Abstract
Purpose
The aim of this study was to assess the uncertainty in estimation of MR tracer kinetic parameters and water exchange rates in T1-weighted dynamic contrast enhanced (DCE)-MRI.
Methods
Simulated DCE-MRI data were used to assess four kinetic models; general kinetic model with a vascular compartment (GKM2), GKM2 combined with the 3S2X model (SSM2), adiabatic approximation of the tissue homogeneity model (ATH), and ATH combined 3S2X model (ATHX).
Results
In GKM2 and SSM2, increase in transfer constant (Ktrans) led to underestimation of vascular volume fraction (vb), and increase in vb led to overestimation of Ktrans. Such coupling between Ktrans and vb was not observed in ATH and ATHX. The precision of estimated intracellular water lifetime (τi) was substantially improved in both SSM2 and ATHX when Ktrans > 0.3 min−1. Ktrans and vb from ATHX model had significantly smaller errors than those from ATH model (p<0.05).
Conclusion
The results of this study demonstrated the feasibility of measuring τi from DCE-MRI data albeit low precision. While the inclusion of the water exchange model improved the accuracy of Ktrans, vb, and the interstitial volume fraction estimation (ve), it lowered the precision of other kinetic model parameters within the conditions investigated in this study.
Keywords: DCE-MRI, tracer kinetics, pharmacokinetic model, water exchange, numerical simulation, uncertainty, adiabatic approximation of the tissue homogeneity model
INTRODUCTION
T1-weighted dynamic contrast enhanced (DCE) MRI data have been used to estimate physiologically relevant MR tracer kinetic parameters, such as transfer constant (Ktrans), vascular volume fraction (vb), and extravascular extracellular volume fraction (ve) (1–3). It has been demonstrated that such kinetic parameters can be used for diagnosis of cancer (4,5) as well as for prediction (6,7) and monitoring (8) of treatment response. However, accurate estimation of the kinetic parameters from DCE-MRI data still remains a challenging problem, particularly when additional model parameters, such as inter-compartmental water exchange rates (9–11), are included.
DCE-MRI data are usually acquired with a short TR where a repetitively pulsed system rapidly approaches a steady state described by Ernest and Anderson (12). For a system of multiple compartments with chemical exchange, Spencer and Fishbein (13) presented an analytical solution for the steady state magnetization of each compartment and demonstrated that the apparent T1 values of multiple compartments deviate from the original T1 values (with no exchange) as TR increases. When TR approaches zero, the apparent T1 values approach the original T1 values. On the other hand, the smaller the flip angle (α) is, the larger the deviation of apparent T1 from the original T1 is. These observations that Spencer and Fishbein (13) reported are closely related to T1-weighted DCE-MRI scans which are usually performed with a short TR (~5 ms) and a small α (~10°) for fast 3D imaging, leading to a question whether the water exchange effect should be included for accurate and robust estimation of the kinetic parameters.
Water exchange effect has also been observed in DCE-MRI studies using inversion recovery prepared 2D gradient echo sequences (14). Landis et al (15) measured the T1 of rat skeletal muscle infused with an MRI contrast agent (CA), gadolinium diethylenetriamine pentaacetic acid (Gd-DTPA), which typically stays in the interstitial space and does not enter the intracellular space. They found that the relationship between the measured T1 and CA concentration was not linear and was better described by a non-linear model that included water exchange between tissue compartments. Several other studies in tumor imaging reported that the influence of water exchange needed to be included based on goodness of fitting of the model to the DCE-MRI data (16–18). However, it has not been shown whether such better model fitting can lead to more accurate and precise estimation of the kinetic model parameters.
Hence, the present study was conducted to assess the uncertainty in kinetic model parameters estimated using two representative MRI CA kinetic models; extended general kinetic model (GKM2) (19) and extended shutter speed model (SSM2) (20). In addition, we also conducted a similar study to investigate the effect of water exchange in adiabatic approximation to the tissue homogeneity model (ATH) (21) which assumes the capillary bed as a plug-flow system and the interstitial space as one compartment, and the ATH model combined with the 3S2X water exchange model (ATHX). Since there is no means to independently and accurately measure the same kinetic parameters measured by DCE-MRI, we used realistic simulation data in this study.
METHODS
Data simulation
Realistic DCE-MRI data were generated in two steps; calculation of the CA concentrations in tissue compartments with a bolus injection and conversion to corresponding MRI signal curves. The CA concentration curve of a single voxel was simulated using a single blood tissue exchange unit provided in the multiple path, multiple tracer, indicator dilution 4 region (MMID4) model (National Simulation Resources, University of Washington) which models capillaries as axially distributed blood-tissue exchange units (22). It was assumed that the voxel of interest for simulation did not contain any large vessel such that optional large vessels in the MMID4 model were not included in the simulation. The blood-tissue exchange unit has four regions: intravascular, endothelial, interstitial, and parenchymal intracellular spaces. Volume fraction of the endothelium was assumed to be negligible in the current study. The capillary length was set to 0.1 cm; the number of segments was 31; the axial diffusion coefficients for plasma and interstitial fluid region were 1e-5 cm2/s. Volume fractions of the intravascular, interstitial, and intracellular spaces were noted as vb, ve and vi, respectively. It was assumed that the three compartments were connected in a linear form such that there was no direct exchange of CA molecules between the intravascular and intracellular compartments. It was also assumed that the permeability of cellular membrane to CA was zero.
The equilibrium 1H2O magnetizations of the intravascular, interstitial and intracellular compartments (Mb0, Me0 and Mi0) were assumed to be proportional to the volume fractions of the three compartments. In this linear form of three tissue compartments, we also assumed that there was no direct exchange of the water molecules between the intravascular and intracellular compartments. The magnetizations of three compartments (Mb, Me and Mi) were described using the three-site-two-exchange (3S2X) model proposed by Spencer and Fishbein (13):
| (1) |
| (2) |
| (3) |
where R1b, R1e and R1i are the longitudinal relaxation rates in three compartments, kAB denotes the rate constant from compartment A to compartment B. The four rate constants used in Eqs. (1), (2) and (3) can also be expressed in terms of water lifetimes (τb, τe and τi) and volume fractions of the three compartments by keeping mass balance: kbe = τb−1; keb = τe−1−(vi/ve)τi−1; kei = τe−1−(vb/ve)τb−1; kie = τi−1. We assumed that the water fraction in each compartment was 1 for simplicity in this study.
