Abstract
The Reelin–Disabled 1 (Dab1)-signaling pathway plays a critical role in neuronal cell positioning in the brain. We have isolated two alternatively spliced variants of Dab1 from chick retina, an early form (chDab1-E) expressed in undifferentiated cells and a late form (chDab1-L) expressed in amacrine and ganglion cells. A key difference between the two forms is the exclusion in chDab1-E of two Src-related tyrosine kinase recognition sites implicated in Reelin-mediated Dab1 tyrosine phosphorylation. Retinal cultures transfected with a chDab1-L expression construct undergo a dramatic change in morphology, accompanied by the formation of numerous thin elongated processes, increased tyrosine phosphorylation, activation of Src family kinase(s) and increased levels of the axonal outgrowth protein growth-associated protein-43. In contrast, chDab1-E transfectants retain an undifferentiated morphology. Mutational analysis implicates a specific tyrosine (tyr-198) in the morphological and biochemical alterations associated with chDab1-L expression. We propose that alternative splicing of chDab1 represents an effective and flexible way of regulating the Reelin–Dab1-signaling pathway in a mixed cell population, by ensuring that secreted Reelin activates the signaling cascade only in target neuronal cells.
Keywords: alternative splicing, Disabled-1, retinal development, Src family kinases, tyrosine phosphorylation
Introduction
Transmission of the visual signal to the brain depends on the proper organization of differentiated retinal cells into distinct nuclear and synaptic layers. Photoreceptors, which capture light photons, are located in the outer nuclear layer (ONL) closest to the retinal pigmented epithelium (RPE). These cells transmit a neural signal via the outer plexiform layer to the inner nuclear layer (INL) where the input signal is further processed by four classes of neuronal cells (amacrine, bipolar, horizontal and interplexiform). The visual signal is then transmitted to the innermost nuclear layer of the retina, called ganglion cell layer (GCL), via the inner plexiform layer. Ganglion cells send their axons through the optic nerve to form connections with specific targets in the brain, resulting in the projection of the visual image to the brain (Dowling, 1987). These six classes of neuronal cells as well as the Müller glial cells, located in the INL, are derived from neuroectodermal precursor cells called retinoblasts, which migrate to their proper location as they differentiate (Turner et al, 1990).
The Reelin–Disabled 1 (Dab1)-signaling pathway has been implicated in the positioning of migrating neurons in the brain (D'Arcangelo et al, 1995; Howell et al, 1997a; Sheldon et al, 1997). Reelin is a secreted extracellular matrix glycoprotein that binds to the very low density lipoprotein receptor (VLDLR) and apolipoprotein E receptor 2 (ApoER2) (D'Arcangelo et al, 1999; Trommsdorff et al, 1999). Dab1, an intracellular adapter protein, binds to NPxY motifs located in the cytoplasmic tails of Reelin receptors. Mice deficient in Reelin (reeler), Dab1 (yotari/scrambler) or both VLDLR and ApoER2 have similar neuronal cell positioning defects in the brain, with inversion of neuronal layers in the cerebral cortex, and laminar defects in the cerebellum and hippocampus (D'Arcangelo et al, 1995; Sheldon et al, 1997; Trommsdorff et al, 1999).
Binding of Reelin to its receptors induces Dab1 tyrosine phosphorylation and stimulates the activation of Src family tyrosine kinases and Akt serine/threonine kinase (Howell et al, 1999; Beffert et al, 2002; Ballif et al, 2003; Benhayon et al, 2003). Src, Fyn and Yes kinases are involved in Dab1 phosphorylation while phosphorylated Dab1 enhances their activity, suggesting a self-regulating tyrosine-signaling cascade (Arnaud et al, 2003; Bock and Herz, 2003). A possible downstream effector of phosphorylated Dab1 is glycogen synthase kinase 3β (GSK-3β), which is downregulated in response to Reelin signaling (Beffert et al, 2002; Ohkubo et al, 2003). GSK-3β modulates the activity of the microtubule-associated protein Tau involved in microtubule assembly during neuronal differentiation (Ishiguro et al, 1993).
The main form of mammalian Dab1 has an open reading frame of 555 amino acids (aa) and consists of an N-terminal domain that associates with Reelin receptors, an internal domain containing Reelin-dependent tyrosine phosphorylation sites and a C-terminal domain implicated in the modulation of Reelin–Dab1 signaling (Herrick and Cooper, 2002). The human and mouse Dab1 genes span 1100 kb and have 14 main coding exons. Alternative internal and 5′ exons have been identified (Bar et al, 2003) and different-size isoforms have been detected by Western blot analysis. Some Dab1 isoforms show developmental- and tissue-specific expression patterns, suggesting a role in embryogenesis and organogenesis.
We have isolated chicken Dab1 (chDab1) cDNA from undifferentiated retinal cells using a differential display strategy. Here, we show that chDab1 undergoes alternative splicing as a function of developmental stage, with early and late forms of the protein expressed in proliferating and differentiating cells, respectively. Transfection of primary retinal cultures with expression constructs containing either the early or the late form of chDab1 results in dramatic changes in cellular morphology, with cells transfected with the late form producing numerous thin and extended processes. This morphological alteration is accompanied by Src family kinase activation and is dependent on tyrosine phosphorylation sites that are spliced out in the early form of chDab1. We propose that the absence of these tyrosine phosphorylation sites in the early form of chDab1 results in uncoupling of the Reelin–Dab1 pathway.
Results
Dab1 cDNA is highly expressed in chick retina compared to the brain
To identify genes enriched in the undifferentiated chick retina, differential display (DD)-PCR analysis was carried out with poly(A)+ RNA from retina at embryonic day (ED)3.5, ED5, ED16 and brain at ED5 and ED16. Using primer pair T11MG and OPA-10, a 532 bp band was generated that was more intense in ED3.5 and ED5 retina than in ED16 retina and the brain. Sequencing of this cDNA revealed a high degree of similarity to the 5′ ends of human and mouse Dab1. The 532 bp cDNA was used to probe a Northern blot of the retina, brain, heart, liver, kidney and gut at different developmental stages. Highest levels of chDab1 RNA were found in ED5 and ED10 retina as well as ED16 kidney, with ∼two- to three-fold lower levels in ED16 retina (Figure 1A). Considerably lower levels of chDab1 RNA were found in the brain, heart, liver and gut.
Figure 1.
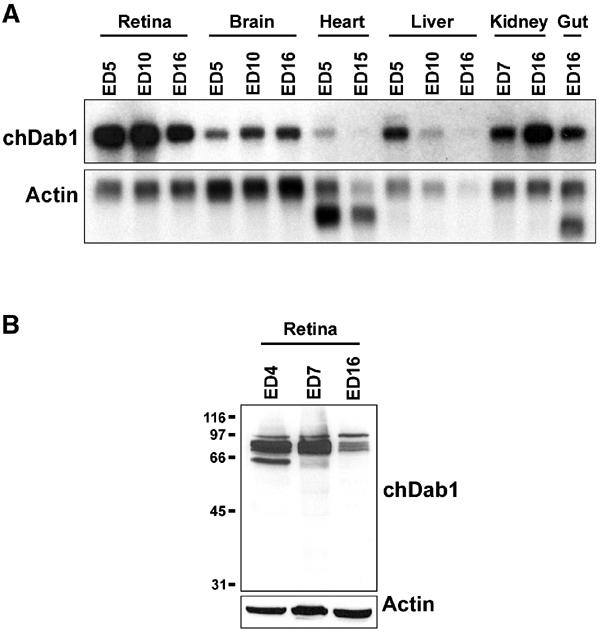
Northern and Western blot analyses of chDab1 expression. (A) Northern blots were prepared using poly(A)+ RNA (2 μg/lane) extracted from the retina (ED5, ED10, ED16), brain (ED5, ED10, ED16), heart (ED5, ED15), liver (ED5, ED10, ED16), kidney (ED7, ED16) and gut (ED16). The filter was sequentially hybridized with 32P-labeled: (i) 532 bp chDab1 cDNA and (ii) actin cDNA. The extra bands obtained with the actin probe in the heart and gut represent tissue-specific actin mRNAs. We consistently find actin RNA to be low in the liver, especially at later developmental stages. (B) Western blots were prepared using ED4, ED7 and ED16 total chick retina extracts (50 μg protein/lane). The filter was sequentially incubated with rabbit anti-Dab1 antibody (1:5000) and goat anti-actin antibody (1:500). Molecular mass standards (in kDa) are indicated on the left.
