Abstract
Gastric cancer including the cardia and non-cardia types is the second frequent cause of cancer-related deaths worldwide. A subset of non-cardia gastric cancer genetic susceptibility loci have been addressed among Asian through genome-wide association studies (GWASs). This study was to evaluate the effects of single nucleotide polymorphisms (SNPs) of long intergenic non-coding RNAs (lincRNAs) on non-cardia gastric cancer susceptibility in Chinese populations. We selected long intergenic noncoding RNAs (lincRNAs) located in non-cardia gastric cancer risk-related loci and identified 10 SNPs located within lincRNA exonic regions. We examined whether genetic polymorphisms in lincRNAs exons are associated with non-cardia gastric cancer risk in 438 non-cardia gastric cancer patients and 727 control subjects in Chinese populations using logistic regression. Functional relevance was further examined by biochemical assays. We found that lincRNA-NR_024015 rs8506AA carrier was significantly associated with risk of non-cardia gastric cancer (adjusted odds ratio [OR] = 1.56, 95%CI = 1.03–2.39, compared with the rs8506 AG or GG genotype. Further stratification analysis showed that the risk effect was more pronounced in subgroups of smokers (P = 0.001). Biochemical analysis demonstrated that the G to A base change at rs8506G>A disrupts the binding site for has-miR-526b, thereby influencing the transcriptional activity of lincRNA-NR_024015 and affecting cell proliferation. Our present study established a robust association between the rs8506G>A polymorphism in the lincRNA-NR_024015 exon and the risk of non-cardia gastric cancer.
Introduction
Gastric cancer (GC) is the second leading cause of cancer death worldwide after lung cancer in 2010, although mortality deaths have decreased slightly from 774,000 in 1990 to about 755,000 in 2010 [1],[2]. Epidemiological studies have showed that environmental factor, including diet, tobacco smoking, alcoholic consumptions and, especially, infection with Helicobacter pylori are associated with a higher risk for GC [3], [4]. Despite of these recognized risk factors, researchers still convinced that genetic factors, particularly single nucleotide polymorphisms (SNPs), are likely to play an essential role in an individual's risk of developing gastric cancer as only a fraction of exposed individuals develop gastric cancer [5].
To date, with the advance of next generation transcriptome sequencing (RNA-Seq), there has been a profound shift in our understanding the entire set of transcriptional aberrations in a disease, including novel transcripts and non-coding RNAs (ncRNAs) not measured by conventional analyses [6]–[9]. Of all of the currently characterized classes of non-coding RNAs molecules, these have been called long intervening ncRNAs (lincRNAs) longer than 200 nucleotides (nt) that are lack an open reading frame and do not overlap protein-coding genes [7], [10]. Groups of lincRNAs have been well characterized to some extent and demonstrated to correlated with important cellular processes such as imprinting, X chromosome inactivation, pluripotency maintenance, and transcriptional regulation [10]–[15]. Furthermore, emerging evidence of dysregulated lincRNA expression in numerous cancers have emerged lincRNA as a new aspect of biology, with evidence suggesting that a major role for involvement of lincRNA in human tumorigenesis and metastasis [16], [17]. Indeed, a well-described example, HOTAIR have been studied the contributions to the stepwise progression of tumorigenesis [11], [18], highlighting the role of lncRNAs in cancer biology. In addition, the long noncoding RNA MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), is frequent misregulation and as a predictive marker for a variety of human cancers of the colon, breast and prostate [19]–[22]. Nevertheless, the mechanisms underlying the specific function of lincRNAs in cancer development has not been fully delineated.
In the past decade, multiple unbiased genome-wide association studies (GWAS) have broadened our understanding of genetic variations related to different types of diseases and cancers by high throughput technologies [23]; however, at least one-third of the identified variants are within non-coding intervals [24]. Recently, two GWAS revealed that several susceptibility risk loci that are associated with non-cardia gastric cancer (NCGC) risk in a Chinese population. Bioinformatics analysis has uncovered numerous lincRNAs close to these loci. Furthermore, several relevant single nucleotide polymorphisms (SNPs) located in the exonic regions of lincRNAs that may associate with NCGC were identified; however, the association between genetic variations in lincRNAs exons and cancer susceptibility has rarely been reported.
In the present study, we hypothesized that SNPs in the exonic region of lincRNAs may altered expression levels and thereby may contribute to NCGC. To test this hypothesis, we conducted a hospital-based case-control study to investigate the associations between these SNPs and susceptibility to NCGC in a Chinese population.
Materials and Methods
Study Subjects
All subjects in the current study were ethnically homogenous Han Chinese including 438 NCGC patients and 727 healthy controls. Patients who underwent surgery at the Affiliated Hospitals of Soochow University (Suzhou) were consecutively recruited from 2003 to 2009, with a response rate of 94%. Patients were from Suzhou city and its surrounding regions, and there were no age, sex, and histology restrictions. Details regarding the clinical features of the patients are summarized in Table 1 . The tumor, node, metastasis (TNM) classification and tumor staging were evaluated according to the 2002 American Joint Committee on Cancer Staging system. Population controls were cancer-free people living in Suzhou region; they were selected from a nutritional survey conducted in the same period as the cases were collected. The control samples were available to us from previous studies which were randomly selected from a database consisting of 3500 individuals based on a physical examination [25]–[27]. The measurement for serum H.pylori immunoglobulin G in NCGC patients and controls was determined by enzyme-linked immunosorbent assay (ELISA). This study was approved by the medical ethics committee of Soochow University. All the participants were genetically-unrelated ethnic Han Chinese and none had blood transfusion in the last 6 months. Having given a written informed consent, each participant was scheduled for an interview with a structured questionnaire to collect selected information, and to donate 5 ml peripheral blood.