MRI signal was calculated for the case of using a spoiled gradient recalled echo sequence. The transverse relaxation rate was assumed not affected within the CA concentrations considered in this study. The longitudinal relaxation rates of the three compartments prior to contrast injection (R1b0, R1e0 and R1i0) were assumed to be 0.67 s−1. The CA relaxivity was held constant as 3.8 mM−1s−1 in all three compartments. The relationship between relaxation rate R1(t) and CA concentration C(t) is assumed linear within each compartment; R1j(t)=r1jCj(t)+R1j0 where j=b, e and i. Then, the observed magnetization of the 3S2X system in Eqs. (1), (2) and (3) for the given TR and α was calculated using the Ernst-Anderson relationship (13,20). Matrix exponential was calculated using a function provided in Matlab (MathWorks, Natick, MA), expm, which was implemented based on a scaling and squaring algorithm with a Pade approximation method (23). Sum of the magnetizations in all three compartments was used as the corresponding MRI signal.
Arterial input function (AIF) for data simulation with the MMID4 was obtained from a DCE-MRI data set measured in one of the external iliac arteries of a patient from our previous study (24) (dots in Figure 1(a)). A bi-exponential clearance function convoluted with a box-shaped function (25) was fitted to the measurement data, in order to generate a noise-free, realistic AIF model that can provide data at any given temporal resolution. The AIF model, Cb(t) as shown with the solid line in Figure 1(a), was defined as
Figure 1.
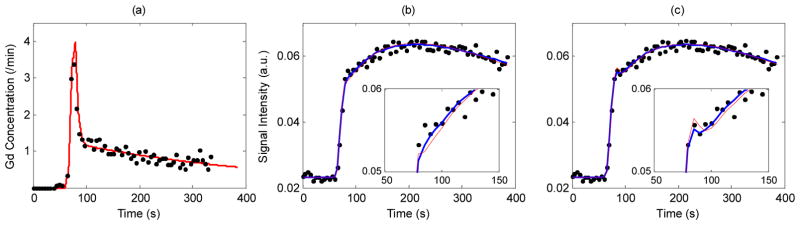
(a) Measured AIF from prostate DCE-MRI data (black dots) and the box-exponential clearance function fit (red solid line) used for simulation. (b) Representative MRI data generated using the MMID4 and the 3S2X model (Ktrans=0.25/min with PS=0.28/min and Fp=1.2/min, vb=0.06, ve=0.45, τb=0.05s, τi=0.2s and Tc=3s) with noise level SNR=20 (black dots) and the model fits using SSM2 (blue thick line) and GKM2 (red thin line). (c) Similar to (b) with ATHX (blue thick line) and ATH (red thin line).
| (4) |
with B(t) = 1 for 8.6 s<t<9.6 s and B(t) = 0 elsewhere, and f(t) = Aexp(−Bt)+Cexp(−Dt) with A = 2.9125, B = 0.4070, C = 0.6447 and D = 0.0028. * is a convolution operator. This AIF model was used to generate all simulation data in this study.
The simulation of DCE-MRI data was initially conducted with a temporal resolution of 0.2 s. Subsequently, the time intensity curve generated from the simulation was re-sampled to a lower resolution of 5 s that is more easily achievable in an MRI experiment. The under-sampling process was repeated to generate 100 data sets with randomly chosen starting point between 0 and 5 s. These data were combined with Rician noise by assuming the simulated MRI signal was the real part of complex MRI data and adding independent Gaussian noise to imaginary and real data. In this simulation study, signal to noise ratio (SNR) was defined as the ratio of the baseline signal to the standard deviation of the Gaussian noise applied to the imaginary and real part of the complex data. Figure 1(b) shows an example of the MRI data generated for SNR=20 with typical SSM2 and GKM2 model fits. Similarly, Figure 1(c) shows the same MRI data generated for SNR=20 with typical ATH and ATHX model fits. Data with SNR=20 were used throughout the study unless specified otherwise.
MR tracer kinetic models
The simulation data with the known tissue parameters described above were used to assess the uncertainty in measurement of parameters of two groups of tissue kinetic models; one based on the general kinetic model (GKM) (1) and another based on the adiabatic approximation to the tissue homogeneity (ATH) model (21). The first group included extended general kinetic model (GKM2) (19) and extended shutter speed model (SSM2) (20). Both GKM2 and SSM2 models are based on Kety model extended to include a vascular compartment, shown in Eq. (5):
| (5) |
where Ctis(t) is the CA concentration of the whole tissue in a voxel. In GKM2, the longitudinal relaxation rate of tissue, R1(t), was calculated using a linear relationship; R1(t) = R10 + r1Ctis(t) where r1 was 3.8 mM−1s−1 and R10 was 0.67 s−1. With given TR and α, the steady-state magnetization was calculated using the Ernst-Anderson relationship for spoiled gradient recalled echo pulse sequence (13,20). Thus, there were 3 free parameters (Ktrans, ve and vb) to estimate in GKM2.
SSM2 takes into account the effect of water exchange between compartments when estimating the longitudinal relaxation rates for given CA concentrations. The two terms in the right hand side of Eq. (5) represent the CA concentrations in the intravascular and interstitial compartments, respectively. The CA concentration in the intracellular space was assumed zero. Then, the steady-state magnetization was calculated using the 3S2X model described in the above Data Simulation section. SSM2 had 5 free parameters (Ktrans, ve, vb, τb and τi). SSM2 parameter estimation was performed with and without fixing τb as a free parameter for parameter estimation.
The second group of kinetic models included ATH and ATHX models. In the tissue homogeneity (TH) model, the CA concentration in the intravascular space is defined as a function of both time and distance along the length of the capillary (26). An approximate closed-form solution in time domain, known as the ATH model, was derived based on the adiabatic approximation that CA concentration in the extravascular space changes slowly compared to that of the intravascular space (21). The ATH model has recently been used in DCE-MRI data analysis (27–29). It is defined as follows:
| (6) |
where Fp is plasma flow and Tc=vb/Fp is mean transit time. Ktrans is a function of both Fp and permeability surface area product (PS), given as Ktrans = [1−exp(−PS/Fp)]Fp. Thus, there were 4 free parameters (PS, Fp, ve and Tc) for the ATH model. In the ATHX model, the two terms on the right hand side of Eq. (6) were assumed to represent the CA concentration in the intravascular and interstitial compartments. Thus, the ATHX model had 6 free parameters (PS, Fp, ve, Tc, τb and τi). Similar to SSM2, ATHX parameter estimation was performed with and without fixing τb as a constant, in order to assess whether it would be necessary to keep τb as a free parameter for estimation.
The model parameters were estimated by minimizing the sum of squares of the difference between the simulation data and the model fit. Parameter estimation was conducted using the Simplex (30) method provided in Matlab (MathWorks, Natick, MA). The result of parameter estimation can be sensitive to initial values (27,31), particularly when the data is noisy. For each noisy time-intensity curve (out of 100 noisy time-intensity curves per each parameter set), we used 50 randomly selected initial guess values and selected the result with the lowest cost function value as the final result. This study was conducted using a Sun Microsystems cluster computer system from New York University high performance computing service which allowed us to use 64 computing threads at a time. Estimation of the kinetic parameters using the numerical integration time step of 1 s took about 0.1 min for GKM2 and ATH models and about 2 min for SSM2 and ATHX.