Dab1 protein was examined by Western blot analysis of whole-cell lysates prepared from ED4, ED7 and ED16 retina. The pattern of expression was similar to that observed by Northern blotting, with higher levels of chDab1 protein in ED4 and ED7 retina compared to ED16 retina (Figure 1B). In addition to quantitative changes, differences were observed in the banding patterns of ED4, ED7 and ED16 retina, with the lowest band disappearing by ED16. The number of bands observed in each lane suggests the presence of multiple isoforms and/or different post-translational modifications.
Developmentally regulated alternative splicing of chDab1
Full-length chDab1 cDNA encoding a predicted open reading frame of 535 aa was obtained by screening an ED7 chick retina cDNA library with the 532 bp DD-PCR DNA fragment (Figure 2). Overall, the chicken Dab1 protein sequence had a high level of similarity to the 555 aa human and mouse sequences (92% identical; 96% similar), with two major differences: (i) a deletion of 105 bp (35 aa) located immediately after aa 186 and corresponding to aa 187–221 of human/mouse Dab1, and (ii) an insertion of 57 bp (19 aa) located after aa 206. Based on the exon/intron structure of human Dab1 genomic DNA, the 105 bp deletion region corresponds to two exons (exons 7 and 8) and the 57 bp insertion region corresponds to a single exon (exon 9-2) (Bar et al, 2003). While alternative splicing of the 105 bp deletion region has not been previously reported, alternative splicing of the 57 bp exon has been described in chicken, as well as in mouse where this exon is duplicated (Bar et al, 2003).
Figure 2.
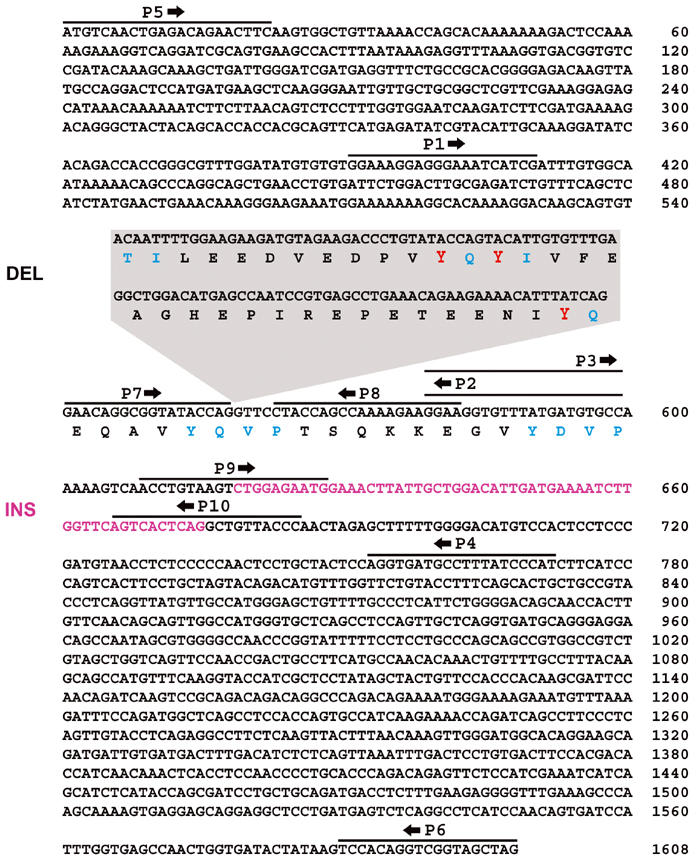
Nucleotide (nt) sequences of the coding regions of early and late chDab1. The early form of chDab1 (ChDab1-E) is 1608 nt long including a 57 nt insertion region (INS) at nt 618 (magenta). ChDab1-L(late) is 1656 nt long and includes a 105 nt region deleted (DEL) in chDab1-E at nt 558 (boxed callout). The 57 nt insertion region is not present in chDab1-L. The amino-acid sequence is shown for the region spanning the tyrosine phosphorylation domains with individual tyrosine phosphorylation motifs indicated in blue and the three tyrosines deleted in chDab1-E indicated in red. The indicated deletion results in the conversion of Y185QTI to Y185QVP. The primers used for RT–PCR (P1–P6) and splice form-specific in situ hybridizations (DEL: P7, P8; INS: P9, P10) are indicated. Sequence data for chDab1-E and chDab1-L have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers AY242122 and AY242123, respectively.
To further investigate the possibility of alternative splicing in chDab1, RT–PCR analysis of the retina and brain at different developmental stages was carried out using primer pairs flanking the deletion region (P1/P2) and the insertion region (P3/P4) (Figure 2). As shown in Figure 3A, RT–PCR of the deletion region generated two DNA bands, of 209 and 314 bp. The 209 bp band was predominant in the ED5 retina, while the 314 bp band was most abundant in the ED10 and ED16 retina, with a barely detectable 209 bp band at ED10. An intermediate banding pattern was observed in the ED7 retina. A similar pattern was seen in the developing brain, with both bands observed at ED3.5 and ED5, and the 314 bp band being predominant at ED16. Next, we studied the insertion region by RT–PCR. Two bands of 194 and 137 bp were observed. The higher band was predominant in ED5 retina and ED3.5 brain. By ED16, the 137 bp band was more intense in both the retina and brain.
Figure 3.

RT–PCR analysis of chDab1 deletion and insertion regions. (A) cDNAs synthesized from poly(A)+ RNA from the retina (ED5, ED7, ED10, ED16) and brain (ED3.5, ED5, ED16) were amplified using P1 and P2 primers for deletion analysis and P3 and P4 primers for insertion analysis. Sizes of amplified bands are indicated. (B) Banding patterns obtained by RT–PCR analysis of ED5, ED10 and ED16 retina using P1 and P4 primers. The lower band is 382 bp and the higher band is 430 bp.
DNA isolated from the 314, 209, 194 and 137 bp bands was sequenced. As expected, the 209 and 194 bp bands, predominant at early stages of retina and brain development, exclude the 105 bp deletion region (DEL) and include the 57 bp insertion region (INS), respectively (Figure 2). This previously uncharacterized form has been labeled chDab1-E. The 314 and 137 bp bands, predominantly found at later stages of development, include the deletion region and exclude the insertion region, respectively. This form, well documented in humans and mice, has been labeled chDab1-L.