Table 1. Distributions of select characteristics among non-cardia gastric cancer patients and controls in Chinese populations.
| Characteristics | Cases | Controls | ||
| N (438) | (%) | N (727) | (%) | |
| Age(years) | ||||
| ≤60 | 156 | 35.6 | 283 | 38.9 |
| >60 | 282 | 64.4 | 444 | 61.1 |
| Sex | ||||
| Male | 322 | 73.5 | 491 | 67.5 |
| Female | 116 | 26.5 | 236 | 32.5 |
| Smoking status | ||||
| Positive | 189 | 43.2 | 221 | 30.4 |
| Negative | 249 | 56.8 | 506 | 69.6 |
| Drinking | ||||
| Positive | 175 | 39.9 | 267 | 36.7 |
| Negative | 263 | 60.1 | 460 | 63.3 |
| Family history | ||||
| Positive | 34 | 7.8 | 51 | 7.0 |
| Negative | 404 | 92.2 | 676 | 93.0 |
| H.pylori status | ||||
| H. pylori positive | 322 | 73.5 | ||
| H. pylori negative | 116 | 26.5 | ||
| TNM stage | ||||
| I | 104 | 23.7 | ||
| II | 90 | 20.6 | ||
| III | 169 | 38.6 | ||
| IV | 75 | 17.1 | ||
SNP Selection
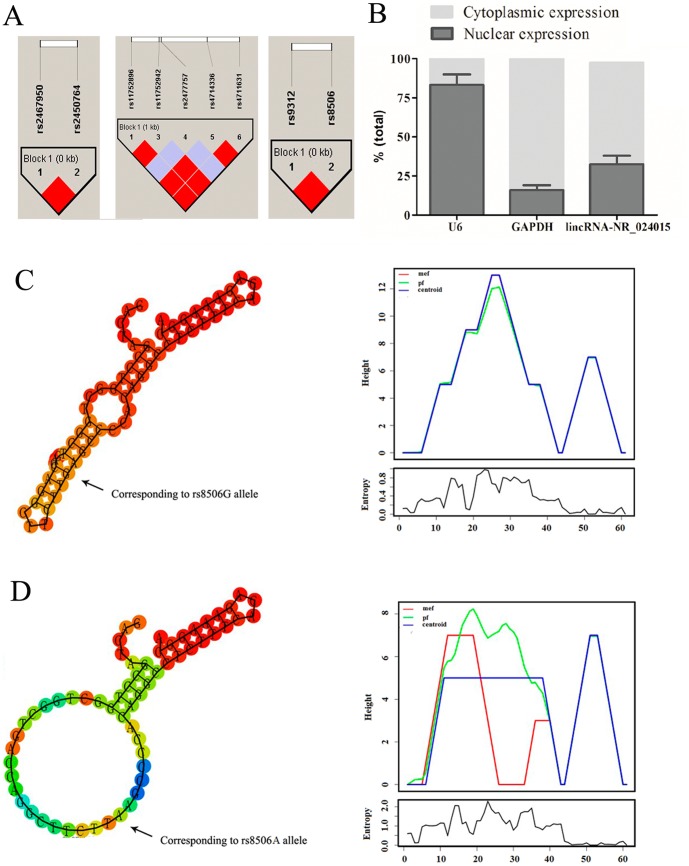
All published literature investigating an association between genetic susceptibility and NCGC risk were eligible. We searched for studies up to August 2013 using the PubMed database and Web of Science. Relevant search terms were “genome-wide association study”, “GWAS”, “NCGC”, “gastric cancer”, “stomach cancer”, and “Asian”. We also manually searched the reference lists in selected articles. We firstly excluded some articles by scanning the titles and abstracts of studies that were not written in English. Then, after reading the full text of the remaining articles, we identified a final set of studies. All the selected studies met the following criteria: (1) the outcome investigated was based on GWAS in relation to NCGC in humans; (2) the articles were published in English; (3) the latest studies were selected among overlapping data and duplicated data; (4) GWAS was conducted using chip technology. The major exclusion criteria were (1) reviews, tutorials, letters, and editorials; (2) duplicate data; (3) not a case-control design; and (4) overlapping data or data dumped by the latest reports. Next, we scoured the susceptibility loci identified by GWAS for NCGC in selected articles, 4 lincRNAs that did not overlap with any recognized genes within a 1-MB range of these loci, in either direction (a total span of 2 MB) were eventually identified from the human lincRNAs database [28]. Furthermore, Haploview software 4.2 was used for bioinformatics analysis of haplotype block based on the Chinese Han Beijing (CHB) population data in HapMap (HapMap Data Release 27 Phase II+III, February 2009, on the NCBI B36 assembly, dbSNP b126). These SNPs with minor allele frequencies of greater than 5% in the Chinese population was extracted. There were three haplotype blocks in the Chinese population ( Figure 1A ). The haplotype-tagging SNPs were selected with the Haploview software 4.2 Tagger program; it was found that the rs11752896, rs11752942, rs4714336 and rs4711631 covered the haplotype in block 1 at a 100% frequency (rs11752896 and rs11752942: D′ = 1.0, r2 = 1.0; rs11752896 and rs4714336: D′ = 1.0, r2 = 1.0; rs11752896 and rs4711631: D′ = 1.0, r2 = 1.0). In addition, rs2467950, rs2450764 and rs9312, rs8506 covered the haplotype in block 1 at a 100%, respectively (rs2467950 and rs2450764: D′ = 1.0, r2 = 1.0; rs9312 and rs8506: D′ = 1.0, r2 = 1.0). SNPs rs2304285 and rs2477757 are outside the blocks. Therefore, the SNPs rs11752896, rs2467950, rs8506, rs2477757 and rs2304285 were chosen as five potential functional SNPs in the exonic of the selected lincRNAs to be analyzed for their associations with risk of gastric cancer.