Effect of MRI scan parameters
As reported by Spencer and Fishbein (13) earlier, MRI scan parameters, such as TR and α, determine how much water exchange affects the apparent T1 of the target tissue with respect to the original T1 values of individual compartments. The kinetic models with and without the water exchange terms were used to demonstrate the difference between two models (GKM2.vs. SSM2 and ATH.vs. ATHX) as a function of TR and α. The kinetic parameters were assumed to be Ktrans = 0.25 min−1, ve = 0.45 and vb = 0.06 as reported for a breast cancer (27). In SSM2 and ATHX, τi and τb were assumed to be 200 ms and 50 ms, respectively. The sum of absolute differences between these two curves (SDC) was used as a global measure of the effect of water exchange in various imaging condition with different TR (0 – 25 ms) and α (0 – 50°). Although not all combinations of TR and α are practically useful, the SDC contour in these ranges was generated to assess the general trend of the water exchange effect. This analysis was performed to provide an overview of how the result in this study (conducted for TR = 5 ms and α = 15°) would change in different MRI scan protocols.
Simulation of different tissue conditions
The baseline tissue parameters for simulation were vb = 0.06, ve =0.15, vi = 0.79, PS = 0.345 ml/(g•min), Fp = 1.2 ml/(g•min), τb = 0.05 s, and τi = 0.2 s. Since there has not been a study, to our best knowledge, that measured all the parameters considered in our study, it was not possible to choose the baseline values from one reference. Instead, the baseline values were selected based on several previous studies (18,20,27). With the given Fp and PS, the baseline transfer constant Ktrans was 0.30 min−1. Data simulation was conducted by changing one parameter at a time as shown in Table 1. Tissue parameters could be related to each other in some degree. But, for the simplicity in the simulation study, we varied one parameter at a time, except the volume fractions. When changing either ve or vb, vi was varied inversely proportional to the change such that the sum of the volume fractions remained 1.
Table 1.
Kinetic model parameters used for the simulation studies.
| Baseline | Ktrans change | ve change | vb change | τi change | |
|---|---|---|---|---|---|
| Ktrans (min−1) | 0.30 | 0.05, 0.15, 0.30, 0.45, 0.60 | 0.30 | 0.30 | 0.30 |
| ve | 0.15 | 0.15 | 0.10, 0.20, 0.30, 0.40, 0.50 | 0.15 | 0.15 |
| vb | 0.06 | 0.06 | 0.06 | 0.02, 0.05, 0.08, 0.11, 0.14 | 0.06 |
| τi (s) | 0.2 | 0.2 | 0.2 | 0.2 | 0.10, 0.30, 0.50, 0.70, 0.90 |
Statistical analysis
Accuracy of the estimated parameters was measured by the difference between the median of estimated value and the true value used for data simulation. Precision was assessed in terms of coefficient of variation (CV = standard deviation/mean). In addition, kinetic models were compared in terms of Bayesian Information Criterion (BIC), defined in Eq. (7)
| (7) |
where RSS is the residual of sum of squares, N is the number of data points, and k is the number of free parameters (27). Wilcoxon signed-rank test was used to test the difference in accuracy or precision between kinetic models with and without the water exchange terms. A p-value of less than 0.05 was considered significant.
RESULTS
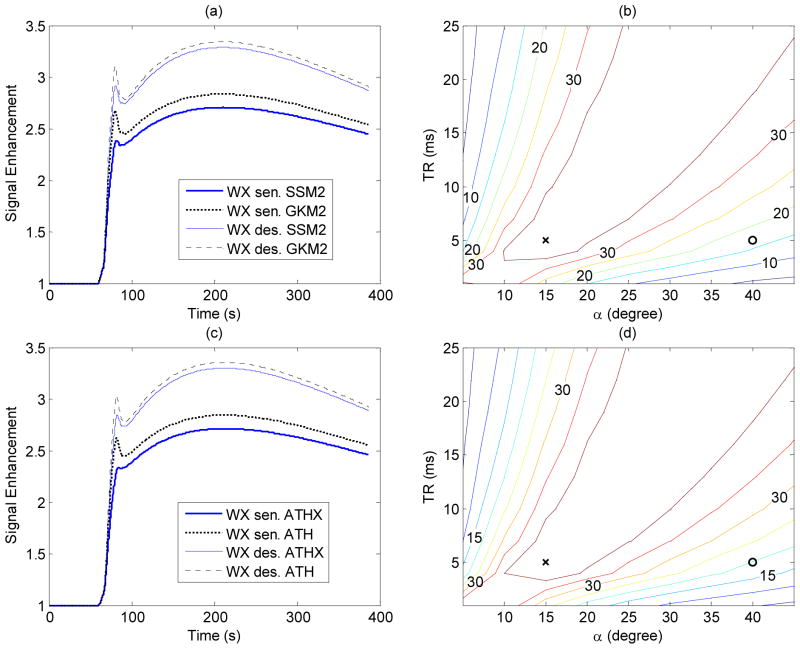
The influence of water exchange on DCE-MRI signal depending on MRI scan parameters, TR and α, is depicted in Figure 2, in terms of difference between the time intensity curves expected from kinetic models with and without the water exchange model. Figure 2(a) shows representative examples of GKM2 and SSM2 time-intensity curves for Ktrans = 0.25 min−1, ve = 0.45, and vb = 0.06 that were reported for a breast cancer (27), when the data acquisition protocol was water-exchange sensitized (TR = 5 ms and α = 15°) and water-exchange desensitized (TR = 5 ms and α = 40°). The SDC for the given TR and α was defined as the sum of absolute differences between these two data sets. Figure 2(b) shows a contour plot of SDC as a function of TR and α. It can be observed that, when α is small (< 10°), the SDC decreases as TR increases. For short TRs (< 5 ms), SDC decreases as α increases. The basic pattern of the SDC did not change noticeably by using different kinetic model parameters. The uncertainty analysis in the present study was conducted for α = 15° and TR = 5 ms, marked by a cross in Figure 2(b). Similar observation was made for ATH and ATHX models as shown in Figure 2(c) and (d).
Figure 2.
The effect of water exchange on DCE-MRI time intensity curve depending on the scan condition. (a) Representative MRI signal curves generated using GKM2 and SSM2 models (Ktrans=0.25/min with PS=0.28/min and Fp=1.2/min, vb=0.06, ve=0.45, τb=0.05s, τi=0.2s and Tc=3s) in two scan conditions; water-exchange (WX) sensitized (TR=5ms, flip angle=15°) and water-exchange desensitized (TR=5ms, flip angle=40°). (b) Contour plot of sum of absolute difference between GKM2 and SSM2 curves (SDC) for different flip angle α and TR values. The cross (x) and circle (o) marks correspond to water-exchange-sensitized acquisition (TR=5ms, flip angle=15°) and water-exchange-desensitized acquisition (TR=5ms, flip angle=40°) respectively. (c) and (d) are same as (a) and (b) but with the ATHX and ATH models.