The conversion from chDab1-E to chDab1-L involves two alternative splicing events. To determine whether these splicing events occur at the same time during retinal development, RT–PCR analysis was carried out using primer set P1 and P4, which spans the deletion/insertion region (Figure 2). Simultaneous splicing events would result in only two amplified bands: (i) a band of 430 bp including the 105 bp deletion region and excluding the 57 bp insertion region, and (ii) a band of 382 bp excluding the deletion region and including the insertion region. If the insertion/deletion events occurred at different developmental stages, intermediate products of 487 or 325 bp would be observed. RT–PCR analysis of ED5, ED10 and ED16 retina generated only two bands of 382 and 430 bp (Figure 3B). The ED10 retina had both bands, while ED5 and ED16 had the 382 bp band and the 430 bp band, respectively. All four bands were isolated and sequenced to confirm our predictions.
Examination of the chDab1-E deletion and insertion regions reveals some important clues as to the function of these domains. The 35 aa deletion region overlaps with three nonreceptor tyrosine kinase (NTK) recognition sites: Y185QTI, Y198QY200I and Y220QVP (Howell et al, 1997b, 2000; Keshvara et al, 2001), with the Y185QTI/Y220QVP motifs converted into YQVP as a result of the deletion (Figure 2). Tyr-198 and tyr-220 have previously been shown to be the major sites for Reelin-induced Dab1 phosphorylation in embryonic neurons (Howell et al, 1999; Keshvara et al, 2001). The 19 aa insertion region has no homology to any known motifs; however, it does show some similarity (11/19 aa) to an exon found in Dab1-related proteins: murine p96/p67 (Xu et al, 1995), also known as Dab2, and the human DOC-2 (Albertsen et al, 1996), suggesting consolidation of some Dab2 function in chDab1-E.
ChDab1-E and chDab1-L RNAs are expressed in undifferentiated and differentiated retinal cells, respectively
Retinal tissue sections were examined by in situ hybridization to determine the distribution pattern of chDab1 RNA. At ED5, when 85% of cells are undifferentiated and proliferating (Dütting et al, 1983), chDab1-positive cells were found throughout the retina (Figure 4A). After 2 days (ED7), chDab1-positive cells were found in the inner two-thirds of the INL, and in the GCL (Figure 4B). At this developmental stage, proliferating cells (∼50% of total) are located in the central portion of the INL. The emerging photoreceptor layer next to the RPE was negative. In the differentiated ED16 retina, chDab1-positive cells were primarily found in the GCL and the innermost third of the INL where amacrine cells are located (Figure 4C). There was also a weak signal in the ONL containing photoreceptor cells as well as in the outermost layer of cells in the INL where horizontal cells are located.
Figure 4.
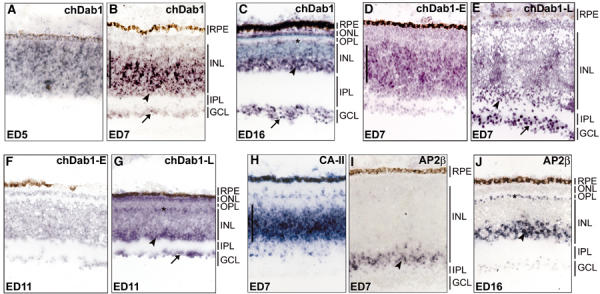
Localization of chDab1 transcripts in the developing chick retina. In situ hybridization was performed to identify the cell types expressing chDab1 mRNA. Frozen sections of ED5 (A), ED7 (B, D, E, H, I), ED11 (F, G) and ED16 (C, J) retina were hybridized with DIG-labeled chDab1 (A–C), splice form-specific chDab1-E (D, F), splice form-specific chDab1-L (E, G), CA-II (H) and AP-2β (I, J) antisense RNA. ChDab1 sense RNA served as the negative control (data not shown). The DIG signal was detected using anti-DIG antibody and AP-coupled secondary antibody. The purple color was generated using BCIP and NBT. Panel C was counterstained with ethyl green to show cell layers. All sections were photographed using a × 20 objective. The arrows point to the GCL, the arrowheads point to the amacrine cells in the INL, the asterisks indicate the horizontal cells in the INL and the vertical bars span the undifferentiated cells in the ED7 retina. RPE, retinal pigment epithelium; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer.
Using primers flanking the deletion and insertion regions, we generated splice form-specific probes to study the pattern of expression of chDab1-E and chDab1-L transcripts in the differentiating retina. The sizes of these probes were 77 nt (insertion region specific to chDab1-E) and 147 nt (deletion region specific to chDab1-L). At ED7, chDab1-E was detected in the central region of the INL where undifferentiated cells are located (Figure 4D), while the chDab1-L-specific probe generated a weak signal in the inner part of the INL and a stronger signal in the GCL (Figure 4E). At ED11, no signal was detected with the chDab1-E-specific probe (Figure 4F); however, the chDab1-L-specific probe generated a signal in the inner part of the INL and in the GCL (Figure 4G). In general agreement with the insertion region (chDab1-E) results, Bar et al (2003) reported elevated expression of the murine equivalent of the insertion region in the proliferating cells of the ventricular zone in E14 brain, with decreased levels at P0.
To identify more conclusively the cells expressing chDab1-E and chDab1-L within the INL of the developing retina, we used two markers: carbonic anhydrase II (CA-II), expressed in proliferating precursor cells and Müller glial cells (Vardimon et al, 1986; Witte and Godbout, 2002), and AP-2β, expressed in amacrine cells and horizontal cells (Bisgrove and Godbout, 1999). In situ hybridization of ED7 sections revealed CA-II mRNA in the central part of the INL (Figure 4H), while AP-2β was detected in the innermost layers of the INL in both ED7 (Figure 4I) and ED16 (Figure 4J) retina, with a positive outermost layer of cells (horizontal cells) also seen at ED16. Taken together, the in situ hybridization and RT–PCR results indicate that chDab1-E is expressed in retinoblasts, while chDab1-L is primarily found in amacrine and ganglion cells.
ChDab1-L modulates cellular morphology and tyrosine phosphorylation of retinal cells
To address the significance of having two chDab1 isoforms in the differentiating retina, the coding regions of chDab1-E and chDab1-L were generated by RT–PCR using poly(A)+ RNA from ED5 and ED16 chick retina, respectively, and cloned in-frame with GFP into the pEGFP-C1 expression vector. After verification of the cDNA sequences, both expression constructs, as well as empty vector, were transfected into primary retinal cultures prepared from ED5 embryos. Western blot analysis confirmed expression of the GFP–chDab1-fusion proteins in transfected cells (Figure 5A).
Figure 5.
