Abstract
Cullin-based E3 ligases target substrates for ubiquitin-dependent degradation by the 26S proteasome. The SCF (Skp1–Cul1–F-box) and ECS (ElonginC–Cul2–SOCS box) complexes are so far the best-characterized cullin-based ligases. Their atomic structure has been solved recently, and several substrates have been described in different organisms. In addition to Cul1 and Cul2, higher eucaryotic genomes encode for three other cullins: Cul3, Cul4, and Cul5. Recent results have shed light on the molecular composition and function of Cul3-based E3 ligases. In these complexes, BTB-domain-containing proteins may bridge the cullin to the substrate in a single polypeptide, while Skp1/F-box or ElonginC/SOCS heterodimers fulfill this function in the SCF and ECS complexes. BTB-containing proteins are evolutionary conserved and involved in diverse biological processes, but their function has not previously been linked to ubiquitin-dependent degradation. In this review, we present these new findings and compare the composition of Cul3-based ligases to the well-defined SCF and ECS ligases.
Keywords: BTB domains, cullin, E3 ligases, ubiquitin and proteasome
Introduction
Most cellular processes require a tightly controlled coordination between synthesis and degradation of proteins. Regulating protein turnover allows cells to adapt rapidly to various internal and external cues, and eliminate proteins that are no longer required or could even be deleterious in the new environment.
Ubiquitin-dependent degradation by the 26S proteasome has emerged as a central mechanism to control protein turnover. Covalent attachment of a polyubiquitin chain on lysine residues of the substrate mediates its recognition and subsequent degradation by the proteasome. The polyubiquitination reaction requires the coordination of three classes of different enzymes E1, E2, and E3. The ubiquitin-activating enzyme (E1) and the ubiquitin-conjugating enzyme (E2) are involved in activating and transferring ubiquitin through thioester bond formation. Ubiquitin ligases (E3) are multiprotein complexes that specifically recognize the substrates and mediate their ubiquitin-dependent degradation. Two classes of E3 ligases have been characterized: HECT-type and RING-H2-type E3s (for review, see Glickman and Ciechanover, 2002). HECT-type E3s display catalytic activity, while RING-H2-type E3s promote ubiquitinylation by positioning the activated E2 in close proximity to the substrate. It is now clear that ubiquitin-dependent degradation pathways regulate many biological processes including cell cycle progression, transcription, and cell-fate specification. Genetic and biochemical studies, mainly in budding yeast, have allowed the identification and characterization of two major types of RING-H2 E3 ligases: the APC/C (anaphase-promoting complex/cyclosome) and the SCF complex (Skp1, Cullin, F-box) (for review, see Deshaies, 1999; Harper et al, 2002). APC/C controls the onset of anaphase and exit from mitosis by degrading securin and cyclin B, while SCF ensures timely entry into S phase by triggering the degradation of the CDK-inhibitor Sic1. SCF is a macromolecular complex containing at least four subunits (Figure 1): the cullin Cul1, the ring-finger protein Hrt1 (also called Roc1 or Rbx1), the linker Skp1, and a member of the F-box protein family (for review, see Deshaies, 1999). In addition to Cul1, most genomes encode four additional cullins (Cul2, Cul3, Cul4, Cul5), whereas yeast genomes encode only two: a Cul3 and Cul4 ortholog in Schizosaccharomyces pombe (Pcu3, Pcu4) and in Saccharomyces cerevisiae a Cul3 ortholog (Ygr003w/Cul3) as well as another cullin (Yjl047c, Rtt101). While all cullin complexes may possess ubiquitin-ligase activity (Furukawa et al, 2002), it is not known if they associate with the same subunits. Available evidence suggests that this might not be the case. For example, Skp1 binds Cul1/Cdc53 but does not interact with the other cullins as shown by two-hybrid and co-immunoprecipitation experiments. Importantly, recent work has revealed that the SCF complex is the prototype of an emerging family of cullin-based ligases including the well-defined ECS complex (ElonginC–Cul2–SOCS) and the newly discovered Cul3-based ligases (Furukawa et al, 2003; Geyer et al, 2003; Pintard et al, 2003; Xu et al, 2003). These three complexes are composed of the same catalytic core (cullin and the ring-finger protein Hrt1/Roc1), but use different substrate recognition modules with striking structural similarities. Below, we first summarize the current findings and compare the composition of these novel cullin-based ligases to the well-defined SCF and ECS complexes. In the second part of this review, we discuss the potential roles of Cul3-based ligases in cell regulation.