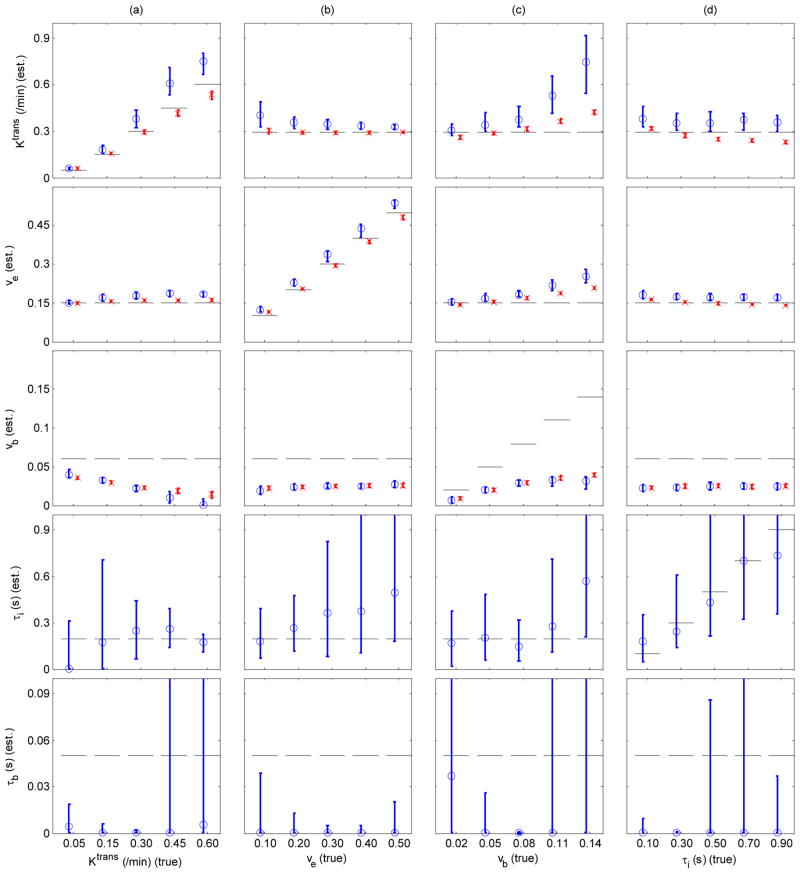
Figure 3 shows that SSM2 and GKM2 model parameters estimated by fitting the models to the simulated data with SNR=20. Each column of plots in Figure 3 represents a set of cases where one parameter was varied while holding the other parameters same. SSM2 Ktrans estimates were higher (4 – 152%) than the true values, particularly for large true Ktrans or vb. SSM2 ve estimates were also higher (1 – 69%) than the true values except for small Ktrans and vb. In all cases, GKM2 Ktrans and ve had smaller errors (1 – 44% for Ktrans and 2 – 38% for ve) than the SSM2 estimates with smaller inter-quartile range (IQR). The IQR of Ktrans increased with increase in Ktrans, while that of τi decreased. There was no particular extremum observed in terms of IQR. vb was largely underestimated in both models; the error became bigger as Ktrans or vb increased. Increase of vb resulted in increase of the estimated Ktrans in both models. The estimated τi in SSM2 has poor precision (CV = 52 – 170%). When τi increased, SSM2 Ktrans did not change noticeably whereas GKM2 Ktrans decreased by 27% (Figure 3(d)). However, the accuracy of SSM2 and GKM2 parameter estimation was substantially improved when a vascular dispersion function (32,33) was used together with a higher temporal resolution of 0.2 s/frame. (Supplemental Figure 1). Figure 3 also shows that τb estimates were close to 0 (≪ 50 ms). Another separate study of SSM2 estimation with τb fixed to 1 ms also produced similar results (Supplemental Figure 2).
Figure 3.
Accuracy and precision of SSM2 (circles and solid blue lines) and GKM2 (crosses and dashed red lines) parameter estimation. Each column represents a case where all model parameters were held constant except one parameter; Ktrans (a), ve (b), vb (c) and τi (d). In each column, the parameter varied during generation of reference data is shown on the x-axis. Plotted are the median and inter-quartile ranges of estimated parameters from 100 noisy data sets for each case. Horizontal dashed lines represent the true values used for the reference data.
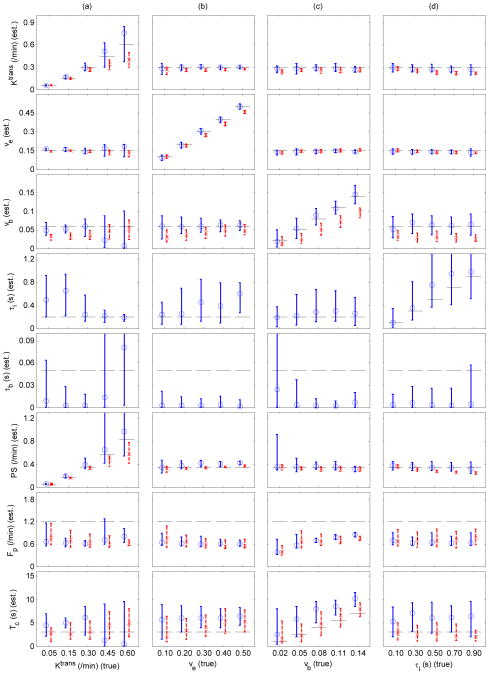
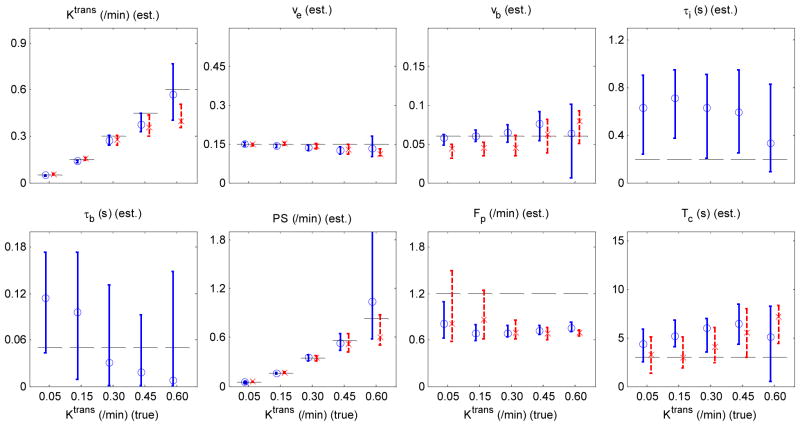
The results of ATH and ATHX parameter estimation are summarized in Figure 4. For Ktrans ≤ 0.3 min−1, both ATH and ATHX had reasonably accurate estimates of Ktrans and ve. But for Ktrans > 0.3 min−1, ATH Ktrans was smaller than the true value (34%) and ATHX Ktrans was bigger than the true value (26 %). The accuracy of vb estimation was substantially improved compared to those of GKM2 and SSM2 and did not show any systematic change due to Ktrans as in GKM2 and SSM2. The accuracy and precision of τi estimation were markedly improved when Ktrans > 0.3 min−1, compared with when Ktrans ≤ 0.3 min−1. It can also be noted that, unlike in GKM2 and SSM2, increase of vb did not induce any noticeable effect on the estimated Ktrans in both ATH and ATHX models. Increase in τi led to underestimation of ATH Ktrans by up to 26%, but much less in ATHX Ktrans (11%). Similar to what was observed in SSM2, τb estimates were close to 0 (≪ 50 ms) and ATHX parameter estimation with τb fixed to 1 ms also produced similar results (Supplemental Figure 3).
