Abstract
We present a systematic study of three bacterial species that belong to the class Mollicutes, the smallest and simplest bacteria, Spiroplasma melliferum, Mycoplasma gallisepticum, and Acholeplasma laidlawii. To understand the difference in the basic principles of metabolism regulation and adaptation to environmental conditions in the three species, we analyzed the metabolome of these bacteria. Metabolic pathways were reconstructed using the proteogenomic annotation data provided by our lab. The results of metabolome, proteome and genome profiling suggest a fundamental difference in the adaptation of the three closely related Mollicute species to stress conditions. As the transaldolase is not annotated in Mollicutes, we propose variants of the pentose phosphate pathway catalyzed by annotated enzymes for three species. For metabolite detection we employed high performance liquid chromatography coupled with mass spectrometry. We used liquid chromatography method - hydrophilic interaction chromatography with silica column - as it effectively separates highly polar cellular metabolites prior to their detection by mass spectrometer.
Introduction
In contrast to the abundance of systems-oriented approaches describing changes on the transcriptome or proteome level, relatively few studies examined the metabolome. Metabolomics studies are necessary to shed light on the link between genotypes and phenotypes. The goal of the presented research is to identify as many intracellular metabolites as possible in bacteria belonging to three species, namely Spiroplasma melliferum, Mycoplasma gallisepticum, and Acholeplasma laidlawii. All these bacteria are members of the class Mollicute, a unique category of bacteria that share the absence of a cell wall, a reduced genome, and simplified metabolic pathways. Mollicutes are widespread organisms in nature engaged in a parasitic lifestyle.
The poultry pathogen M. gallisepticum is a small bacterium that causes chronic respiratory disease in chickens and turkeys. S. melliferum is an insect pathogen with the capability to infect the honey bee [1]. A. laidlawii is a common contaminant of cell culture and has a broad variety of hosts, including bovines, the human oral cavity, birds and plants; it was originally isolated from sewage [2], [3], [4], [5], [6].
The mycoplasmic metabolome is significantly reduced, possibly due to its parasitic nature. Like all Mycoplasma, the three bacterial species lack some general metabolic pathways, including the TCA, amino acids synthesis, as well as purine and pyrimidine synthesis [7]. Genome-reduced organisms have limited adaptability to external factors [8]. We believe that comparison of the metabolomes of the three Mollicutes may reveal the primary adaptive features of the simplest metabolomes.
Metabolite identification in our study was performed by flow injection time-of-flight mass spectrometry (Q-TOF). Metabolite identity was confirmed by fragmentation of previously detected ions using targeted mass spectrometry. The liquid chromatography approach applied here, hydrophilic interaction chromatography (HILIC) with silica columns, effectively separates highly polar cellular metabolites for their subsequent detection using a high accuracy mass spectrometer in positive and negative acquisition modes [9].
Here we present reliable measurements of a hundred metabolites, including components of sugar, amino acid, and nucleotide metabolisms. We have identified about a third of all possible intracellular metabolites predicted by genome annotation and compared the metabolite content of three Mollicute species. Metabolomic data analysis is based on a previous proteogenomic study performed in our lab for all three bacterial species [3], [10], [11], [12]. The high-throughput-omics analysis, including transcriptomics, proteomics, and complemented with the metabolomic study performed in this work, will improve our comprehensive understanding how the three closely-related Mollicute species are regulated under different conditions [13].
Materials and Methods
Bacteria strains and growth conditions
Spiroplasma melliferum KC3 and Acholeplasma laidlawii PG-8A were kindly provided by Prof. G. Wroblewsky of the University of Rennes, France. The Mycoplasma gallisepticum S6 strain was provided by Prof. S.N. Borkhsenius, Institute of Cytology,St. Petersburg, Russian Academy of Science. S. melliferum was grown in SP4 medum without any specific aeration and with mixing at 30°C for 20 hours after 1∶10 dilution, as described earlier [14]. Acholeplasma laidlawii PG-8A and Mycoplasma gallisepticum S6 were grown in a modified Edward's medium (Tryptose 20 g/L, NaCl 5 g/l, NaOAc 5 g/l, KCl 1.3 g/l, Tris 3 g/l, yeast dialysate 5%, horse serum 6%, glucose 0.5%, pH 7.6) at 37°C for 20 hours after 1∶100 dilution [15]. Bacteria were cultured in covered flasks, and the time of growth and the pH of the solution were controlled [11]. Bacteria were harvested by centrifugation (10 min, 16,000 g) in the log phase, which was determined by pH values (6.5–6.6 for both, S. melliferum and M. gallisepticum). The metabolic profiles were obtained from cultures grown from a single clone for less than five passages. Each metabolome experiment was independently repeated at least seven times; 50 ml cultures from the same strain and with the same sample preparation procedure were used for each experiment.
Determination of pH tolerance
We used the “color test” method to determine whether Mollicutes survive in acidic conditions and to estimate the critical pH value for each species. Aliquots of M. gallisepticum, A. laidlawii and S. melliferum culture were exposed to decreased pH (hydrochloric acid was added) in steps of ΔpH = 0.1 for 1 hour starting at pH 7.5. Each aliquot was then subjected to a series of dilutions (up to 109) in medium (pH 7.5) containing pH indicator (Phenol Red), and the presence of living cells was detected by the medium color change. The tolerated acidity was defined as the minimum acidity at which 10% of cells survived.