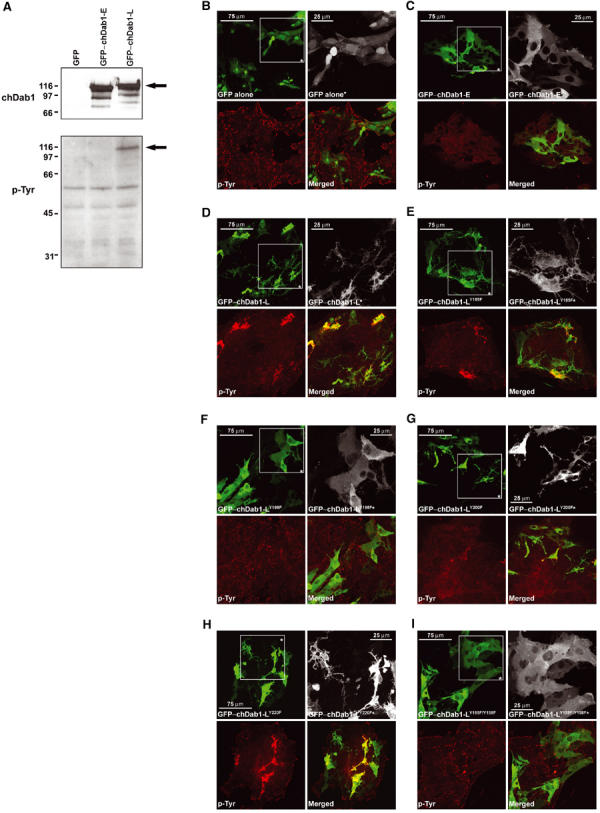
Analysis of chDab1-transfected primary retinal cultures. (A) Western blot analysis of GFP-, GFP-chDab1-E- and GFP-chDab1-L-transfected retinal cells. Cell extracts (30 μg/lane) were electrophoresed in a 10% SDS–PAGE gel and blotted onto a nitrocellulose filter. GFP–chDab1 expression was detected using anti-Dab1 antibody (1:5000) while phosphotyrosine was detected using anti-phosphotyrosine antibody (1:1000). GFP–chDab1 and tyrosine phosphorylated GFP–chDab1 proteins are indicated by an arrow. Molecular mass standards (in kDa) are shown on the left. (B–I) GFP expression and phosphotyrosine (p-Tyr) in GFP–chDab1-transfected retinal cultures. Cells were transfected with expression constructs encoding GFP (B), GFP–chDab1-E (C), GFP–chDab1-L (D), GFP–chDab1-LY185F (E), GFP–chDab1-LY198F (F), GFP–chDab1-LY200F (G), GFP–chDab1-LY220F (H) and GFP–chDab1-LY185F/Y198F (I). Cells were fixed with 4% paraformaldehyde, permeabilized and stained with anti-phosphotyrosine antibody and Alexa 555 goat anti-mouse secondary antibody. The GFP signal in transfected cells was detected by epifluorescence. Boxed regions (*) are enlargements to show morphological details of transfected cells.
The subcellular localization of the GFP-fusion proteins was studied by confocal microscopy. Both isoforms of chDab1 had a cytoplasmic pattern (Figure 5C and D). In contrast, GFP protein was found throughout the cell (Figure 5B). Transfection of the chDab1-L construct was accompanied by striking morphological differences, with numerous thin elongated processes extending from most GFP–chDab1-L-positive cells (Figure 5D). In addition, many of the transfected cells appeared smaller and more stringy. Cells transfected with GFP (Figure 5B) and GFP–chDab1-E (Figure 5C) were generally more spherical and neuroblastic, with fewer and less elongated processes.
As a major difference between the late and early forms of chDab1 is the lack of tyrosine phosphorylation sites predicted to be necessary for Dab1 activation and relaying of the Reelin–Dab1 signal, we next looked at tyrosine phosphorylation in the transfected cells. As shown in Figure 5D, a strong phosphotyrosine signal was observed in GFP–chDab1-L-positive retinal cells. In contrast, no phosphotyrosine signal above background was detected in either GFP–chDab1-E-transfected cells (Figure 5C) or GFP control (Figure 5B). Phosphorylation of chDab1-L was confirmed by Western blotting of cellular extracts derived from transfected cells with anti-phosphotyrosine antibody (Figure 5A). These results suggest a specific role for the chDab1-L isoform in relaying tyrosine phosphorylation signals that affect cellular morphology.
To study the role of the four tyrosine residues located within or in the immediate vicinity of the two exons deleted in chDab1-E, we mutated each of these residues in the GFP–chDab1-L expression construct by converting the tyrosine into a phenylalanine. Retinal cells transfected with wild type and mutant constructs were analyzed by Western blotting to ensure that full-length GFP-fusion proteins accumulated to the same levels in transfected cells (see Supplementary Figure 1), as well as by confocal microscopy. As shown in Figure 5E, mutation of tyr-185 produced cells with an appearance similar to those transfected with wild-type chDab1-L. Mutation of tyr-198 abolished tyrosine phosphorylation and produced cells similar in appearance to chDab1-E-transfected cells (Figure 5F). Mutation of tyr-200 generated an intermediate phenotype, with some tyrosine phosphorylation detected above background and a moderate number of processes in transfected cells (Figure 5G). The fourth mutation, tyr-220, had no effect on either the morphology or phosphorylation status of transfected cells (Figure 5H). As expected, cells transfected with an expression construct mutated at both tyr-185 and tyr-198 were similar in appearance to those transfected with chDab1-E (Figure 5I).
ChDab1-L expression results in Src activation and induction of growth-associated protein (GAP)-43
Dab1 tyrosine phosphorylation activates Src family kinases. To address whether expression of chDab1-L results in Src family kinase activation, GFP–chDab1-E and GFP–chDab1-L transfectants were stained with an antibody to phosphorylated Src (tyr-416) predicted to recognize all Src family kinases phosphorylated at this residue. GFP–chDab1-L-transfected cells showed strong staining with this antibody compared to GFP–chDab1-E-transfected cells (Figure 6A–D), suggesting that at least some of the proteins detected with the antibody to phosphotyrosine represent activated Src family kinases. Analysis of chDab1-L mutants generated the expected results; that is, activation of Src family kinases in cells transfected with chDab1-L(Y185F), chDab1-L(Y220F) and to a lesser extent chDab1-L(Y200F). However, no induction of phospho-Src(416) was observed in cells transfected with the chDab1-L(Y198F) mutant (data not shown).
Figure 6.

Phospho-Src and GAP-43 analysis of chDab1-transfected primary retinal cultures. GFP–chDab1-E- (A, B, E, F) and GFP–chDab1-L- (C, D, G, H) transfected retinal cultures were immunostained with anti-phospho-Src(416) (A–D) or anti-GAP-43 (E–H) antibodies followed by Alexa 555 goat anti-mouse secondary antibody. (A, C, E, G)A low magnification view of the transfected cultures taken with a × 2.5 objective to demonstrate the overall extent of p-Src(416) and GAP-43 staining. (B, D, F, H) A high magnification view of individual clumps of cells taken with a × 40 objective.
Neuronal maturation is accompanied by neurite outgrowth. To determine whether the enhanced formation of processes observed in chDab1-L transfectants was associated with a more differentiated state, transfected cells were stained with an antibody to neurofilament-H, the heavy subunit of the major intermediate filaments found in the axons of mature neurons. Neurofilament-H was expressed in a significant proportion of both transfected and nontransfected retinal cells, with no obvious differences in staining patterns observed between chDab1-E and chDab1-L transfectants (data not shown).
GAP-43 is a neuronal phosphoprotein associated with axonal outgrowth during development, regeneration and sprouting. In the retina, GAP-43 is primarily found in the axons of ganglion cells, but has also been reported in the processes of amacrine cells (Kapfhammer et al, 1997). A difference in staining pattern was observed between chDab1-E and chDab1-L transfectants using anti-GAP-43 antibody, with a considerably stronger signal observed in GFP–chDab1-L-expressing cells (Figure 6E–H). These results are in agreement with Western blot analysis of whole-cell extracts isolated from GFP–chDab1-transfected cells (Supplementary Figure 2). The intensity of the GAP-43 band was ∼two-fold stronger in chDab1-L transfectants than in chDab1-E and GFP (control) transfectants. In contrast, the signal intensity in all three transfectants was virtually identical for GSK-3β and actin.