Figure 1.
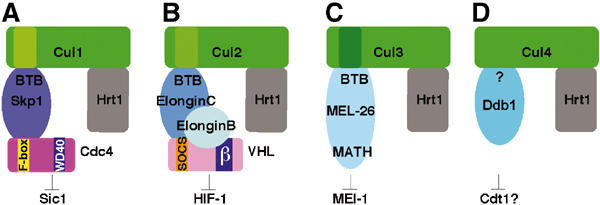
Cullin-based ligases display striking similarities. The known cullin-based ligases including the recently identified Cul3-based E3 ligases display similar overall composition and structure. In SCF (A) and ECS complexes (B), Skp1 and ElonginC bridge the interaction between the cullin and the substrate-recognition protein (F-box and SOCS-box proteins, respectively). These adaptors bind substrates through a distinct protein–protein interaction domain (e.g. WD40 in the case of the F-box protein Cdc4, and β-domain in the case of VHL). In Cul3-based complexes (C), BTB proteins incorporate features of Skp1/F-box or ElonginC/SOCS-box dimers, and are thought to bridge cullin and the substrate in a single polypeptide (e.g. MEL-26 bridges the interaction between CUL-3 and MEI-1 in C. elegans). (D) Cul4 has recently been implicated in the control of DNA replication and DNA repair in C. elegans, humans, and S. pombe. While no interaction of Cul4 with Skp1, ElonginC, or BTB proteins could be observed, it was shown recently that Cul4 binds Ddb1. It is thus possible that Cul4-like ligases may use a novel adaptor family to interact with its substrates (Wertz et al, 2004).
The SCF complex
SCF has been shown to target many substrates for degradation both in yeast and higher eucaryotes. The resolution of the crystal structures of Skp1 in a complex with the F-box protein Skp2 and more recently the entire SCF complex was an important step towards understanding its molecular composition and function (Schulman et al, 2000; Zheng et al, 2002). Within the SCF complex, Cul1 (Cdc53 in budding yeast) serves a scaffolding function: it interacts through its N-terminal helical region with the linker protein Skp1, which bridges Cul1 to the substrate-specific adapter, the F-box protein (Figure 1A). F-box proteins are characterized by a conserved 40-amino-acid F-box motif that binds Skp1, followed by distinct protein–protein interaction modules such as leucine-rich repeats (LRRs) or WD40 repeats that bind substrates (for review, see Deshaies, 1999). Through its C-terminal part, Cul1 interacts with Hrt1 and they together form the catalytic core of the complex that recruits the ubiquitin-conjugating enzyme Cdc34.
Introduction of a flexible region in Cul1 or mutations changing the orientation of the F-box protein abolish the activity of the SCF complex, suggesting that the overall structure of the SCF is rigid and that this is essential for its function (Zheng et al, 2002; Orlicky et al, 2003). Recent in vitro experiments suggest that E2 only transiently associates with Hrt1 and gets released in proximity to the substrate. This mechanism introduces some flexibility into the system, which might explain why several lysine residues of the substrate are polyubiquitinated (Deffenbaugh et al, 2003).
The ECS complex
The ECS complex is best known for its role in degradation of the hypoxia-inducible factor HIF-1, involved in oxygen homeostasis (Maxwell et al, 1999; Kile et al, 2002). The ECS complex displays striking overall similarities to the SCF complex (Figure 1) (for review, see Kile et al, 2002). Skp1 and ElonginC share 30% sequence identity between their amino-terminal two-thirds of the proteins (Stebbins et al, 1999). This sequence similarity indicated that these two proteins might fold into a similar overall structure. Indeed, crystal structure determination revealed that Skp1 and ElonginC adopt an α/β structure (Figure 2; Schulman et al, 2000). However, despite their striking sequence and structural similarity, it is important to note that ElonginC specifically binds the amino-terminal domain of Cul2 but not Cul1 (Pause et al, 1999), while conversely Skp1 interacts with Cul1 but not Cul2 (Michel and Xiong, 1998). Several residues located in the β-strand 3 (S3) or in the α-helix 5 (H5) of Skp1 were shown to contact Cul1, and are thus likely to contribute to binding specificity (Figure 2).
Figure 2.