Figure 1. Cellular and molecular characterization of lincRNA-NR_024015.
(A) Linkage disequilibrium (LD) map for selected SNPs of lincRNA-NR_024015 exon. It showed that rs11752896, rs11752942, rs4714336 and rs4711631 are in LD with D′ = 1; rs2467950 and rs2450764 are in LD with D′ = 1; rs9312 and rs8506 are in LD with D′ = 1. (B) The levels of nuclear control transcript (U6), cytoplasmic control transcript (GAPDH mRNA), and lincRNA-NR_024015 were assessed by RT-qPCR in nuclear and cytoplasmic fractions. Data are mean±standard error of the mean. Data are presented as a percentage of U6, GAPDH and lincRNA-NR_024015 levels and total levels for each were taken to be 100%. (C–D) In-silico prediction of folding structures induced by rs8506G>A in lincRNA-NR_024015. The mountain plot is an xy-graph that represents a secondary structure including MFE structure, the thermodynamic ensemble of RNA structures (pf), and the centroid structure in a plot of height versus position. “mfe” represents minimum free energy structure; “pf” indicates partition function; “centroid” represents the centroid structure.
Genotyping Analysis
Genome DNA was extracted from peripheral blood lymphocytes of the study subjects. Allele-specific MALDI-TOF mass spectrometry was used to genotype the markers used in the association analyses, as previously described [27], [29]. A total of 60 samples were randomly selected for direct sequencing to confirm the genotyping results from the mass spectrometric analysis, and the results were in 100% agreement. Approximately, 10% of the samples were also randomly selected for a blinded repeat of the genotyping without prior knowledge of the previous genotyping result or the status of being a case and control, and the results were in 100% agreement.
Cell Culture
The 293T or HGC-27 cells were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, Shanghai Institute of Cell Biology, and were passaged for fewer than 6 months. The 293T or HGC-27 cells were maintained in DMEM with high glucose (Gibco-BRL, Gaithersburg, MD, USA) or RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (Gibco-BRL, Gaithersburg, MD, USA) and 50 µg/ml streptomycin (Gibco-BRL, Gaithersburg, MD, USA) at a 37°C in the presence of 5% CO2.
Subcellular Fractionation
HGC-27 cells were cultured in a humidified incubator for 2 days. For subcellular fractionation experiments, up to 2×106 cells were used. Cytosolic and nuclear extracts from breast cancer cells were collected using a Nuclear/Cytosol Fractionation kit (Biovision, USA) according to the manufacturer's instructions.
In-silico Prediction of Folding Structures Induced by Rs8506G>A in LincRNA-NR_024015
As certain structures are more likely to play key roles in biological functions; thus we used RNAfold and SNPfold algorithms to predict the putative influence of rs8506G>A on the local folding structures of lincRNA-NR_024015 by analyzing the 61-bp regions flanking the polymorphism.
Construction of Reporter Plasmids
Two reporter plasmids containing 160 bp lincRNA-NR_024015 exon region fragment flanking the rs8506G or rs8506A allele were synthesized by the Genewiz Company (Suzhou, China) and then cloned into the psiCHECK-2 basic vector (Promega, Madison, WI) (Figure 1B). The resulting construct with lincRNA-NR_024015 rs8506 SNP (psiCHECK-2-lincRNA-rs8506G and psiCHECK-2-lincRNA- rs8506A) was confirmed by sequencing.