Figure 4.
Accuracy and precision of ATHX (circles and solid blue lines) and ATH (crosses and dashed red lines) parameter estimation. Each column represents a case where all parameters were held constant except one parameter; Ktrans (a), ve (b), vb (c) and τi (d). In each column, the parameter varied for generation of reference data is shown on the x-axis. Plotted are the median and inter-quartile ranges of the estimated parameters from 100 noisy data for each case. Horizontal dashed lines represent the true values used for the reference data.
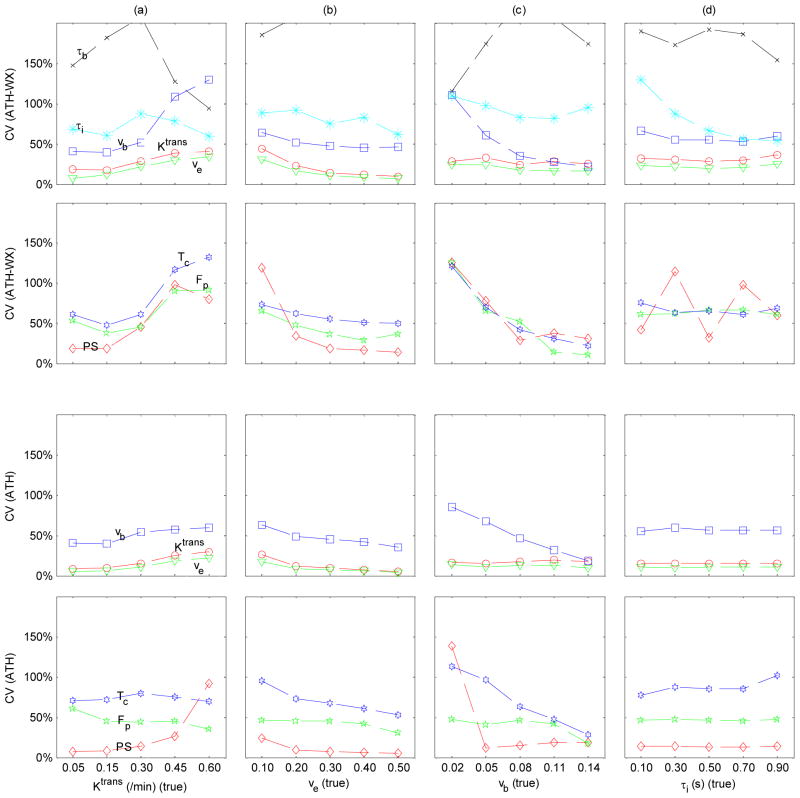
Precisions of the estimated parameters for ATHX and ATH model were assessed in terms of the CV as shown in Figure 5 for SNR = 20. For ATHX model in the top two rows, ve (triangles) had smallest CVs in all cases presented. The CV of ve increased substantially (339 %) as Ktrans increased from 0.05 to 0.6 min−1. Ktrans (circles) had up to 142 % higher CV than ve and showed similar pattern of increased CV for large Ktrans values. The CV of vb (squares) also increased substantially (219 %) by increase in Ktrans or decreased substantially (81 %) by decrease in vb. The bottom two rows of Figure 5 show the CV of ATH model parameters. Table 2 presents a summary of the uncertainty of kinetic parameters estimated by the two models. Kinetic parameters (Ktrans and vb) estimated by ATHX had significantly smaller errors than those of ATH (p < 0.05). ATH Ktrans, ve, PS and Fp had significantly smaller CVs than those of ATHX (p < 0.05).
Figure 5.
Coefficient of variation (CV) of estimated ATHX (top 2 rows) and ATH (bottom 2 rows) model parameters (red circles for Ktrans, green triangles for ve, blue squares for vb, black crosses for τb, cyan asterisks for τι, red diamonds for PS, green pentagrams for Fp and blue hexagram for Tc) depending on the change in one parameter as noted in the x-axis. Each column represents a case of changing only one parameter; Ktrans (a), ve (b), vb (c) and τi (d).
Table 2.
Comparison between ATH and ATHX in terms of accuracy and precision (SNR=20). Accuracy was defined as the percent difference from the true value and precision as coefficient of variance. Accuracy and precision of each parameter were measured from the data sets shown in Figure 4 and Figure 5 where the selected parameter was at the nominal value (Ktrans = 0.30 min−1, ve = 0.15, vb = 0.06, and τi = 0.2 s).
| ATH
|
ATHX
|
|||
|---|---|---|---|---|
| Error (%) | CV (%) | Error (%) | CV(%) | |
| Ktrans (min−1) | −13.14 ± 10.37 | 15.27 ± 6.22 | −0.11 ± 9.64* | 27.08 ± 9.35* |
| ve | −5.15 ± 5.38 | 10.79 ± 4.50 | −1.73 ± 8.29 | 19.26 ± 7.55* |
| vb | −37.29 ± 13.25 | 50.68 ± 14.37 | −7.04 ± 25.02* | 58.26 ± 27.65 |
| τi (s) | — | — | 54.22 ± 67.95 | 80.63 ± 19.00 |
| τb (s) | — | — | −81.94 ± 35.37 | 179.38 ± 36.19 |
| PS | −5.63 ± 11.85 | 23.43 ± 32.80 | 9.50 ± 8.98* | 55.50 ± 37.95* |
| Fp | −44.81 ± 7.66 | 43.17 ± 8.30 | −45.55 ± 8.03 | 55.79 ± 26.76* |
| Tc | 14.29 ± 34.14 | 74.85 ± 19.55 | 79.97 ± 58.48* | 66.45 ± 28.09* |
represents a significant difference (p < 0.05) between ATH and ATHX.
Performances of kinetic models were also compared in terms of BIC (data not shown). Out of 20 different tissue parameter sets included in this study (5 different values/parameter × 4 parameters as shown in Table 1), the median BICs of SSM2 in 17 cases were significantly smaller than those of GKM2. In contrast, the median BICs of ATHX were significantly larger (p < 0.05) than those of ATH in 14 cases, as there was no improvement in goodness of fitting by using the water exchange in those cases.