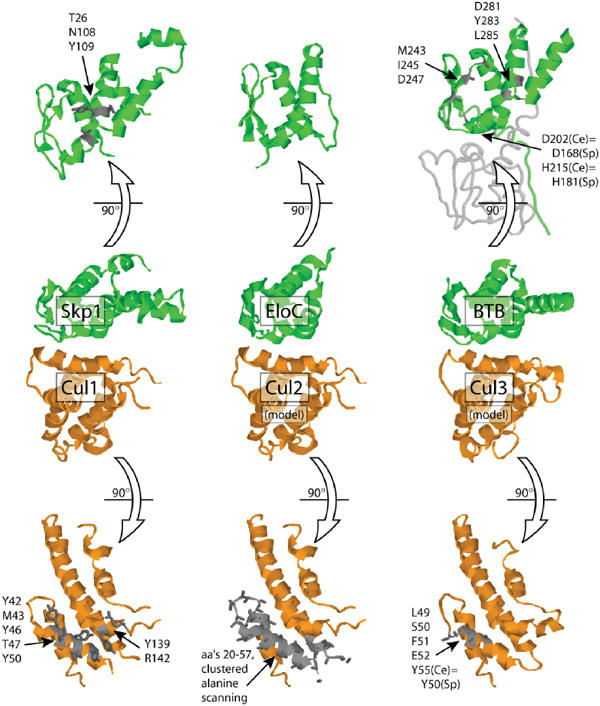
Structural analysis of the cullin–BTB interaction. The CUL-1/SKP1 co-crystal structure (Zheng et al, 2002) served as the basis to model the interface between CUL-2 and CUL-3 with human ElonginC (Stebbins et al, 1999) and C. elegans MEL-26 (based on the human PLZF BTB domain structure (Ahmad et al, 1998). The highlighted residues have been shown by mutagenesis to be important for the cullin–BTB interaction (Pause et al, 1999; Zheng et al, 2002; Pintard et al, 2003; Geyer et al, 2003; Xu et al, 2003). ‘Sp' indicates corresponding positions in S. pombe Btb3 and Pcu3. Note that two mutations in Btb3 that disrupt its interaction with Pcu3 (D168A, H181L) are not found at the predicted BTB/cullin interface but instead are located near the putative BTB/BTB dimer interface as seen in PLZF.
Similar to the Skp1/F-box module, ElonginC bridges the interaction between Cul2/5 and the substrate-specific adapter the SOCS-box-containing proteins (Kamura et al, 1998). Besides the SOCS-box motif, SOCS-box proteins contain various protein–protein interaction domains such as β-domains, WD40 repeats, or ankyrin motifs that bind substrates (Figure 1B). Thus, the F-box and SOCS-box domains function as binding motifs, which link the substrate to the cullin through their interaction with Skp1 and ElonginC, respectively. ElonginC also forms a complex with the ubiquitin-like protein ElonginB, but the function of ElonginB in the ECS complex is not understood.
The Cul3-based complexes
Skp1 and ElonginC display structural similarity to the BTB-domain fold (Schulman et al, 2000). BTB domains were originally found in the Drosophila melanogaster transcription factors Bric à Brac, Tramtrack, and Broad Complex (Zollman et al, 1994). All eucaryotic species express a large variety of proteins that contain a BTB domain (Table I). The structural homology between BTB proteins and Skp1 or ElonginC led to the hypothesis that BTB proteins might directly interact with cullins. This prediction has now been experimentally confirmed by several recent studies. Interestingly, the available results suggest that several BTB proteins interact with Cul3, but not with other cullins. For example, a two-hybrid screen aimed at identifying binding partners of Caenorhabditis elegans CUL-3 led to the isolation of 11 BTB proteins, which were confirmed by biochemical methods (Xu et al, 2003). A similar yeast two-hybrid approach using human Cul3 as bait identified 13 BTB-containing proteins, which specifically interact with Cul3 in vitro (Furukawa et al, 2003). Finally, Geyer et al (2003) extended this concept and found that S. pombe Cul3-homolog Pcu3 not only co-purifies with the Hrt1-homolog Pip1, but also with all three BTB proteins of S. pombe (termed Btb1, Btb2, Btb3). These results raise the possibility that BTB proteins may generally function as subunits of Cul3-based E3 ligases.
Table 1.