Transient Transfections and Luciferase Assays
Bioinformatics analysis revealed that the rs8506G>A polymorphism locate at the binding site of microRNA has-miR-526b (http://snpinfo.niehs.nih.gov/). Thereby, the mimics and inhibitors of has-miR-526b (GenePharma Co, Shanghai) were applied to analyze the effect of has-miR-526b on psiCHECK-2-lincRNA-rs8506 reporter genes in vitro. The 293T or HGC-27 cells were seeded in 24-well plates (1×105 cells per well) and cultured to 60–70% confluence before transfection; cells were then transfected with the reporter plasmids described above using Lipofectamine 2000 (Invitrogen, CA, USA) as previously described [25]. In each well, co-transfection was performed using 800 ng of constructed plasmid DNA and 0, 1, or 40 pmol microRNA has-miR-526b mimics (Shanghai GenePharma Co., Ltd.), and with or without 40 pmol has-miR-526b inhibitor, according to the manufacturer's instructions. The luciferase activity was measured with the Dual-Luciferase Reporter assay system (Promega, Madison, WI, USA) using a TD-20/20 luminometer (Turner Biosystems, Sunnyvale, CA, USA), and the results were normalized against the activity of the Renilla luciferase gene. Each group included 6 replicates, and independent triplicate experiments were performed.
Expression Vector Construction
To further study the role of lincRNA-NR_024015 in cancer progression, the full-length cDNA of lincRNA-NR_024015 harboring rs8506G and rs8506A alleles were synthesized by the Genewiz Company (Suzhou, China) and then cloned into the pcDNA3.1 vectors. The resulting construct with lincRNA-NR_024015 rs8506 SNP (pcDNA-lincRNA-rs8506G and pcDNA-lincRNA-rs8506A) was confirmed by sequencing.
RNA Isolation and Quantitative RT-PCR Analysis
Thirty-two NCGC tissue specimens were obtained from biopsies of individual patients and stored at −80°C before analysis. Total RNA was obtained from these cancerous tissues with TRIzol reagent (Molecular Research Center, Inc). cDNA was generated from mRNA using the random primer and Superscript II (Invitrogen) according to the manufacturer's protocol. Real-time quantitative polymerase chain reaction (RT-PCR) was carried out to quantify the relative gene expression of lincRNA-NR_024015, using an ABI Prism 7500 sequence detection system (Applied Biosystems) based on the SYBR-green method, and GAPDH was used as an internal reference gene in each reaction.
Cell Visibility Assay
In 96-well, flat-bottomed plates (BD Biosciences, Bedford, MA), 100 µL HGC-27 cells cotransfected with pcDNA-lincRNA-rs8506SNP and has-miR-526b or control were aliquoted into each well. Cell viability was measured by Cell Counting Kit-8 (CCK-8) (Dojindo Laboratory, Kumamoto, Japan) based on the manufacturer's instructions.
Statistical Analysis
The differences in the distributions of selected demographic variables between cases and controls, as well as the allele and genotype frequencies were assessed by two-sided chi-squared tests. Unconditional logistic regression models were used to estimate the associations of genotypes of SNPs with risk of gastric cancer by odds ratios (OR) and their 95% confidence intervals (CIs), followed by stratification analysis by age, sex, smoking and drinking status. Logistic regression modeling was used in the trend test, as well as to evaluate the potential multiplicative and additive gene-gene and gene-environmental factor interactions. Furthermore, the data were further stratified by sub-groups of the clinic-pathological variables. Statistical power was computed by applying the PS software (http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize, accessed Dec 14, 2010). One-way ANOVA test was used to evaluate the effect of various SNPs on the lincRNA-NR_024015 transcript expression. All tests were two-sided by using the SAS software (version 9.1; SAS Institute, Cary, NC, USA). A P<0.05 was used as the criterion for statistical significance.
Results
Genotypes and Risk of Non-cardia Gastric Cancer
In the present study, five SNPs were genotyped between cases and controls. As shown in Table 2 , a significant association with NCGC risk was observed for rs8506G>A. Specifically, the results of genotyping showed that compared with the rs8506GG or AG genotype, lincRNA-NR_024015 rs8506AA carrier was significantly associated with risk of non-cardia gastric cancer (adjusted odds ratio [OR] = 1.56, 95%CI = 1.03–2.39). In addition, no significant differences were examined in the other four SNPs (P>0.05). Thus, we could conclude that genetic variant rs8506G>A polymorphism in lincRNA-NR_024015 plays a significantly role in mediating the risk of NCGC.
Table 2. Associations between five SNPs in lincRNAs and risk of non-cardia gastric cancer in Chinese population.
| Genotype | Patients (438) | Controls (727) | Adjusted OR (95% CI)a | P trend | ||
| No. | (%) | No. | (%) | |||
| rs11752896 | ||||||
| AA | 222 | (50.7) | 354 | (48.7) | 1.00 (Reference) | |
| AG | 183 | (41.8) | 319 | (43.9) | 0.90 (0.69–1.16) | |
| GG | 33 | (7.5) | 54 | (7.4) | 0.96 (0.61–1.60) | 0.25 |
| rs2467950 | ||||||
| AA | 17 | (3.9) | 24 | (3.3) | 1.00 (Reference) | |
| AG | 93 | (21.2) | 182 | (25.0) | 0.71 (0.32–1.46) | |
| GG | 328 | (74.9) | 521 | (71.7) | 0.88 (0.64–1.75) | 0.67 |
| rs2477757 | ||||||
| CC | 300 | (68.5) | 516 | (71.0) | 1.00 (Reference) | |
| CT | 138 | (31.5) | 211 | (29.0) | 1.11 (0.84–1.45) | 0.37 |
| rs2304285 | ||||||
| CC | 249 | (56.8) | 432 | (59.4) | 1.00 (Reference) | |
| CT | 189 | (43.2) | 295 | (40.6) | 1.10 (0.83–1.39) | 0.39 |
| rs8506 | ||||||
| GG | 226 | (51.6) | 418 | (57.5) | 1.00 (Reference) | |
| AG | 164 | (37.4) | 256 | (35.2) | 1.17 (0.89–1.51) | |
| AA | 48 | (11.0) | 53 | (7.3) | 1.66 (1.06–2.60) | 0.01 |
| GG+AG | 390 | (89.0) | 674 | (92.7) | 1.00 (Reference) | |
| AA | 48 | (11.0) | 53 | (7.3) | 1.56 (1.03–2.39) | |
Data were calculated by logistic regression analysis with adjusted for age, sex, smoking, and drinking status.