ChDab1 isoform switching corresponds to increased levels of phosphotyrosine, activated Src and GAP-43
To determine whether expression of chDab1-L correlates in vivo with the presence of phosphotyrosine, activated Src family kinases and GAP-43, ED5 and ED16 retinal tissue sections were immunostained with antibodies to Dab1, phosphotyrosine, phospho-Src(Y416) and GAP-43. At ED5 (when chDab1-E is predominant), a strong Dab1 cytoplasmic signal was observed throughout the retina (Figure 7A–C). Phosphotyrosine (Figure 7A), phospho-Src (Figure 7B) and GAP-43 (Figure 7C) were mainly found in the emerging NFL containing the axons of ganglion cells. At ED16 (when chDab1-L is predominant), Dab1 was primarily expressed in the NFL and IPL, with a substantial expression in ganglion cells and in the inner half of the INL where the elongated stringy staining pattern was consistent with expression in the cytoplasm and processes of amacrine cells (Figure 7D–F). Phosphotyrosine had a similar distribution pattern, although the signal was comparatively stronger in the OPL (Figure 7D). Phospho-Src expression was strong in ganglion and amacrine cells, although an equally strong signal was also detected in the INL cells (presumably horizontal) adjacent to the OPL (Figure 7E). GAP-43 was predominant in the NFL and in the IPL immediately adjacent to the amacrine cells (Figure 7F). These results indicate a strong correlation between Dab1, phosphotyrosine, activated Src and GAP-43 expression at ED16, with all four expressed in ganglion and amacrine cells and/or their processes.
Figure 7.
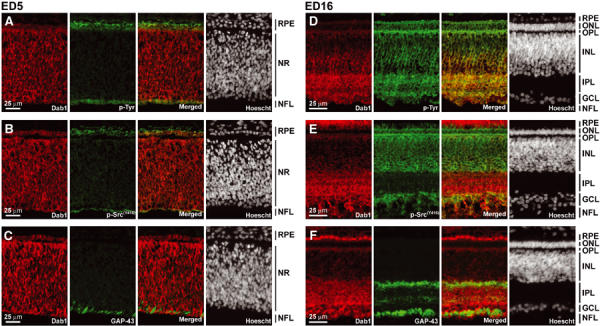
Immunofluorescence analysis of chDab1, phosphotyrosine, phosphorylated Src family kinases and GAP-43 in the developing chick retina. Sections were double stained with anti-Dab1 and anti-phosphotyrosine, anti-phospho-Src(416) [p-Src(416))] or anti-GAP-43 antibodies, followed by counterstaining with the fluorescent dye Hoescht 33258 to label the nuclei. Retinal tissue sections were prepared from ED5 (A–C) and ED16 (D–F) embryos. RPE, retinal pigment epithelium; NR, neural retina; NFL, nerve fiber layer; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer.
Discussion
We report the isolation of two alternatively spliced forms of Dab1 from the developing chick retina: an early form expressed in the undifferentiated precursor cells of the retina and a late form found in amacrine and ganglion cells. Although more abundant in the retina, these two forms are also observed in the developing brain. ChDab1-L encodes a predicted protein of 551 aa and is highly similar throughout its length to human and murine Dab1. In contrast, chDab1-E encodes a predicted protein of 535 aa that contains a deletion of 35 aa and an insertion of 19 aa. While the 19 aa insertion has no recognizable features, the 35 aa deletion results in the loss of tyrosine phosphorylation sites implicated in Reelin–Dab1-mediated signal transduction (Howell et al, 1997b; Keshvara et al, 2001; Arnaud et al, 2003; Bock and Herz, 2003). To date, mammalian counterparts of chDab1-E containing both the 35 aa deletion and the 19 aa insertion have not been described; however, Bar et al (2003) have shown by RT–PCR analysis that exons encoding the 19 aa insertion are transcribed in ED11/12 mouse brain as well as in ED6 chick brain. These exons were not transcribed in later stage embryos or in primary cortical neuron cultures.
Reelin and Dab1 have been shown to play a central role in the positioning of migrating neurons in the cerebral cortex, cerebellum and hippocampus (D'Arcangelo et al, 1995; Howell et al, 1997a; Sheldon et al, 1997). Like the brain, the retina is a highly organized laminated structure characterized by migration of neuronal cells during development, their positioning into specific layers and communication from one nuclear layer to the next through specific synaptic circuitry. Detailed structural analysis of the retina in Reelin-deficient mice has revealed a number of abnormalities, including a decrease in the density of rod bipolar cells, a reduced density of dendrites in the IPL, which could not be attributed to reduced amacrine cell density, and alteration in the layering of amacrine cell processes in the IPL (Rice and Curran, 2000; Rice et al, 2001).
In support of a role for the Reelin–Dab1-signaling pathway in the retina, Reelin expression is elevated in the ganglion cells of the mouse, chicken, turtle and lizard retina (Schiffmann et al, 1997; Bernier et al, 1999, 2000; Goffinet et al, 1999). Dab1 has been detected in the mouse retina shortly after birth and is primarily found in a single layer of amacrine cells, called glycinergic type AII, located immediately next to the IPL (Rice and Curran, 2000). The IPL, containing the processes of Dab1-positive amacrine cells, is also strongly positive for Dab1, with two distinct zones corresponding to the On and Off sublayers. Similar to the mouse retina, we have found chDab1-L expression in amacrine cells, with particularly high levels in the IPL. However, there are a number of differences between mouse and chicken: (i) chick retinal precursor cells express elevated levels of chDab1-E, (ii) chDab1-L is expressed in ganglion cells and (iii) rather than a single layer of amacrine cells, chDab1-L extends through several layers of amacrine cells. Our results suggest a more widespread role for Dab1 in the chick retina than in mouse retina, involving retinoblasts and ganglion cells as well as amacrine cells.
To address the role of chDab1 alternative splicing in chick retina development, we overexpressed chDab1-E and chDab1-L in primary retinal cultures. Our results showed strong induction of tyrosine phosphorylation and Src phosphorylation, specific to chDab1-L-transfected cells. These phosphorylation events were associated with the formation of numerous thin elongated cellular processes reminiscent of neurite outgrowth, a hallmark of differentiated neuronal cells. No significant differences in neurofilament-H staining patterns were observed between chDab1-E and chDab1-L transfectants; however, levels of the axonal protein GAP-43 were significantly increased in chDab1-L transfectants. GAP-43 expression correlates with neurite outgrowth and is postulated to play a fundamental role in elongation and/or guidance of axons by regulating the response of growing axons to intra- and extracellular signals (Benowitz and Routtenberg, 1997). Overexpression and depletion of GAP-43 have previously been associated with the formation of neurite-like filopodia, and reduction in the size of neurites and growth cones, respectively (Aigner and Caroni, 1993, 1995). Our transfection data support a role for GAP-43 in neurite formation, and suggest a link between the Reelin–Dab1-signaling pathway and GAP-43, mediated through activated Src family kinases. A relationship between GAP-43 and the Reelin–Dab1 pathway is supported by the fact that chDab1-L, GAP-43 and phospho-Src are found in the same differentiated cell types in retinal tissue (ganglion and amacrine and/or their processes). A role for Reelin–Dab1 in the formation of processes is in general agreement with the reduced density of dendrites observed in Reelin-deficient mice (Rice et al, 2001) and with a recent report by Niu et al (2004), indicating that Reelin and Dab1 affect dendritic outgrowth from normal hippocampal neurons.