Characteristics and functions of selected BTB proteins
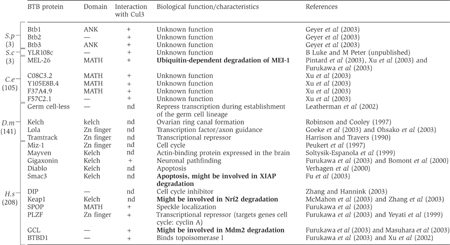 |
| BTB proteins are involved in diverse biological processes. To date, a functional link to ubiquitin-dependent degradation has only been established in C. elegans for the CUL-3/MEL-26 complex. However, it has recently been shown that CUL-3 interacts with several other BTB proteins (+: positive interaction with Cul3; nd: not determined) in all species. The numbers in parentheses indicate the total number of BTB proteins in the following species: S.p, Schizosaccharomyces pombe; S.c, Saccharomyces cerevisiae; C.e, Caenorhabditis elegans; D.m, Drosophila melanogaster; H.s, Homo sapiens. Bold: BTB proteins for which a link to ubiquitin-dependent degradation is established or emerging. |
Importantly, the functional relevance of a Cul3–BTB protein complex was demonstrated in C. elegans. In this system, a complex containing CUL-3 and the BTB protein MEL-26 is required for the ubiquitin-dependent degradation of the microtubule-severing protein MEI-1 in early embryos. Degradation of MEI-1 by the CUL-3/MEL-26 complex after meiosis is essential for the assembly of the mitotic spindle (Figure 3). Indeed, mel-26(RNAi), cul-3(RNAi), or mel-26 temperature-sensitive mutant embryos fail to degrade MEI-1, leading to a failure to assemble the mitotic spindle. Inactivation of mei-1 in a mel-26(-) or cul-3(-) background suppresses the defects in mitotic spindle assembly, implying that degradation of MEI-1 is the critical event during this process (Kurz et al, 2002; Pintard et al, 2003). Finally, reconstituted CUL-3/MEL-26 complex can sustain polyubiquitination of MEI-1 in vitro (Furukawa et al, 2003). Taken together, these results identify a novel and conserved class of cullin-based E3 ligases containing BTB proteins (Figure 1C).
Figure 3.
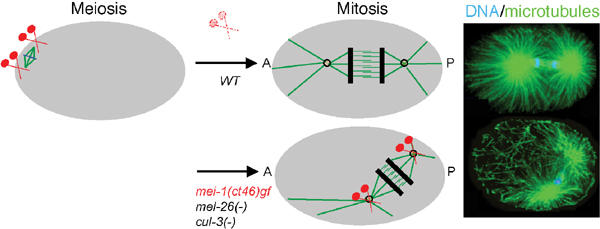
The CUL-3-based complex is required for MEI-1 degradation at the meiosis-to-mitosis transition in C. elegans embryos. In C. elegans embryos, two different microtubule-based structures assemble within a 20 min interval in the same cytoplasm, for example, the meiotic and the mitotic spindle. Contrary to the anastral meiotic spindle that forms close to the cell cortex, the mitotic spindle assembles and elongates along the antero-posterior axis of the embryo (A–P). Long arrays of astral microtubules and the formation of a robust spindle are required during mitosis for correct spindle positioning, elongation, and cytokinesis. The microtubule-severing complex MEI-1/2 is essential for formation of the meiotic spindle (Clark-Maguire and Mains, 1994a, 1994b; Srayko et al, 2000). However, inactivation of this complex through MEI-1 (in red) degradation by the CUL-3-based ligase is a prerequisite for the assembly of a functional mitotic spindle. The regulation of MEI-1 degradation is not known.
The structure of this novel Cul3–BTB complex is not known at present, but based on mutational analysis it is likely to resemble ECS and SCF complexes (Figure 2). As in Skp1, mutations in the BTB domain of MEL-26 that are located in the β-strand 3 (S3) or in the α-helix 5 (H5) abolish the interaction with CUL-3 (Xu et al, 2003). How can the different cullins interact specifically with the BTB folds of Skp1, ElonginC, and BTB proteins? Although the amino-terminal domains of cullins on the whole are poorly conserved between paralogs, several residues are common to distinct orthologs including Cul1, Cul2/5, and Cul3 (Pintard et al, 2003). Based on the structure of Cul1, these conserved residues are all located on the surface where Skp1 binds Cul1 (Zheng et al, 2002). Mutations of the highly conserved ‘LSFE' cluster in CUL-3 abolish its interaction with MEL-26 (Pintard et al, 2003; Xu et al, 2003). Likewise, several conserved residues are also present on the corresponding surface of Cul2 and Cul5, and they are good candidates to mediate a specific interaction with ElonginC. Taken together, these observations suggest that conserved residues contribute to the specificity of the interaction between cullins and the corresponding BTB partner. In the future, structure determination of this novel complex will help to understand the interaction between the N-terminal domain of CUL-3 and BTB proteins.