Stratification Analysis of Rs8506G>A Genotypes and Risk of NCGC
We further performed a stratification analysis of the associations between variant genotypes and risk of NCGC by subgroups of clinicopathological features of NCGC in this study. As shown in Table 3, a significant association between the variant genotypes and the risk of NCGC was observed in subjects with smoking (adjusted OR = 2.48, 95%CI = 1.63–3.78, homogeneity test P = 0.001), suggesting that smoking modulates the association between the lincRNA-NR_024015 rs8506G>A variant genotypes and the risk of NCGC. No significant association was found in other subgroups.
Table 3. Stratification analysis of the lincRNA-NR_024015 rs8506G>A genotypes by selected variables in non-cardia gastric cancer patients and controls.
| Cases (N = 438) | Controls (N = 727) | Adjusted OR (95% CI)a | P valueb | |||||||
| GG | (%) | AG+AA | (%) | GG | (%) | AG+AA | (%) | AG+AA vs. GG | ||
| Age(years) | ||||||||||
| ≤60 | 91 | (20.8) | 65 | (14.8) | 194 | (26.7) | 89 | (12.2) | 1.56 (1.04–2.34) | |
| >60 | 135 | (30.8) | 147 | (33.6) | 224 | (30.8) | 220 | (30.3) | 1.11 (0.82–1.49) | 0.19 |
| Sex | ||||||||||
| Male | 148 | (33.8) | 174 | (39.7) | 242 | (33.3) | 249 | (34.2) | 1.14 (0.86–1.51) | |
| Female | 78 | (17.8) | 38 | (8.7) | 176 | (24.2) | 60 | (8.3) | 1.43 (0.88–2.32) | 0.44 |
| Smoking Status | ||||||||||
| Positive | 106 | (24.2) | 83 | (19.0) | 168 | (23.1) | 53 | (7.3) | 2.48 (1.63–3.78) | |
| Negative | 120 | (27.4) | 129 | (29.5) | 250 | (34.4) | 256 | (35.2) | 1.05 (0.77–1.42) | 0.001 |
| Drinking | ||||||||||
| Positive | 118 | (26.9) | 57 | (13.0) | 186 | (25.6) | 81 | (11.1) | 1.11 (0.74–1.67) | |
| Negative | 108 | (24.7) | 155 | (35.4) | 232 | (31.9) | 228 | (31.4) | 1.46 (1.08–1.98) | 0.29 |
| Family history | ||||||||||
| Positive | 15 | (3.4) | 19 | (4.3) | 22 | (4.0) | 29 | (4.0) | 0.96 (0.40–2.30) | |
| Negative | 211 | (48.2) | 193 | (44.1) | 396 | (54.5) | 280 | (38.5) | 1.29 (1.01–1.66) | 0.52 |
| H.pylori status | ||||||||||
| H. pylori positive | 162 | (37.0) | 160 | (36.5) | 418 | (57.5) | 309 | (42.5) | 1.37 (1.05–1.78) | |
| H. pylori negative | 64 | (14.6) | 52 | (11.9) | 418 | (57.5) | 309 | (42.5) | 1.66 (1.12–2.47) | 0.42 |
| TNM stage | ||||||||||
| I | 54 | (12.3) | 50 | (11.5) | 418 | (57.5) | 309 | (42.5) | 1.25 (0.83–1.89) | |
| II | 43 | (9.8) | 47 | (10.7) | 418 | (57.5) | 309 | (42.5) | 1.48 (0.95–2.29) | |
| III | 86 | (19.6) | 83 | (19.0) | 418 | (57.5) | 309 | (42.5) | 1.31 (0.93–1.83) | 0.71 |
| IV | 43 | (9.8) | 32 | (7.3) | 418 | (57.5) | 309 | (42.5) | 1.01 (0.62–1.63) | |
ORs were adjusted for age, sex, smoking, and drinking status of non-cardia gastric cancer in a logistic regression model.
P value of the test for multiplicative interaction between stratum-related variables and lincRNA-NR_024015 (rs8506G>A AG+AA vs.GG genotypes).
Cellular Characterization of LincRNA-NR_024015
The levels of nuclear control transcript (U6), cytoplasmic control transcript (GAPDH mRNA), and lincRNA-NR_024015 were assessed by RT-qPCR in nuclear and cytoplasmic fractions of HGC-27 cells, respectively. The results showed that GAPDH mRNA was exclusively detected in the cytoplasmic fraction, while nucleus-retained U6 was predominantly found in the nuclear fraction. And lincRNA-NR_024015 expression was predominantly cytoplasmic ( Figure 1B ).