Figure 6 shows a comparison between ATHX and ATH in a water-exchange-desensitized acquisition scheme (TR = 5ms, α = 40°), as opposed to the water-exchange-sensitized scheme (TR = 5 ms, α = 15°) used for the above results. The ATH parameters from the water-exchange-desensitized scan in Figure 6 were not noticeably different from those from the water-exchange-sensitized scan in Figure 4. In contrast, the ATHX parameters showed more noticeable difference, particularly for τi, as its IQR becomes substantially large even with high Ktrans (> 0.3 min−1). However, inclusion of the water exchange terms, as in the ATHX model, did not lower the accuracy of other kinetic parameters even in the water-exchange-desensitized scan condition.
Figure 6.
Comparison between ATHX (blue circles and solid lines) and ATH (red crosses and dashed lines) when a water-exchange-desensitized protocol (TR = 5ms, flip angle = 40°) was used. Horizontal dashed lines represent the true values used for the reference data.
DISCUSSION
Tracer kinetic models for DCE-MRI data provide a non-invasive means to probe tissue microenvironment. It remains, however, challenging to assess the accuracy and precision of the estimated model parameters as there is no gold standard method to independently validate the measured kinetic parameters. When more complex models, such as SSM2 and ATHX, are employed for data analysis, goodness of fitting alone may not be sufficient to ensure that the estimated parameters accurately reflect the corresponding biophysical properties of the tissue. Hence, in this study, we used simulation data generated from well established tissue models, MMID4 (22) and 3S2X (13), and assessed the uncertainty of kinetic model parameters including intravascular and intracellular water lifetimes (τb and τi). We found that the Ktrans, ve and vb values estimated using ATHX had higher accuracy than those from other kinetic models. The accuracy of τi was relatively high for high Ktrans, but the precision of τi estimation varied substantially depending on Ktrans. Although our findings are based on simulation data, they can be helpful in interpreting the result from a kinetic model analysis of actual data.
One of the main findings in the present study is a good agreement of Ktrans and ve between ATH and ATHX for small Ktrans (≤0.3 min−1), as well as in between GKM2 and SSM2. This observation is consistent with what Buckley et al reported earlier from their DCE-MRI study of the skeletal muscle (34). Buckley et al found that there was no significant difference between Ktrans values estimated using no exchange (NX) assumption and fast exchange limit (FXL) assumption, implying that the influence of intracellular-interstitial water exchange is negligible (34). The Ktrans values reported in that study were typically smaller than 0.1 min−1 in which we made a similar observation that the influence of water exchange was small, if not negligible. Note that there are some differences between our models and the ones Buckley et al used, as we included a vascular compartment in the kinetic models and also used a full 3S2X model instead of using only a part of the solution. Nevertheless, the small effect of water exchange with low Ktrans was consistently observed.
For large Ktrans (> 0.3 min−1), however, we found that the ATH Ktrans was substantially underestimated, while ATHX Ktrans was estimated with a better accuracy (error < 26 % for the whole range of Ktrans values tested). This observation is consistent with previous reports comparing GKM and SSM (both without vascular components) which demonstrated that GKM could not follow the early fast rising portion of the enhancement curve (16–18). Thus, our results substantiate the previous findings that the effect of water exchange between tissue compartments should be considered when Ktrans is relatively high as in the leaky vessels of a tumor.
The median of τi estimation from multiple noisy data (SNR=20) approached the true value used for simulation data in most of the cases, however, the precision was poor in general. The accuracy of τi decreased substantially with smaller Ktrans values. This observation is also consistent with the report by Buckley et al where they found poor precision of τi in skeletal muscles with low Ktrans (34). The poor precision of τi could also be related to a considerably large overlap of τi values of malignant and benign breast lesions (5,35), as well as in between complete responders and partial responders to chemoradiation therapy in head and neck cancer patients (7). τi has been a parameter of interest because it could potentially represent cell membrane viability. But there has been no study to substantiate its potential as a useful biomarker in a DCE-MRI study. When a higher CA dose injection (higher magnitude of AIF CA concentration) was used for simulation data, we found that the precision of τi was noticeably improved (data not shown). Another possibility of improving the precision is to reduce the number of fitting parameters. Since τb is difficult to estimate as shown in this study, it could rather be fixed to a small value. As shown in Supplemental Figures 2 and 3, we also assessed ATHX and SSM2 analyses with τb fixed to 1 ms. We found the overall trend of both accuracy and precision did not change noticeably by using a fixed τb. Further study is warranted to investigate a means to improve the precision of τi measurement.
While our study was conducted with the SSM2 model based on the 3S2X water exchange model, an approximation of the SSM2 model, commonly known as the fast exchange regime (FXR) edited model, has been used more often in previous studies (16–18). The FXR model is an approximation of the full model for the case where the long relaxation term in the biexponential solution of the two-site exchange model is the dominant term when the CA concentration is approximately less than 5 mM (15) The contribution of the short relaxation term, ignored in the FXR model, can vary along the contrast enhancement curve and also depending on the different kinetic parameters included in this study. The FXR model also assumes that the vascular contribution is negligible. These two assumptions were not used in the SSM2 model used in our study. It may not be straightforward to apply the results of this study to the FXR model. But it can be expected that the FXR model would give somewhat similar result if the two assumptions for the FXR model are met.
The poor SNR commonly seen with fast imaging methods could be another limiting factor in utilizing the benefit of more comprehensive models, such as the ATHX model. We investigated the uncertainty of ATH and ATHX model parameters using the simulated data with noise level at SNR=5 for limited cases (data not shown). The increased noise reduced the accuracy and precision of parameter estimation substantially for both models as expected. Although the accuracy of ATHX was still better than that of ATH at SNR=5, the difference would become negligible when the noise level increases furthermore.
Given the temporal resolution and SNR used in the current study, we believe our results of the ATH model are compatible with the observations in other studies using the ATH model. Buckley (27) performed a simulation study with the MMID4 model (similar to our current study) and showed that using GKM leads to systematic overestimation of Ktrans and underestimation of vb, and the ATH model provides more accurate estimation. Nash et al (29) assessed different kinetic models for lung cancer DCE-MRI data and found that the ATH model was the best model based on Akaike’s information criterion. In both ATH and ATHX models, Fp was underestimated and Tc had a poor precision. This is consistent with the observation by Kershaw and Cheng (36) in which they investigated the accuracy of ATH parameters depending on temporal resolution. Kershaw and Cheng reported that a low temporal resolution, such as 4.5 s/frame or 6.5 s/frame, lead to underestimation of Fp and a poor precision of Tc, which is consistent with what we observed in this study. Kershaw and Cheng also showed that the accuracy of Fp and the precision of Tc improved substantially with a high temporal resolution of 1.5 s. In the current study, we found that the ATHX model parameters had better accuracy than the ATH model parameters. Although the ATHX parameters had lower precision than the ATH parameters, please note that the CV values reported in our study (Table 2 for both ATH and ATHX) are similar to the CV values reported in the study by Kershaw and Buckley (28) on the precision of ATH parameter estimation. Hence, we believe our simulation study results are within the expected range of outcomes from using the ATH or ATHX models.