BTB proteins function as substrate-specific adaptors in Cul3-based ligases
Available evidence strongly suggests that BTB proteins function as substrate-specific adaptors of Cul3 ligases, although this notion is currently mainly based on the analysis of the CUL-3/MEL-26 complex in C. elegans. MEL-26 interacts with MEI-1 in yeast two-hybrid assays and in vitro binding experiments (Pintard et al, 2003; Xu et al, 2003), suggesting that this interaction is likely to be direct. Importantly, MEL-26 fails to interact with the product of gain-of-function allele of mei-1 (MEI-1(gf)), which is not degraded at the meiosis-to-mitosis transition of C. elegans embryos (Figure 3) (Clark-Maguire and Mains, 1994a; Pintard et al, 2003). Moreover, mel-26 loss-of-function mutations phenocopies the mei-1 gain-of-function mutations (Dow and Mains, 1998), strongly suggesting that the recognition of MEI-1(gf) by MEL-26 is also impaired in vivo (Figure 3).
Interestingly, the BTB proteins MEL-26 in C. elegans and Btb3 in S. pombe accumulate when the function of CUL-3/Pcu3 is impaired, suggesting that MEL-26 and Btb3 are degraded by a mechanism that depends on the Cul3 complex (Geyer et al, 2003; Pintard et al, 2003). Several F-box or SOCS proteins are similarly degraded by an autocatalytic mechanism (Zhou and Howley, 1998; Galan et al, 1999; DeRenzo et al, 2003). However, the physiological relevance of this regulation is still poorly understood.
Several lines of evidence suggest that dimerization of substrate-specific adaptor proteins may be of functional importance for E3-ligase activity. For example, heterodimers of the F-box proteins Pop1 and Pop2 are observed in vivo, and appear to be required for the degradation of Cdc18 in S. pombe (Kominami et al, 1998; Wolf et al, 1999; Seibert et al, 2002). Similarly, it has been shown that the BTB protein MEL-26 forms dimers at least by yeast two-hybrid assay. Interestingly, the product of the mel-26(sb45) allele interacts with MEL-26 but not with MEI-1 (Pintard et al, 2003). This allele acts in a dominant-negative manner in vivo, suggesting that dimerization of BTB proteins may be important for the activity of the CUL-3 complex (Dow and Mains, 1998). However, the functional significance of dimerization of BTB proteins and substrate-specific adaptors, in general, deserves further analysis.
A single polypeptide containing a BTB and a protein–protein interaction domain may bridge Cul3 to the substrate and thus fulfill the function of Skp1/F-box or ElonginC/SOCS-box heterodimers
Skp1 and ElonginC are both small proteins, while most of the BTB proteins contain an additional protein–protein interaction domain. For example, five out of 11 BTB-containing proteins picked as CUL-3 interactors contain a Meprin and TRAF homology (MATH) domain, suggesting that they could bind substrates through this domain and therefore bridge in a single polypeptide the substrate to CUL-3. In support of this prediction, Xu et al (2003) have shown that MEL-26 indeed interacts with MEI-1 through its MATH domain. The S. pombe BTB proteins Btb1 and Btb3 contain ankyrin repeats in addition to the BTB domains, and the human BTB proteins found to interact with Cul3 contain MATH, KELCH, or Zn-finger domains that may similarly serve as binding sites for substrates. It is tempting to speculate therefore that BTB proteins containing such additional protein–protein interaction domains fulfill in one polypeptide the function of the Skp1/F-box dimer. It is interesting to note that the plant protein ZTL or the human protein KIAA1332 both contain an F-box and Kelch repeats, which thus makes the organization of the Skp1/ZTL heterodimer similar to the BTB–Kelch complex, except that the two complexes are predicted to use distinct cullins (Somers et al, 2000). However, several Cul3-interacting proteins appear to contain only a BTB domain (Xu et al, 2003), and are thus unlikely to possess the ability to interact simultaneously with potential substrates. Therefore, heterodimerization of such proteins with a single BTB domain with another BTB protein containing an additional protein–protein interaction domain may be necessary to bring a substrate recognition motif into the Cul3 complex. Alternatively, these proteins might contain uncharacterized protein–protein interaction motifs that are able to bind substrates.
Are all BTB proteins substrate-specific subunits of Cul3-based complexes?