The expression patterns of chDab1-E and chDab1-L in the developing retina and brain, and the specific effects of chDab1-L in primary retinal cultures suggest that both forms of Dab1 may be important in retinal/brain development and differentiation. Although there are no recognizable motifs within the 19 aa insertion in chDab1-E, this insertion may alter some aspect of Dab1 function in precursor cells. The 35 aa deletion specific to chDab1-E spans a region containing three tyrosine phosphorylation sites. One of these sites, tyr-198, and the adjacent tyr-220 have been shown to undergo Reelin-induced tyrosine phosphorylation (Keshvara et al, 2001). In support of a critical role for tyrosine phosphorylation in Reelin-mediated Dab1 activity, mice mutated at all five tyrosines in the tyrosine phosphorylation motifs, including tyr-198 and tyr-220, display phenotypes identical to those of reeler and yotari (Howell et al, 2000). Furthermore, mouse Dab1 protein is phosphorylated specifically in the developing brain as opposed to the adult brain, at a time when cells are expanding and axonal networks are developing (Howell et al, 1997b).
By mutational analysis, we have shown that tyr-198 is required for the formation of elongated processes in our transfected cells. Mutation of tyr-220 and tyr-185 residues had little or no effect on the retinal cells. The partial effect observed upon mutation of tyr-200 suggests that this residue is required for optimal recognition of the tyr-198 phosphorylation motif. In agreement with this, Keshvara et al (2001) observed reduced reactivity of their anti-phospho-tyr198/tyr-200 Dab1 antibody when tyr-200 was mutated. Tyr-185 and tyr-198/tyr-200 are part of two YQXI motifs that bind to Src-like SH2 domains. Tyr-220, along with tyr-232, which resides outside the deletion region, are part of two YXVP motifs that bind to Abl/Nck/Crk-like SH2 domains (Songyang et al, 1993; Howell et al, 1997b). In chDab1-E, Y185-Q becomes linked to V-P, thereby converting this motif from an Src-like SH2-binding domain to an Abl/Nck/Crl SH2-binding domain. Thus, in chDab1-E, the two YQXI motifs are lost, while two YXVP motifs are retained. The alternative splicing event underlying deletion of this region therefore appears to target Src family kinases specifically. We propose that loss of YQXI phoshorylation sites in chDab1-E represents a novel way of uncoupling the Reelin–Dab1 pathway, to ensure that this signaling cascade is not prematurely induced in undifferentiated retinal and brain cells by secreted Reelin. This uncoupling may be especially important in the brain and retina as there is a mixture of proliferating and differentiating cells until relatively late in development. Of note, Reelin has been found to be expressed in the ganglion cells of chick retina as well as in many parts of the brain by ED6, the earliest stage tested (Bernier et al, 2000). An estimated 75% of retinal cells are still in the proliferative stage at ED6 (Dütting et al, 1983)
In summary, we have identified two developmentally regulated alternatively spliced forms of chicken Dab1. We propose that Reelin-responsive chDab1-L plays a role in the formation of neurite extensions mediated through Src family kinase activation and GAP-43 expression, while chDab1-E uncouples transduction of the Reelin signal. ChDab1 isoform switching ensures specific and appropriate responses of different cell types to secreted Reelin during development. The discovery and characterization of the two chDab1 isoforms reveals a novel mechanism for regulating the Reelin–Dab1-signaling pathway and links the function of a particular splice variant to a specific developmental stage.
Materials and methods
DD-PCR analysis
DD-PCR analysis was performed as described (Liang and Pardee, 1992; Godbout and Andison, 1996). Poly(A)+ RNA was extracted from chick retinas at ED3.5, ED5 and ED16, and from chick brains at ED5 and ED16. The cDNAs were generated by reverse transcription of 1 μg poly(A)+ RNA using T11MN primers (where M represents G, A or C and N represents any nucleotide). The cDNAs were PCR-amplified in the presence of 35S-dATP with Taq polymerase (Stratagene) using a T11MN primer and a random decamer oligonucleotide primer (OPA) (Operon Biotechnology Inc.). Amplified cDNAs were electrophoresed on a 6% polyacrylamide-urea gel and visualized by autoradiography. Selected bands were then reamplified and ligated into a pBluescript vector (Stratagene) with a T overhang at the EcoRV site.
Screening of an ED7 chick retina cDNA library
The cDNA library was prepared as previously described (Godbout, 1993), except that the cDNA was produced from ED7 poly(A)+ RNA. Approximately 2 × 105 bacteriophage were filter-lifted and hybridized with the DD-PCR-isolated 532 bp chDab1 cDNA fragment. Purified clones were obtained after three rounds of screening. An ABI 310 automated sequencer was used for sequencing of the cDNAs using a combination of sequential deletion and sequence-specific oligomers.
Northern blot analysis
Poly(A)+ RNA was isolated from the retina, brain, heart, liver, kidney and gut at the developmental stages indicated in Figure 1 legend. In all, 2 μg of each poly(A)+ RNA were electrophoresed in a 6% formaldehyde–1.5% agarose gel in MOPS buffer and transferred to nitrocellulose. The 532 bp chDab1 band, obtained using T11MG and OPA-10 (5′-GTGATCGCAG-3′), was used to probe the blot. Filters were washed at 55°C in 0.1 × SSC, 0.1% SDS and visualized by autoradiography. Hybridization to mouse actin cDNA was used as a control for mRNA level variation.
Western blot analysis
Whole-cell lysates were electrophoresed in a 10% polyacrylamide–SDS gel followed by electroblotting onto nitrocellulose. Blots were incubated with either rabbit anti-Dab1 antibody (1:5000 dilution) (Rockland Immunochemicals), goat anti-actin antibody (I-19) (1:500) (Santa Cruz Biotechnology), mouse anti-phosphotyrosine antibody (P-Tyr-1000) (1:1000) (Cell Signaling Technologies), mouse anti-GAP-43 antibody (GAP-7B10) (1:500) (Sigma) or mouse anti-GSK-3β antibody (1:2500) (BD Biosciences). Primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) using the ECL detection system (Amersham Pharmacia Biotech).
RT–PCR analysis
In all, 1 μg poly(A)+ RNA from the retina (ED5, ED7, ED10, ED16) and brain (ED3.5, ED5, ED16) was reverse transcribed using an oligo d(T) primer and Superscript reverse transcriptase (Stratagene). PCR was carried out using 1/20 of the cDNA generated. Primer set P1 and P2 was used for the analysis of the deletion region, primer set P3 and P4 was used for the analysis of the insertion region, and primer set P1 and P4 was used for the analysis of both the deletion and insertion regions (Figure 2). DNA fragments were run in 10% (P1/P2 and P3/P4 primer sets) and 15% (P1/P4 primer set) polyacrylamide gels. For sequencing, amplified DNAs were cloned into the pBluescript-T overhang vector as described in‘DD-PCR analysis'.