In-silico Analysis of the Effect of Rs8506G>A in LincRNA-NR_024015 Folding
Using RNAfold and SNPfold algorithms in in-silico analysis, we predicted local structural changes of lincRNA-NR_024015 caused by the rs8506G>A polymorphism located within the exonic region of lincRNA-NR_024015. As shown in Figure 1C and D , the results suggested that the G to A base change of rs8506G>A directly affects the folding of lincRNA-NR_024015, which may affect the binding site for the microRNA. This may then influence the lincRNA-NR_024015 gene expression.
Rs8506G>A Genotypes Influence the LincRNA-NR_024015 Expression by Disrupting the Binding of Has-miR-526b in vitro
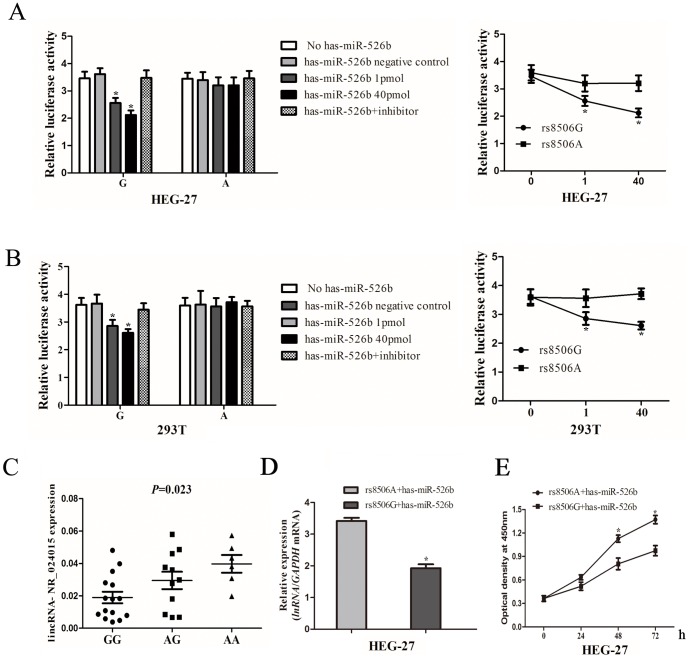
Two luciferase reporter gene constructs contained rs8506G or A allele were assayed by transiently co-transfecting with mimic and inhibitor of has-miR-526b that were predicated binding to rs8506G>A polymorphic site by bioinformatics analysis. The result of luciferase activity showed that the HEG-27 cells transiently co-transfected has-miR-526b mimics and construct containing the rs8506G allele exhibited significantly reduced luciferase activity, in a concentration-dependent manner, and the has-miR-526b inhibitors significantly reversed and upregulated their activities. However, no evident change was observed for reporter gene with A allele treated with has-miR-526b mimics or inhibitors (P>0.05) ( Figure 2A ). The same results were also observed when these experiments were repeated using 293T cells ( Figure 2B ).
Figure 2. The rs8506G>A genotypes affect lincRNA-NR_024015 expression.
Representative graph of luciferase activity of variant allele on luciferase reporter genes bearing the lincRNA-NR_024015 exonic region spanning 160 bp flanking the rs8506G>A polymorphism segments in HEG-27 (A) and 293T cells (B). Results are shown as percentage relative to luciferase activity (Renilla luciferase activity was measured and normalized to firefly luciferase). Relative luciferase activity of the psiCHECK-2-lincRNA-NR_024015-G-allele and psiCHECK-2- lincRNA-NR_024015-A-allele constructs cotransfected with has-miR-526b mimic and inhibitor. Six replicates for each group and the experiment repeated at least three times. Data are mean ± SEM. Asterisk indicates a significant change (P<0.001). (C) LincRNA-NR_024015 expression levels in thirty-two non-cardia gastric cancer patients for different rs8506G>A genotypes (15 rs8506GG, 11 rs8506AG and 6 rs8506AA); data are mean±standard error of the mean. (D) HEG-37 cells were cotransfected pcDNA-lincRNA-rs8506G or pcDNA-lincRNA-rs8506A with has-miR-526b. After 24 h, cells were collected, RNA extracted and real time PCR performed. Data are mean±standard error of the mean. “asterisk” represents P<0.05. (E) Cells' proliferation rate was significantly inhibited when cells cotranfected lincRNA-NR_024015 haboring rs8506G allele and has-miR-526b. Cell proliferation was performed by the cell viability assay and the effect became obvious from day 2. Six replicates for each group and the experiment repeated at least three times. Data are mean±standard error of the mean. “asterisk” represents P<0.05.
Association of Rs8506G>A Genotypes with LincRNA-NR_024015 Expression
We collected 32 tumor tissues from the untreated NCGC patients with different genotypes and performed real-time PCR to evaluate the effects of lincRNA-NR_024015 rs8506G>A on lincRNA-NR_024015 expression. The result showed that patients with the rs8506AG and rs8506AA genotypes expressed significantly higher lincRNA-NR_024015 mRNA levels (mean±SEM) compared to carriers of the rs8506GG genotype (AG: 0.029±0.005; AA: 0.040±0.005; GG: 0.019±0.004; P = 0.023), as shown in Figures 2C .