The discovery that BTB proteins can assemble into a novel class of E3 ligases is of general importance, given the high number of BTB-containing proteins that are present in the genome of different organisms (Table I). Some of these BTB proteins are characterized and are known to be involved in a variety of biological processes, including regulation of microtubules and microfilament dynamics, transcription, and apoptosis. However, based on the published results, connections to ubiquitin-dependent degradation pathways can currently only be made for a few BTB proteins (Table I), and it thus remains to be demonstrated how many BTB proteins indeed function in E3-ligase complexes in vivo. For example, a recent study revealed that the BTB–Kelch protein Keap1 might target Nrf2 for ubiquitin-dependent degradation. Nrf2 is a transcription factor that activates the expression of a battery of genes encoding drug metabolism enzymes and antioxidant proteins. In response to oxidative stress, Nrf2 translocates into the nucleus and binds to the anti-oxidant responsive elements (AREs) present in the promoter region of its target genes. In the absence of stress, Keap1 sequesters Nrf2 in the cytoplasm and triggers its ubiquitin-dependent degradation by the proteasome (McMahon et al, 2003; Zhang and Hannink, 2003). A plausible link to degradation is also emerging for the BTB protein Smac3, a member of the Diablo family. In response to an apoptotic signal, Diablo is released from the mitochondria and inactivates caspase inhibitors (Verhagen et al, 2000). Although the exact molecular mechanism by which Diablo proteins accomplish this function is not clear, it has been proposed that Smac3 may promote ubiquitin-dependent degradation of the apoptotic inhibitor XIAP (Fu et al, 2003). Finally, the conserved BTB protein mouse germ cell-less-1 (mgcl-1) originally found in D. melanogaster might be involved in modulating the activity of the tumor suppressor p53 by enhancing degradation of Mdm2 (Masuhara et al, 2003). Importantly, GCL has been shown to interact with Cul3 by two-hybrid and co-immunoprecipitation assays (Furukawa et al, 2003). However, as in the other cases, although a tantalizing connection to ubiquitin-dependent degradation pathways can be made, the requirement for Cul3 for the degradation of Mdm2 has not been experimentally documented.
Several transcriptional regulators contain BTB domains including tramtrack, the c-Myc regulator Miz1, and the promyelocytic leukemia zinc-finger protein (PLZF). All these transcriptional regulators play a key role during development, cell cycle progression, or cell differentiation. For example, in human cells, Miz1 modulates the activity of the transcription factor Myc (Peukert et al, 1997), while PLZF may control cell cycle progression by preventing expression of cell cycle regulators such as cyclin A (Yeyati et al, 1999). Importantly, PLZF interacts with CUL-3 in a two-hybrid assay, suggesting that a CUL3-based complex may indeed regulate transcription by controlling the stability of PLZF (Furukawa et al, 2003).
Several BTB proteins including Mayven and Kelch appear to regulate the microtubule and microfilament networks. Of particular interest in the present context, mutations in the BTB–kelch protein gigaxonin lead to the accumulation of giant axons resulting in neuropathy (Bomont et al, 2000). Many mutations are located within the BTB domain, thus highlighting its functional importance. Gigaxonin was recently shown to control microtubule dynamics through its interaction with the microtubule-associated protein MAP1B. This MAP promotes neuronal stability by maintaining the integrity of cytoskeletal structures and ensuring axonal transport over long distances (Ding et al, 2002). Importantly, Gigaxonin is able to interact with human Cul3 (Furukawa et al, 2003), raising the possibility that a Cul3-based/Gigaxonin E3 ligase may target MAP1B for degradation. However, at present, no evidence suggests that the stability of MAP1B is indeed regulated in vivo. Clearly, further work is now necessary to investigate the possible link between BTB proteins and ubiquitin-dependent degradation. However, despite the exciting connections to Cul3-based E3-ligase complexes, we should not ignore the possibility that some BTB proteins may serve functions independent of their role as subunits of Cul3-based E3 ligases. For example, the well-characterized CUL-3 based subunit MEL-26 is likely to control cortical activities in early C. elegans embryos in a manner that may not involve CUL-3 (S Glaser, L Pintard, and M Peter, unpublished results). There is also precedence for such cases for F-box and SOCS-box proteins. For example, it has been shown that yeast Skp1 forms three complexes that lack the cullin Cdc53, including the RAVE, the Ctf13/Skp1, and Rcy1/Skp1 complexes (Seol et al, 2001). The F-box protein Rcy1 is involved in recycling of membrane proteins in budding yeast (Galan et al, 2001), while the RAVE complex is involved in the function of the vacuole. Genetic evidence confirms that the recycling function of Rcy1 requires Skp1, but is independent of Cdc53 or the E2 Cdc34. Moreover, it was recently shown that VHL functions as a microtubule stabilizer and mutational analysis implies that this function is distinct from its role as subunit of the ECS complex (Hergovich et al, 2003). Together, these observations suggest that the substrate-recognition modules of cullin-based ligases fulfill cellular functions in addition to their role in ubiquitin-dependent degradation.