In situ hybridization
Digoxigenin (DIG)-labeled sense and antisense riboprobes were generated by in vitro transcription of linearized plasmids using T3 or T7 polymerase (Roche). Riboprobes were prepared from the 532 bp chDab1 cDNA fragment isolated by DD-PCR, a 77 bp cDNA generated by PCR amplification of chDab1-E using primer set P7 and P8 flanking the insertion region (Figure 2), a 147 bp cDNA fragment generated by PCR amplification of chDab1-L using primer set P9 and P10 flanking the deletion region (Figure 2), a 2.0 kb CA-II cDNA fragment (Godbout, 1993) and a 1.2 kb AP-2β cDNA fragment (Bisgrove and Godbout, 1999). Tissues were fixed in 4% phosphate-buffered saline (PBS)-buffered paraformaldehyde at 4°C, cryoprotected with 12, 16 and 18% sucrose and embedded in OCT (Tissue-Tek, Miles Inc., Elkhart, IN, USA). Frozen sections (6–8 μm) were prehybridized at 50–55°C in 40% formamide, 10% dextran sulfate, 1 × Denhardt's solution, 4 × SSC, 10 mM DTT, 1 mg/ml yeast tRNA and 1 mg/ml heat denatured herring testis sperm DNA. Riboprobes were heat-denatured and hybridized to tissue sections overnight at 50–55°C. Tissue sections were washed as described (Belecky-Adams et al, 1997) and incubated with alkaline-phosphatase (AP)-conjugated anti-DIG antibody. The signal was detected with BCIP/NBT after polyvinyl alcohol enhancement (Jowett, 1997).
Transfection analysis
For the construction of GFP-fusion constructs, full-length chDab1-E and -L cDNAs were generated by RT–PCR using poly(A)+ RNAs from ED5 and ED16 chick retinas, respectively. The cDNAs were amplified with a mixture of Taq/Pfu polymerase (100:1) using the P5/P6 primer set spanning the entire open reading frame. BamHI and EcoRI restriction endonuclease sites were added to the ends of the oligonucleotides to allow in-frame cloning. cDNAs were purified, digested with EcoRI and BamHI and cloned into the pEGFP-C1 vector (Clontech). Both cDNAs were sequenced to ensure that they were error-free. Primary retinal cultures were prepared from ED5 chick retinas trypsinized prior to plating onto glass coverslips (one-twelfth of a retina per 12 mm coverslip). Cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum and incubated in a 5% CO2 humidified chamber. Cells were transfected by calcium phosphate DNA precipitation and the DNA was removed after 16 h. After 30 h, the cells adhering to coverslips were fixed with 4% paraformaldehyde in PBS for 10 min and permeabilized for 5 min in 0.5% Triton X-100/PBS. Cells were incubated overnight with mouse anti-phosphotyrosine antibodies (PT-66 (1:250) (Sigma), P-Tyr-1000 (1:250) (Cell Signaling Technologies)), anti-phospho-Src(Y416) (9A6) (1:50) (Upstate) or anti-GAP-43 antibody (GAP-7B10) (1:250) (Sigma) followed by Alexa 555 goat anti-mouse secondary antibody (Molecular Probes) (1:200) for 1 h. The coverslips were mounted on slides using glycerol containing 1 mg/ml p-phenylenediamine+1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI). Cells were viewed on a Zeiss LSM 510 confocal microscope.
Site-directed mutation analysis
Site-directed mutagenesis of chDab1-L tyr-185, tyr-198, tyr-200, tyr-220 and tyr-185/tyr-198 was carried out by sequential PCR (Cormack and Castano, 2002). Partially complementary primers containing a point mutation corresponding to a tyr → phe substitution (TA(T/C) → TT(T/C)) were used in conjunction with pEGFP-C1 vector primers located upstream of the EcoRI site and downstream of the BamHI site to generate DNA fragments corresponding to full-length chDab1-L, each mutated at a specific tyrosine residue. DNA fragments were annealed, extended and amplified using pEGFP-C1 vector primers. The DNA was digested with EcoRI and BamHI and cloned into pEGFP-C1. Constructs were sequenced to ensure that they were error-free. Expression of full-length GFP–chDab1-L mutant proteins was confirmed by transfection and Western blot analysis.
Immunofluorescence of retinal sections
Retinal tissue sections were prepared as described in ‘In situ hybridization'. Frozen sections were rehydrated in PBS, fixed in 4% PBS-buffered paraformaldehyde and permeabilized in 1% PBS-buffered NP-40. Sections were double-stained with rabbit polyclonal anti-Dab1 (1:500) and either mouse anti-phosphotyrosine (1:500), anti-phospho-Src(Y416) (1:25) or anti-GAP-43 (1:250) antibodies, followed by fluorescent secondary antibodies (Alexa 555 goat anti-rabbit and Alexa 488 goat anti-mouse) (1:150). Sections were counterstained with the Hoescht 33258 fluorescent nuclear stain (Molecular Probes) and mounted with Fluorosave (Calbiochem). Images were collected with a Zeiss-Axioplan II microscope (Carl Zeiss) equipped with a cooled charge-coupled device camera (Cooke Corporation).
Supplementary Material
Supplemental data on-line
Acknowledgments
We are grateful to Mary Packer and Randy Andison for excellent technical assistance, Laith Dabbagh for his invaluable assistance with tissue sectioning and Dr Xuejun Sun, Manager of the Cell Imaging Facility, for his help with the preparation of figures. We thank Dr Jonathan A Cooper for an initial aliquot of anti-Dab1 antibody. We also thank Drs Gordon Chan and D Alan Underhill for critical reading of the paper. This work was supported by the Canadian Institutes of Health Research. SK was supported by a graduate studentship award from the Alberta Cancer Foundation.