The Effect of MiRNA-dependent Regulation of LincRNA-NR_024015 Expression on Cell Proliferation
We further investigated whether the lincRNA-NR_024015 rs8506G>A genotypes have effects on cell proliferation in vitro. As showed in Figure 2D , lincRNA-NR_024015 expression decreased after 24 h transfection in cells transiently co-transfected with pcDNA-lincRNA-rs8506G and has-miR-526b compared with those co-transfected with pcDNA-lincRNA-rs8506A and has-miR-526b (P<0.001). Cells with decreased expression of lincRNA-NR_024015 had a weak cell growth rate in comparison with cells transfected with pcDNA-lincRNA-rs8506A and has-miR-526b from day 2 (P = 0.004) ( Figure 2E ).
Dicussion
In the present hospital-based case-control study containing a total of 438 patients and 727 healthy controls, our group found the rs8506G>A is associated with risks of NCGC. Our data showed that subjects carrying the rs8506AG and rs8506AA genotypes had a significant increased risk for NCGC compared with the GG genotype (P<0.05). Additionally, it appeared that a high risk effect of this polymorphism was more pronounced in smoking subjects. To our knowledge, this is the first study to comprehensively evaluate the association between the variants in exonic of lincRNA and risk of NCGC.
As another class of regulatory noncoding RNAs, lincRNAs, have recently moved to the forefront of noncoding RNA study. These mRNA-like molecules, lack a significant open reading frame, are generally capped, spliced and polyadenylated have been implicated in a wide range of cellular processes such as nuclear architecture, regulation of gene expression, immune surveillance, or embryonic stem cell pluripotency. A handful of studies have demonstrated lincRNAs emerged as a new aspect of biology in a variety of disease states, and changes in expression levels of lincRNAs may contribute to cancer biology at transcriptional, post-transcriptional and epigenetic levels [18], [30]–[32]. One prominent lincRNA, ANRIL, had been functionally implicated in cancer progression, typically repressing epigenetic gene expression via binding to and recruiting chromatin modifying complexes [33], [34]. Another famous example is the lincRNA, GAS5; genetic aberrations at this lincRNA locus have been found in many types of tumors, including melanoma, breast, and prostate cancers [35]–[37]. These lines of evidence supported the importance of lincRNAs in cellular biology and oncogenesis. Recently, two GWASs have reported a subset of NCGC susceptibility loci (5p13.1, 3q13.31, 8q24.3, 6p21.1 and 7p15.3) that associated with the development of NCGC. Bioinformatics analysis revealed several lincRNAs closed to these loci. Furthermore, several relevant single nucleotide polymorphisms (SNPs) located in the exonic regions of lincRNAs that may associate with NCGC were identified. As we all known, over the past decade, with the availability of large-scale RNA sequencing, it is now becoming remarkably clear that thousands of disease-related genetic variants resided outside of genes or even in non-coding transcripts have already been obtained by genome project in mammals [24]. Recent emerging evidences have indicated the association between SNPs resided in lincRNAs and human cancers. For example, based on the 1000 Genomes data, Guangfu Jin and colleages have found that regions of lncRNA had a SNPs density similar to protein-coding regions and further annotated the phenotype-related SNPs reported by GWAS at lncRNA region may contribute to prostate risk [38]. One recent study also reported that a genetic polymorphism in lincRNA-uc003opf.1gene is associated with an increased risk of developing esophageal squamous cell carcinoma in Chinese populations [39]. Collectively, our present study is consistent with previous findings showed that lincRNA-NR_024015 was moderately more abundant in the cytoplasm than in the nucleus of fractionated gastric cancer cells, suggesting that the function of this lincRNAs is exerted in the cytoplasm. Our results provided a strong evidence supporting a hypothesis for cytoplasmic regulation, in which the lincRNA-NR_024015 rs8506G>A SNP may affect the expression of this lincRNA by modifying the binding site for the has-miR-526b.
In the present study, our result of association between a genetic polymorphism in the exonic regions of a lincRNA and susceptibility to NCGC was firstly obtained in Chinese populations. The relatively large sample sizes used decreased the size of the ORs that can be detected statistically. Moreover, we have achieved a study power of over 90% (two-sided test, α = 0.05) in detecting an OR of 1.28 for the rs8506AG+AA genotypes (occurring at a frequency of 42.5% amongst the controls), when compared with the rs8506GG genotype. Notably, the association is biologically plausible and is consistent with the findings of our functional studies.
In conclusion, the present study provided the first evidence that genetic polymorphisms in the exonic regions of lincRNAs play a vital role in mediating individual susceptibility to NCGC. Our results further support the hypothesis that genetic variants in lincRNA exonic regions may affect microRNA-mediated regulation and associated with the risk of GC. Our findings warrant validation in larger, preferably population-based, case-control studies, as well as by well-designed mechanistic studies.