Concluding remarks
During the past few years, considerable progress has been made on the characterization of cullin-based ligases, first, with the discovery of SCF and ECS complexes and more recently with the identification of the Cul3-based ligases. This cullin-based E3-ligase family may further expand, if Cul4 indeed uses yet another substrate-specific adaptor module (Wertz et al, 2004). Several different names have already been proposed to define cullin-based ligases. For example, SCF2, CBC, and ECS (Kamura et al, 2002; Kile et al, 2002; DeRenzo et al, 2003; Xu et al, 2003) have been used to describe Cul2 complexes, while SCF3 or BCR (Furukawa et al, 2003; Xu et al, 2003) have been proposed for Cul3-based ligases. With the rapid expansion of cullin-based ligases, the use of a unified nomenclature now becomes an important issue.
In summary, these exciting discoveries highlight the extraordinary possibility of cullin-based ligases to target a very large number of substrates for ubiquitin-dependent degradation. If all BTB-containing proteins can assemble into Cul3-based complexes, we estimate the number of cullin-based E3 ligase in humans to more than 500. However, further work is needed to elucidate which BTB proteins are able to assemble into functional CUL3-based complexes, and to identify their in vivo substrates. Nevertheless, since BTB-containing proteins are mutated in some human diseases, re-investigating their function with regard to a possible role in ubiquitin-dependent degradation pathways may be rewarding.
Acknowledgments
We thank S Glaser, H Meyer, T Kurz, B Luke, M Tyers, and W Krek for critical reading of the review. T Kurz provided the immunostaining presented in Figure 3. LP is supported by a Long-Term Fellowship from the Federation of European Biochemical Societies (FEBS) and from a Fellowship from Roche, and MP by the ETHZ and the Swiss National Science Foundation.
References
- Ahmad KF, Engel CK, Prive GG (1998) Crystal structure of the BTB domain from PLZF. Proc Natl Acad Sci USA 95: 12123–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomont P, Cavalier L, Blondeau F, Ben Hamida C, Belal S, Tazir M, Demir E, Topaloglu H, Korinthenberg R, Tuysuz B, Landrieu P, Hentati F, Koenig M (2000) The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet 26: 370–374 [DOI] [PubMed] [Google Scholar]
- Clark-Maguire S, Mains PE (1994a) Localization of the mei-1 gene product of Caenorhabditis elegans, a meiotic-specific spindle component. J Cell Biol 126: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire S, Mains PE (1994b) mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics 136: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffenbaugh AE, Scaglione KM, Zhang L, Moore JM, Buranda T, Sklar LA, Skowyra D (2003) Release of ubiquitin-charged Cdc34-S-Ub from the RING domain is essential for ubiquitination of the SCF(Cdc4)-bound substrate Sic1. Cell 114: 611–622 [DOI] [PubMed] [Google Scholar]
- DeRenzo C, Reese KJ, Seydoux G (2003) Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424: 685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol 15: 435–467 [DOI] [PubMed] [Google Scholar]
- Ding J, Liu JJ, Kowal AS, Nardine T, Bhattacharya P, Lee A, Yang Y (2002) Microtubule-associated protein 1B: a neuronal binding partner for gigaxonin. J Cell Biol 158: 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow MR, Mains PE (1998) Genetic and molecular characterization of the Caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of mei-1, a meiotic-specific spindle component. Genetics 150: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Jin Y, Arend LJ (2003) Smac3, a novel Smac/DIABLO splicing variant, attenuates the stability and apoptosis-inhibiting activity of XIAP. J Biol Chem 278: 52660–52672 [DOI] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y (2003) Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nat Cell Biol 5: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Furukawa M, Ohta T, Xiong Y (2002) Activation of UBC5 ubiquitin-conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J Biol Chem 277: 15758–15765 [DOI] [PubMed] [Google Scholar]
- Galan JM, Peter M (1999) Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA 96: 9124–9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Wiederkehr A, Seol JH, Haguenauer-Tsapis R, Deshaies RJ, Riezman H, Peter M (2001) Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol Cell Biol 21: 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R, Wee S, Anderson S, Yates J, Wolf DA (2003) BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell 12: 783–790 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Harper JW, Burton JL, Solomon MJ (2002) The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev 16: 2179–2206 [DOI] [PubMed] [Google Scholar]