The present study was conducted to investigate the uncertainty of tracer kinetic parameters in a realistic situation with noise. However, we could not include all important factors that could affect pharmacokinetic data analysis of DCE-MRI data, such as temporal resolution, baseline T1 measurement, B1 field inhomogeneity, and motion artifacts. Further simulation studies on these factors can provide useful insights that investigators using similar DCE-MRI and kinetic model analysis should be aware of. This study was focused on investigating the uncertainty of contrast kinetic model parameters in a wide range of values, in order to assess whether the model parameters are adequate and robust to represent changes in tissue microenvironment, although it might have not include all the parameter combinations possible in reality. Considering different kinetic models have their own limitations in either accuracy or precision, we reckon that a simpler model with a higher precision could be preferred to a more comprehensive model with a compromised precision, depending on the need specific to applications. The current study was performed with 100 noisy data sets in each case. Precision of the estimated parameter can depend on the number of data sets used. However we expect that the relative levels of precision between parameters would remain similar to our results.
The uncertainty of estimated kinetic model parameters can also be affected by cross-correlations, if any, between the estimated parameters. Although our current study was not specifically designed to assess the parameter correlations, the results can be used to take a cursory look at some possible parameter correlations within the limited conditions investigated in our study. For instance, the first column of Figure 3 shows that, in both GKM2 and SSM2, the estimated vb monotonically decreased as both true and estimated Ktrans values increased. Note that such negative association of the two parameters was not observed when other parameters were varied as shown in other columns of Figure 3. Another example can be found with vb change. When true vb increased as in the third column of Figure 3, both Ktrans and ve estimates increased, showing a positive correlation. Again note that these two parameters did not show such correlation when other parameters were varied as shown in other columns of Figure 3. These two examples demonstrate that correlation between kinetic model parameter estimates may exist in certain cases. However, it is not a simple, fixed behavior of model parameters, but rather a complex phenomena with different behaviors depending on the parameter values. It may also depend on experimental conditions, such as temporal resolution and SNR. Furthermore, a presence of parameter correlation does not necessarily indicate over-parameterization. These correlations among three parameters, Ktrans, ve, and vb, were not observed in ATH and ATHX models, which have one more parameter than GKM2 and SSM2, respectively, as shown in Figure 4. On the other hand, simultaneous estimation of PS and Fp in ATH or ATHX model was found challenging; the precisions of PS and Fp were low when the true values of one or both parameters were high, and the distribution of the estimates show different patterns of trends when the precision was low. Investigation of relationship between estimated parameters was beyond the scope of this study. However, it is an important issue that deserves more thorough investigation in future studies.
In conclusion, the present study demonstrated that ATHX model parameters could be used to probe complex tissue microenvironment. It was also shown that SSM2 model with a vascular dispersion function can provide the parameter accuracy similar to that of ATHX which has the same number of model parameters. The results from the present simulation study are consistent with previous observations with actual data analyses and provide helpful information for data interpretation. It has been demonstrated that τi can be estimated from DCE-MRI data albeit poor precision and the trend of τi change can also be detected. Inclusion of τi parameter in the ATHX model resulted in more accurate estimation of Ktrans, ve and vb, but with lower precisions. Further investigation is warranted to improve the accuracy and precision of τi measurement, while keeping the accuracy and precision of other parameters same or better, in order to use this parameter for practical application.
Supplementary Material
Acknowledgments
The authors would like to thank the New York University High Performance Computing center for providing wonderful support to conduct this study. This study was partly supported by NIH 1R01 CA160620.
References
- 1.Tofts PS, Brix G, Buckley DL, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10(3):223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 2.Choyke PL, Dwyer AJ, Knopp MV. Functional tumor imaging with dynamic contrast-enhanced magnetic resonance imaging. J Magn Reson Imaging. 2003;17(5):509–520. doi: 10.1002/jmri.10304. [DOI] [PubMed] [Google Scholar]
- 3.Eyal E, Degani H. Model-based and model-free parametric analysis of breast dynamic-contrast-enhanced MRI. NMR Biomed. 2009;22(1):40–53. doi: 10.1002/nbm.1221. [DOI] [PubMed] [Google Scholar]
- 4.Cha S. Update on brain tumor imaging: from anatomy to physiology. AJNR Am J Neuroradiol. 2006;27(3):475–487. [PMC free article] [PubMed] [Google Scholar]
- 5.Huang W, Li X, Morris EA, et al. The magnetic resonance shutter speed discriminates vascular properties of malignant and benign breast tumors in vivo. Proc Natl Acad Sci U S A. 2008;105(46):17943–17948. doi: 10.1073/pnas.0711226105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Laarhoven HW, Klomp DW, Rijpkema M, et al. Prediction of chemotherapeutic response of colorectal liver metastases with dynamic gadolinium-DTPA-enhanced MRI and localized 19F MRS pharmacokinetic studies of 5-fluorouracil. NMR Biomed. 2007;20(2):128–140. doi: 10.1002/nbm.1098. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Loevner LA, Quon H, et al. Prediction of response to chemoradiation therapy in squamous cell carcinomas of the head and neck using dynamic contrast-enhanced MR imaging. AJNR Am J Neuroradiol. 2010;31(2):262–268. doi: 10.3174/ajnr.A1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Connor JP, Jackson A, Parker GJ, Jayson GC. DCE-MRI biomarkers in the clinical evaluation of antiangiogenic and vascular disrupting agents. Br J Cancer. 2007;96(2):189–195. doi: 10.1038/sj.bjc.6603515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YR, Rebro KJ, Schmainda KM. Water exchange and inflow affect the accuracy of T-1-GRE blood volume measurements: Implications for the evaluation of tumor angiogenesis. Magnet Reson Med. 2002;47(6):1110–1120. doi: 10.1002/mrm.10175. [DOI] [PubMed] [Google Scholar]