Funding Statement
This study was supported by the Suzhou Social Development Fund Project (SS08047), Jiangsu Province's Key Medical Department in 2011. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, et al. (2012) Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380: 2095–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Valenti V, Hernandez-Lizoain JL, Beorlegui MC, Diaz-Gozalez JA, Regueira FM, et al. (2011) Morbidity, mortality, and pathological response in patients with gastric cancer preoperatively treated with chemotherapy or chemoradiotherapy. J Surg Oncol 104: 124–129. [DOI] [PubMed] [Google Scholar]
- 3. de Martel C, Forman D, Plummer M (2013) Gastric cancer: epidemiology and risk factors. Gastroenterol Clin North Am 42: 219–240. [DOI] [PubMed] [Google Scholar]
- 4. Kelley JR, Duggan JM (2003) Gastric cancer epidemiology and risk factors. J Clin Epidemiol 56: 1–9. [DOI] [PubMed] [Google Scholar]
- 5. Milne AN, Carneiro F, O'Morain C, Offerhaus GJ (2009) Nature meets nurture: molecular genetics of gastric cancer. Hum Genet 126: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Metzker ML (2010) Sequencing technologies - the next generation. Nat Rev Genet 11: 31–46. [DOI] [PubMed] [Google Scholar]
- 7. Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, et al. (2010) Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martin JA, Wang Z (2011) Next-generation transcriptome assembly. Nat Rev Genet 12: 671–682. [DOI] [PubMed] [Google Scholar]
- 10. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, et al. (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159. [DOI] [PubMed] [Google Scholar]
- 13. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, et al. (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, et al. (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, et al. (2011) Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43: 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huarte M, Rinn JL (2010) Large non-coding RNAs: missing links in cancer? Hum Mol Genet 19: R152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gibb EA, Brown CJ, Lam WL (2011) The functional role of long non-coding RNA in human carcinomas. Mol Cancer 10: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ji P, Diederichs S, Wang W, Boing S, Metzger R, et al. (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22: 8031–8041. [DOI] [PubMed] [Google Scholar]
- 20. Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, et al. (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, et al. (2006) Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology 44: 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guffanti A, Iacono M, Pelucchi P, Kim N, Solda G, et al. (2009) A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics 10: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chung CC, Chanock SJ (2011) Current status of genome-wide association studies in cancer. Hum Genet 130: 59–78. [DOI] [PubMed] [Google Scholar]
- 24. Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 106: 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng J, Jiang L, Zhang L, Yang L, Deng J, et al. (2012) Functional genetic variations in the IL-23 receptor gene are associated with risk of breast, lung and nasopharyngeal cancer in Chinese populations. Carcinogenesis 33: 2409–2416. [DOI] [PubMed] [Google Scholar]
- 26. Zheng J, Liu B, Zhang L, Jiang L, Huang B, et al. (2012) The protective role of polymorphism MKK4-1304 T>G in nasopharyngeal carcinoma is modulated by Epstein-Barr virus' infection status. Int J Cancer 130: 1981–1990. [DOI] [PubMed] [Google Scholar]
- 27. Jiang L, Zhang C, Li Y, Yu X, Zheng J, et al. (2011) A non-synonymous polymorphism Thr115Met in the EpCAM gene is associated with an increased risk of breast cancer in Chinese population. Breast Cancer Res Treat 126: 487–495. [DOI] [PubMed] [Google Scholar]
- 28. Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang L, Deng J, Zhu X, Zheng J, You Y, et al. (2012) CD44 rs13347 C>T polymorphism predicts breast cancer risk and prognosis in Chinese populations. Breast Cancer Res 14: R105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, et al. (2008) Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451: 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song X, Wang B, Bromberg M, Hu Z, Konigsberg W, et al. (2002) Retroviral-mediated transmission of a mouse VL30 RNA to human melanoma cells promotes metastasis in an immunodeficient mouse model. Proc Natl Acad Sci U S A 99: 6269–6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang F, Li X, Xie X, Zhao L, Chen W (2008) UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett 582: 1919–1927. [DOI] [PubMed] [Google Scholar]
- 33. Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, et al. (2011) Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30: 1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, et al. (2010) Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morrison LE, Jewell SS, Usha L, Blondin BA, Rao RD, et al. (2007) Effects of ERBB2 amplicon size and genomic alterations of chromosomes 1, 3, and 10 on patient response to trastuzumab in metastatic breast cancer. Genes Chromosomes Cancer 46: 397–405. [DOI] [PubMed] [Google Scholar]
- 36. Smedley D, Sidhar S, Birdsall S, Bennett D, Herlyn M, et al. (2000) Characterization of chromosome 1 abnormalities in malignant melanomas. Genes Chromosomes Cancer 28: 121–125. [DOI] [PubMed] [Google Scholar]
- 37. Nupponen NN, Carpten JD (2001) Prostate cancer susceptibility genes: many studies, many results, no answers. Cancer Metastasis Rev 20: 155–164. [DOI] [PubMed] [Google Scholar]
- 38. Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, et al. (2011) Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis 32: 1655–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu H, Zheng J, Deng J, Hu M, You Y, et al. (2013) A genetic polymorphism in lincRNA-uc003opf.1 is associated with susceptibility to esophageal squamous cell carcinoma in Chinese populations. Carcinogenesis [DOI] [PubMed] [Google Scholar]