- Hergovich A, Lisztwan J, Barry R, Ballschmieter P, Krek W (2003) Regulation of microtubule stability by the von Hippel–Lindau tumour suppressor protein pVHL. Nat Cell Biol 5: 64–70 [DOI] [PubMed] [Google Scholar]
- Kamura T, Brower CS, Conaway RC, Conaway JW (2002) A molecular basis for stabilization of the von Hippel–Lindau (VHL) tumor suppressor protein by components of the VHL ubiquitin ligase. J Biol Chem 277: 30388–30393 [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Haque D, Liu L, Kaelin WG Jr, Conaway RC, Conaway JW (1998) The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev 12: 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile BT, Schulman BA, Alexander WS, Nicola NA, Martin HM, Hilton DJ (2002) The SOCS box: a tale of destruction and degradation. Trends Biochem Sci 27: 235–241 [DOI] [PubMed] [Google Scholar]
- Kominami K, Ochotorena I, Toda T (1998) Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1–Cullin-1–F-box) ubiquitin ligase. Genes Cells 3: 721–735 [DOI] [PubMed] [Google Scholar]
- Kurz T, Pintard L, Willis JH, Hamill DR, Gönczy P, Peter M, Bowerman B (2002) Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 295: 1181–1412 [DOI] [PubMed] [Google Scholar]
- Masuhara M, Nagao K, Nishikawa M, Kimura T, Nakano T (2003) Enhanced degradation of MDM2 by a nuclear envelope component, mouse germ cell-less. Biochem Biophys Res Commun 308: 927–932 [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399: 271–275 [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Hayes JD (2003) Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem 278: 21592–21600 [DOI] [PubMed] [Google Scholar]
- Michel JJ, Xiong Y (1998) Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ 9: 435–449 [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems A, Tyers M, Sicheri F (2003) Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112: 243–256 [DOI] [PubMed] [Google Scholar]
- Pause A, Peterson B, Schaffar G, Stearman R, Klausner RD (1999) Studying interactions of four proteins in the yeast two-hybrid system: structural resemblance of the pVHL/elongin BC/hCUL-2 complex with the ubiquitin ligase complex SKP1/cullin/F-box protein. Proc Natl Acad Sci USA 96: 9533–9538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peukert K, Staller P, Schneider A, Carmichael G, Hanel F, Eilers M (1997) An alternative pathway for gene regulation by Myc. EMBO J 16: 5672–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, Peter M (2003) The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316 [DOI] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1–Skp2 complex. Nature 408: 381–386 [DOI] [PubMed] [Google Scholar]
- Seibert V, Prohl C, Schoultz I, Rhee E, Lopez R, Abderazzaq K, Zhou C, Wolf DA (2002) Combinatorial diversity of fission yeast SCF ubiquitin ligases by homo- and heterooligomeric assemblies of the F-box proteins Pop1p and Pop2p. BMC Biochem 3: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Shevchenko A, Deshaies RJ (2001) Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat Cell Biol 3: 384–391 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Srayko M, Buster DW, Bazirgan OA, McNally FJ, Mains PE (2000) MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev 14: 1072–1084 [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG Jr, Pavletich NP (1999) Structure of the VHL–ElonginC–ElonginB complex: implications for VHL tumor suppressor function. Science 284: 455–461 [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL (2000) Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102: 43–53 [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM (2004) Human de-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303: 1371–1374 [DOI] [PubMed] [Google Scholar]
- Wolf DA, McKeon F, Jackson PK (1999) F-box/WD-repeat proteins pop1p and Sud1p/Pop2p form complexes that bind and direct the proteolysis of cdc18p. Curr Biol 9: 373–376 [DOI] [PubMed] [Google Scholar]
- Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW (2003) BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425: 316–321 [DOI] [PubMed] [Google Scholar]
- Yeyati PL, Shaknovich R, Boterashvili S, Li J, Ball HJ, Waxman S, Nason-Burchenal K, Dmitrovsky E, Zelent A, Licht JD (1999) Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene 18: 925–934 [DOI] [PubMed] [Google Scholar]
- Zhang DD, Hannink M (2003) Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol 23: 8137–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP (2002) Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]
- Zhou P, Howley PM (1998) Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell 2: 571–580 [DOI] [PubMed] [Google Scholar]
- Zollman S, Godt D, Prive GG, Couderc JL, Laski FA (1994) The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA 91: 10717–10721 [DOI] [PMC free article] [PubMed] [Google Scholar]