References
- Aigner LS, Caroni P (1993) Depletion of 43-kD growth associated protein in primary sensory neurons leads to diminished formation and spreading of growth cones. J Cell Biol 123: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner LS, Caroni P (1995) Absence of persistent spreading, branching, and adhesion in GAP-43-depleted growth cones. J Cell Biol 128: 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen HM, Smith SA, Melis R, Williams B, Holik P, Stevens J, White R (1996) Sequence, genomic structure, and chromosomal assignment of human DOC-2. Genomics 33: 207–213 [DOI] [PubMed] [Google Scholar]
- Arnaud L, Ballif BA, Förster E, Cooper JA (2003) Fyn tyrosine kinase is a critical regulator of disabled-1 during brain development. Curr Biol 13: 9–17 [DOI] [PubMed] [Google Scholar]
- Ballif BA, Arnaud L, Cooper JA (2003) Tyrosine phosphorylation of Disabled-1 is essential for Reelin-stimulated activation of Akt and Src family kinases. Mol Brain Res 117: 152–159 [DOI] [PubMed] [Google Scholar]
- Bar I, Tissir F, Lambert de Rouvroit C, De Backer O, Goffinet AM (2003) The gene encoding disabled-1 (Dab1), the intracellular adaptor of the reelin pathway, reveals unusual complexity in human and mouse. J Biol Chem 278: 5802–5812 [DOI] [PubMed] [Google Scholar]
- Beffert U, Morfini G, Bock HH, Reyna H, Brady ST, Herz J (2002) Reelin-mediated signaling locally regulates protein kinase B/Akt and glycogen synthase kinase 3β. J Biol Chem 277: 49958–49964 [DOI] [PubMed] [Google Scholar]
- Belecky-Adams T, Tomarev S, Li H, Ploder L, McInnes RR, Sundin O, Adler R (1997) Pax-6, Prox 1 and Chx10 homeobox gene expression correlates with phenotypic fate of retinal precursor cells. Invest Opthalmol Vis Sci 38: 1293–1303 [PubMed] [Google Scholar]
- Benhayon D, Magdaleno S, Curran R (2003) Binding of purified Reelin to ApoER2 and VLDLR mediates tyrosine phosphorylation of Disabled-1. Mol Brain Res 112: 33–45 [DOI] [PubMed] [Google Scholar]
- Benowitz L, Routtenberg A (1997) GAP-43: an intrinsic determinant of neuronal development and plasticity. Trends Neurosci 20: 84–91 [DOI] [PubMed] [Google Scholar]
- Bernier B, Bar I, D'Arcangelo G, Curran T, Goffinet AM (2000) Reelin mRNA expression during embryonic brain development in the chick. J Comp Neurol 422: 448–463 [PubMed] [Google Scholar]
- Bernier B, Bar I, Pieau C, Lambert de Rouvroit C, Goffinet AM (1999) Reelin mRNA expression during embryonic brain development in the turtle Emys orbicularis. J Comp Neurol 413: 463–479 [DOI] [PubMed] [Google Scholar]
- Bisgrove DA, Godbout R (1999) Differential expression of AP-2α and AP-2β in the developing chick retina: repression of R-FABP promoter activity by AP-2. Dev Dyn 214: 195–206 [DOI] [PubMed] [Google Scholar]
- Bock HH, Herz J (2003) Reelin activates Src family tyrosine kinases in neurons. Curr Biol 13: 18–26 [DOI] [PubMed] [Google Scholar]
- Cormack B, Castano I (2002) Introduction of point mutations into cloned genes. Meth Enzymol 350: 199–218 [DOI] [PubMed] [Google Scholar]
- D'Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T (1999) Reelin is a ligand for lipoprotein receptors. Neuron 24: 471–479 [DOI] [PubMed] [Google Scholar]
- D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T (1995) A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374: 719–723 [DOI] [PubMed] [Google Scholar]
- Dowling JE (1987) The Retina: An Approachable Part of the Brain Cambridge: The Belknap Press of Harvard University Press [Google Scholar]
- Dütting D, Giere A, Hansmann G (1983) Self-renewal of stem cells and differentiation of nerve cells in the developing chick retina. Dev Brain Res 10: 21–32 [DOI] [PubMed] [Google Scholar]
- Godbout R (1993) Identification and characterization of transcripts present at elevated levels in the undifferentiated chick retina. Exp Eye Res 56: 95–106 [DOI] [PubMed] [Google Scholar]
- Godbout R, Andison R (1996) Elevated levels of cyclin D1 mRNA in the undifferentiated chick retina. Gene 182: 111–115 [DOI] [PubMed] [Google Scholar]
- Goffinet AM, Bar I, Bernier B, Trujillo C, Raynaud A, Meyer G (1999) Reelin expression during embryonic brain development in the lacertilian lizard. J Comp Neurol 414: 533–550 [DOI] [PubMed] [Google Scholar]
- Herrick TM, Cooper JA (2002) A hypomorphic allele of Dab1 reveals regional differences in reelin–Dab1 signaling during brain development. Development 129: 787–796 [DOI] [PubMed] [Google Scholar]
- Howell BW, Gertler FB, Cooper JA (1997b) Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO J 16: 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BW, Hawkes R, Soriano P, Cooper JA (1997a) Neuronal position in the developing brain is regulated by mouse disabled-1. Nature 389: 733–737 [DOI] [PubMed] [Google Scholar]
- Howell BW, Herrick TM, Cooper JA (1999) Reelin-induced tyrosine phosphorylation of disabled 1 during neuronal positioning. Genes Dev 13: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BW, Herrick TM, Hildebrand JD, Zheng Y, Cooper JA (2000) Dab1 tyrosine phosphorylation sites relay positional signals during mouse brain development. Curr Biol 10: 877–885 [DOI] [PubMed] [Google Scholar]
- Ishiguro K, Shiratsuchi A, Sato S, Omori A, Arioka M, Kobayashi S, Uchida T, Imahori K (1993) Glycogen synthase kinase 3 beta is identical to tau protein kinase I generating several epitopes of paired helical filaments. FEBS Lett 325: 167–172 [DOI] [PubMed] [Google Scholar]
- Jowett T (1997) Alternative enzyme substrates. In Tissue In Situ Hybridization: Methods in Animal Development, Jowett T (ed), pp 29–32. New York, USA: John Wiley & Sons, Inc [Google Scholar]
- Kapfhammer JP, Christ F, Schwab ME (1997) The growth-associated protein GAP-43 is specifically expressed in tyrosine hydroxylase-positive cells of the rat retina. Dev Brain Res 101: 257–264 [DOI] [PubMed] [Google Scholar]
- Keshvara L, Benhayon D, Magdaleno S, Curran T (2001) Identification of reelin-induced sites of tyrosyl phosphorylation on disabled-1. J Biol Chem 276: 16008–16014 [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257: 967–971 [DOI] [PubMed] [Google Scholar]
- Niu S, Renfro A, Quattrocchi CC, Sheldon M, D'Arcangelo G (2004) Reelin promotes hippocampal dendrite development through the VLDLR/ApoER2-Dab1 pathway. Neuron 41: 71–84 [DOI] [PubMed] [Google Scholar]
- Ohkubo N, Lee YD, Morishima A, Terashima T, Kikkawa S, Tohyama M, Sakanaka M, Tanaka J, Maeda N, Vitek MP, Mitsuda N (2003) Apolipoprotein E and Reelin ligands modulate tau phosphorylation through an Apolipoprotein E receptor/disabled-1/glycogen synthase kinase-3beta cascade. FASEB J 17: 295–307 [DOI] [PubMed] [Google Scholar]
- Rice DS, Curran T (2000) Disabled-1 is expressed in type AII amacrine cells in the mouse retina. J Comp Neurol 424: 327–338 [DOI] [PubMed] [Google Scholar]
- Rice DS, Nusinowitz S, Azimi AM, Martinez A, Soriano E, Curran T (2001) The reelin pathway modulates the structure and function of retinal synaptic circuitry. Neuron 31: 929–941 [DOI] [PubMed] [Google Scholar]
- Schiffmann SN, Bernier B, Goffinet AM (1997) Reelin mRNA expression during mouse brain development. Eur J Neurosci 9: 1055–1071 [DOI] [PubMed] [Google Scholar]
- Sheldon M, Rice DS, D'Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell BW, Cooper JA, Goldowitz D, Curran T (1997) Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature 389: 730–733 [DOI] [PubMed] [Google Scholar]
- Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, Neel BG, Birge RB, Fajardo JE, Chou MM, Hanafusa H, Schaffhausen B, Cantley LC (1993) SH2 domains recognize specific phosphopeptide sequences. Cell 72: 767–778 [DOI] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J (1999) Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 97: 689–701 [DOI] [PubMed] [Google Scholar]
- Turner DL, Snyder EY, Cepko CL (1990) Lineage-independent determination of cell type in the embryonic mouse retina. Neuron 4: 833–845 [DOI] [PubMed] [Google Scholar]
- Vardimon L, Fox LE, Moscona AA (1986) Developmental regulation of glutamine synthetase and carbonic anhydrase II in neural retina. Proc Natl Acad Sci USA 83: 9060–9064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte RL, Godbout R (2002) Expression of spermidine/spermine N(1)-acetyltransferase in the Müller glial cells of the developing chick retina. Exp Eye Res 74: 605–613 [DOI] [PubMed] [Google Scholar]
- Xu XX, Yang WN, Jackowski S, Rock CO (1995) Cloning of a novel phosphoprotein regulated by colony-stimulating factor 1 shares a domain with the Drosophila disabled gene product. J Biol Chem 270: 14184–14191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data on-line