- 10.Bailey C, Giles A, Czarnota GJ, Stanisz GJ. Detection of Apoptotic Cell Death In Vitro in the Presence of Gd-DTPA-BMA. Magnet Reson Med. 2009;62(1):46–55. doi: 10.1002/mrm.21972. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Priest RA, Woodward WJ, et al. Cell membrane water exchange effects in prostate DCE-MRI. Journal of Magnetic Resonance. 2012;218:77–85. doi: 10.1016/j.jmr.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst RR, Anderson WA. Application of Fourier transform spectroscopy to magnetic resonance. Rev Sci Instrum. 1966;37:93–102. [Google Scholar]
- 13.Spencer RG, Fishbein KW. Measurement of spin-lattice relaxation times and concentrations in systems with chemical exchange using the one-pulse sequence: breakdown of the Ernst model for partial saturation in nuclear magnetic resonance spectroscopy. J Magn Reson. 2000;142(1):120–135. doi: 10.1006/jmre.1999.1925. [DOI] [PubMed] [Google Scholar]
- 14.Larsson HBW, Rosenbaum S, Fritz-Hansen T. Quantification of the effect of water exchange in dynamic contrast MRI perfusion measurements in the brain and heart. Magnet Reson Med. 2001;46(2):272–281. doi: 10.1002/mrm.1188. [DOI] [PubMed] [Google Scholar]
- 15.Landis CS, Li X, Telang FW, et al. Equilibrium transcytolemmal water-exchange kinetics in skeletal muscle in vivo. Magn Reson Med. 1999;42(3):467–478. doi: 10.1002/(sici)1522-2594(199909)42:3<467::aid-mrm9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 16.Yankeelov TE, Rooney WD, Li X, Springer CS., Jr Variation of the relaxographic “shutter-speed” for transcytolemmal water exchange affects the CR bolus-tracking curve shape. Magn Reson Med. 2003;50(6):1151–1169. doi: 10.1002/mrm.10624. [DOI] [PubMed] [Google Scholar]
- 17.Zhou R, Pickup S, Yankeelov TE, Springer CS, Jr, Glickson JD. Simultaneous measurement of arterial input function and tumor pharmacokinetics in mice by dynamic contrast enhanced imaging: effects of transcytolemmal water exchange. Magn Reson Med. 2004;52(2):248–257. doi: 10.1002/mrm.20143. [DOI] [PubMed] [Google Scholar]
- 18.Kim S, Quon H, Loevner LA, et al. Transcytolemmal water exchange in pharmacokinetic analysis of dynamic contrast-enhanced MRI data in squamous cell carcinoma of the head and neck. J Magn Reson Imaging. 2007;26(6):1607–1617. doi: 10.1002/jmri.21207. [DOI] [PubMed] [Google Scholar]
- 19.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging. 1997;7(1):91–101. doi: 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Rooney WD, Springer CS., Jr A unified magnetic resonance imaging pharmacokinetic theory: intravascular and extracellular contrast reagents. Magn Reson Med. 2005;54(6):1351–1359. doi: 10.1002/mrm.20684. [DOI] [PubMed] [Google Scholar]
- 21.St Lawrence KS, Lee TY. An adiabatic approximation to the tissue homogeneity model for water exchange in the brain: I. Theoretical derivation. J Cerebr Blood F Met. 1998;18(12):1365–1377. doi: 10.1097/00004647-199812000-00011. [DOI] [PubMed] [Google Scholar]
- 22.Kroll K, Wilke N, Jerosch-Herold M, et al. Modeling regional myocardial flows from residue functions of an intravascular indicator. Am J Physiol. 1996;271(4 Pt 2):H1643–1655. doi: 10.1152/ajpheart.1996.271.4.H1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higham NJ. The scaling and squaring method for the matrix exponential revisited. Siam J Matrix Anal A. 2005;26(4):1179–1193. [Google Scholar]
- 24.Zhang J, Amarosa A, Rosenkrantz AB, Kim S. Estimation of reference tissue based arterial input function using neural network. ISMRM 20th Annual Meeting; Melbourne, Australia. 2012; 1971. [Google Scholar]
- 25.Weinmann HJ, Laniado M, Mutzel W. Pharmacokinetics of GdDTPA/dimeglumine after intravenous injection into healthy volunteers. Physiol Chem Phys Med NMR. 1984;16(2):167–172. [PubMed] [Google Scholar]
- 26.Johnson JA, Wilson TA. A Model for Capillary Exchange. American Journal of Physiology. 1966;210(6):1299. doi: 10.1152/ajplegacy.1966.210.6.1299. [DOI] [PubMed] [Google Scholar]
- 27.Buckley DL. Uncertainty in the analysis of tracer kinetics using dynamic contrast-enhanced T1-weighted MRI. Magn Reson Med. 2002;47(3):601–606. doi: 10.1002/mrm.10080. [DOI] [PubMed] [Google Scholar]
- 28.Kershaw LE, Buckley DL. Precision in measurements of perfusion and microvascular permeability with T-1-weighted dynamic contrast-enhanced MRI. Magnet Reson Med. 2006;56(5):986–992. doi: 10.1002/mrm.21040. [DOI] [PubMed] [Google Scholar]
- 29.Naish JH, Kershaw LE, Buckley DL, Jackson A, Waterton JC, Parker GJM. Modeling of Contrast Agent Kinetics in the Lung Using T-1-Weighted Dynamic Contrast-Enhanced MRI. Magnet Reson Med. 2009;61(6):1507–1514. doi: 10.1002/mrm.21814. [DOI] [PubMed] [Google Scholar]
- 30.Nelder JA, Mead R. A Simplex-Method for Function Minimization. Comput J. 1965;7(4):308–313. [Google Scholar]
- 31.Henderson E, Sykes J, Drost D, Weinmann HJ, Rutt BK, Lee TY. Simultaneous MRI measurement of blood flow, blood volume, and capillary permeability in mammary tumors using two different contrast agents. J Magn Reson Imaging. 2000;12(6):991–1003. doi: 10.1002/1522-2586(200012)12:6<991::aid-jmri26>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 32.Calamante F, Gadian DG, Connelly A. Delay and dispersion effects in dynamic susceptibility contrast MRI: simulations using singular value decomposition. Magn Reson Med. 2000;44(3):466–473. doi: 10.1002/1522-2594(200009)44:3<466::aid-mrm18>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 33.Annet L, Hermoye L, Peeters F, Jamar F, Dehoux JP, Van Beers BE. Glomerular filtration rate: assessment with dynamic contrast-enhanced MRI and a cortical-compartment model in the rabbit kidney. J Magn Reson Imaging. 2004;20(5):843–849. doi: 10.1002/jmri.20173. [DOI] [PubMed] [Google Scholar]
- 34.Buckley DL, Kershaw LE, Stanisz GJ. Cellular-interstitial water exchange and its effect on the determination of contrast agent concentration in vivo: dynamic contrast-enhanced MRI of human internal obturator muscle. Magn Reson Med. 2008;60(5):1011–1019. doi: 10.1002/mrm.21748. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Huang W, Morris EA, et al. Dynamic NMR effects in breast cancer dynamic-contrast-enhanced MRI. Proc Natl Acad Sci U S A. 2008;105(46):17937–17942. doi: 10.1073/pnas.0804224105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kershaw LE, Cheng HLM. Temporal Resolution and SNR Requirements for Accurate DCE-MRI Data Analysis Using the AATH Model. Magnet Reson Med. 2010;64(6):1772–1780. doi: 10.1002/mrm.22573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.