasulu N, Harshavardhan VT, Govind G, Seiler C, Kohli A (2012) Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506: 265–273. [DOI] [PubMed] [Google Scholar]
- 53. Rana RM, Dong S, Tang H, Ahmad F, Zhang H (2012) Functional analysis of OsHSBP1 and OsHSBP2 revealed their involvement in the heat shock response in rice (Oryza sativa L.). J Exp Bot 63: 6003–6016. [DOI] [PubMed] [Google Scholar]
- 54. Almoguera C, Rojas A, Diaz-Martin J, Prieto-Dapena P, Carranco R, et al. (2002) A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. J Biol Chem 277: 43866–43872. [DOI] [PubMed] [Google Scholar]
- 55. Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, et al. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. von Koskull-Doring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457. [DOI] [PubMed] [Google Scholar]
- 57. Busch W, Wunderlich M, Schoffl F (2005) Identification of novel heat shock factor- dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J 41: 1–14. [DOI] [PubMed] [Google Scholar]
- 58. Lohmann C, Eggers-Schumacher G, Wunderlich M, Schoffl F (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics 271: 11–21. [DOI] [PubMed] [Google Scholar]
- 59. Ikeda M, Mitsuda N, Ohme-Takagi M (2011) Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol 157: 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Begum T, Reuter R, Schoffl F (2013) Overexpression of AtHsfB4 induces specific effects on root development of Arabidopsis. Mech Dev 130: 54–60. [DOI] [PubMed] [Google Scholar]
- 61. Ren Y, Liu Y, Chen H, Li G, Zhang X, et al. (2012) Type 4 metallothionein genes are involved in regulating Zn ion accumulation in late embryo and in controlling early seedling growth in Arabidopsis. Plant Cell Environ 35: 770–789. [DOI] [PubMed] [Google Scholar]
- 62. Morrow G, Inaguma Y, Kato K, Tanguay RM (2000) The small heat shock protein Hsp22 of Drosophila melanogaster is a mitochondrial protein displaying oligomeric organization. J Biol Chem 275: 31204–31210. [DOI] [PubMed] [Google Scholar]
- 63. Low D, Brandle K, Nover L, Forreiter C (2000) Cytosolic heat-stress proteins Hsp17.7 class I and Hsp17.3 class II of tomato act as molecular chaperones in vivo. Planta 211: 575–582. [DOI] [PubMed] [Google Scholar]
- 64. Sun W, Bernard C, van de Cotte B, Van Montagu M, Verbruggen N (2001) At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J 27: 407–415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of the Hsf protein family in barley with corresponding members of rice. Different classes of the HSF numbers correspond to the order of the alignment. The multiple alignment results clearly show the highly conserved DBD domains among all the Hsf genes which are marked with dotted boxes. The secondary structure elements of DBD (α1-β1- β2-a2-a3- β3-β4) are shown above the alignment. These were predicted based on the PSIPRED protein structure prediction server. The scheme at the top depicts the locations and boundaries of the HR-A core, insert and HR-B regions within the HR-A/B regions which are marked with thick boxes. Positions of the other identified motifs nuclear localization signals (NLS) are highlighted with yellow colour, nuclear export signal (NES) highlighted in yellow colour with underline and activator (AHA) motif sequences are shown in the red colour and each motif name is mentioned above the alignment. Alignment was performed by using ClustalW (DNAstar) program.
(PDF)
Multiple alignment of different sub-classes of Hv sHsps. The conserved α- crystallin domain was labeled with dotted box. The defined consensus regions I and II are marked with underline below the sequences. Highly conserved and semi-conserved regions are shown in “*” and “.”, respectively. Small Hsp region specific to respective subclasses was labeled with thick boxes. The chloroplast localized proteins have transit sequences that are specific for organelle and is labeled with red colour. The conserved Arg is displayed in red colour in the β7 strand. The secondary structure assignments for all classes of HvsHsps were labeled above the sequences. The predicted β-strands depicted by thick lines above the alignment are based on their position in known secondary structure of Hsp16.9 from T. aestivum (van Montfort et al. 2001). The IXI/V motif in the C-terminal extension is shown in green. The SXXFD motif and interacting residues in the conserved alpha crystallin domain are in pink colour. Alignment was performed by using ClustalW (DNAstar) program.
(PDF)
Position of putative cis -elements present in the promoter regions of barley Hsf genes. The analysis was performed using PlantCARE and PLACE databases. The “rectangle mark” shows the relative position of the different motifs.
(PDF)
Position of putative cis -elements present in the promoter regions of barley sHsp genes. The analysis was performed using PlantCARE and PLACE databases. The “rectangle mark” shows the relative position of the different motifs.
(PDF)
Subcellular localisation of HvHsfB2c in transient transformed Arabidopsis thaliana mesophyll protoplasts for the the N-terminal CFP-fusion derivatives. scale = 10 µM. A, N-terminal CFP-fusion; B, C-terminal CFP fusion.
(PDF)
Barley sHsp and Hsf Orthologous genes in Arabidopsis .
(PDF)
Additional file 5: Detailed list of functional annotations of genes represented in the network of HvHsfB2c/HvHsp17.5-CI .
(PDF)