Reagents
The following chemicals were used as standards: sodium pyruvate (100 mg/ml, disodium salt hydrate), D-fructose 6-phosphate, hydrate of sodium phospho(enol)pyruvate (97% purity, enzyme quality), dehydrated disodium salt of D-ribose 5-phosphate, di-glyceraldehyde 3-phosphate (46.1 mg/ml). Purified (98%) amino acids, nucleotides, nucleosides (adenosine, deoxyadenosine, inosine, cytosine monophosphate, and thymidine) from Sigma-Aldrich (USA) were used as standards as well. The following reagents were used for extraction and solution preparation: absolute methanol (HPLC grade) from Biosolve (The Netherlands), ammonium acetate (ultra clean grade) from Helicon (Russia), formic acid (98–100%) from Riedel-de Haen (Germany), ammonium hydroxide solution (29.73%) from Fisher Scientific (USA), water (HPLC-MS) and acetonitrile (HPLC-MS) from Panreac (Spain).
Extraction of metabolites
For metabolite analysis, S. melliferum, M. gallisepticum, or A. laidlawii (50 ml) were grown in a liquid medium. Cells were harvested by centrifugation at 16,000 g at 20°C for 10 min and washed with the washing solution (150 mM NaCl/50 mM Tris). The pH of the washing solution was 6.5 (the acidity in the logarithmic phase of Mycoplasma growth), while for acid stress studies the pH of washing solution was about 5.5 (depending on the type of culture and the selected conditions of acid stress). Thus, the culture was under the influence of selected acidity conditions during the washing procedure. The cell pellet was resuspended in 3 ml of washing solution (the pellet volume was 30 µl) and precipitated again. Then cells were washed twice with 150 µl of washing solution.
The metabolism was rapidly quenched by cold methanol extraction. A cold methanol extraction method was developed on the basis of a previously reported cold methanol extraction protocol [16], [17] as described below. The metabolites were extracted by adding 1000 µl of methanol (−77°C) to 75 ul of the resuspended cell pellet; the sample was vigorously shaken (1 min) and then kept at −77°C for 15 min. The sample was warmed for 3 min and then thoroughly shaken again. The resulting sample was centrifuged for 30 min at 16,000 g at 4°C. The supernatant was separated into aliquots and lyophilized. The dry extract was kept no longer than 4 days prior to analysis. The dry extract was dissolved in a mixture consisting of 20% acetonitrile and 80% water and analyzed immediately.
Until recently there was no published record of an LC-MS method of metabolome analysis of the studied bacteria (S. melliferum, M. gallisepticum, and A. laidlawii). Pre-testing a few other options (such as extraction by acetonitrile, methanol/water, acetonitrile/water/acid and others), we have chosen the cold methanol extraction method, as it has been demonstrated to be the most effective. The cold methanol extraction protocol described in [16] is the basis of the protocol used here. A similar modification has been successfully used recently for M. pneumoniae studies [18].
Apparatus and HPLC/MS method
Mass spectrometry analysis was performed on a Q-TOF 6520 series time-of-flight mass spectrometer (Agilent Technologies). The flow from the analytical column was introduced directly into the electrospray ion source of the mass spectrometer. Prior to the experiment, the mass spectrometer was calibrated to 2 ppm accuracy of the m/z value. The ionizing spray voltage was 3500 V in both positive and negative ionization modes. Nitrogen of various degrees of purity was used as a dry gas at a pressure of 20 psi with a flow rate of 6 l/min and as a gas in the collision cell. The temperature of the quartz capillary was 325°C. HPLC-MS and HPLC-MS/MS analyses were carried out using the series 1200 high performance liquid chromatograph (Agilent Technologies), coupled with the mass spectrometer. The following chromatographic analytical column was used in the study: Zorbax RX-SIL Narrow-Bore (150 mm×2.1 mm×5 µm) from Agilent Technologies. To protect the separating phase of these highly effective analytical columns from chemical damage, as a safety cartridge we used the Zorbax RX-SIL 4-Pack analytical guard column (4.6 mm×12.5 mm×5 µm) purchased from Agilent Technologies.
Chromatographic analysis was performed with the following parameters: auto-sampling temperature, 8°C; analytical column temperature, 18°C; injection volume, 2 µl; solvent flow rate, 50 µl/min. The following solvents were used as eluting solutions: eluent A was 20 mM ammonium acetate/0.25 mM ammonium hydroxide in water/acetonitrile mixture of 95∶5 ratio, pH 8.02; eluent B was pure acetonitrile. The gradient of the solvent transition was as follows: for positive ionization mode t = 0, 100% B; t = 30 min, 0% B; t = 32 min, 0% B; t = 35 min, 100% B; t = 60 min, 100% B. In negative ionization mode t = 0, 100% B; t = 30 min, 0% B; t = 32 min, 0% B; t = 35 min, 100% B; t = 60 min, 100% B. Metabolite identification was confirmed by fragmentation spectra of the detected ions. The collision energy was fixed at 20 eV, and MS/MS spectra were recorded in the range of 30–1000 m/z with the minimal width of the ion isolation window at 1.3 m/z.
mzXML spectra results are available at: https://dl.dropboxusercontent.com/u/73633783/Spectra.rar
Identification of metabolites and data processing software
Metabolite search and data processing were performed using the Metabolomic Analysis and Visualization Engine (MAVEN) [19] software and the Trans-Proteomic Pipeline (TPP) resource available online, where the data were converted into the mzXML format (MAVEN compatible). S. melliferum, M. gallisepticum, and A. laidlawii metabolites were identified using the list of all possible metabolites for these bacteria, garnered from all the protein annotation data that were previously reported [3], [11]. A list of all theoretically possible metabolites of S. melliferum, M. gallisepticum, and A. laidlawii was prepared in accordance with the KEGG database [20] and contained all the metabolites associated with the proteins that were annotated for these bacteria. The following parameters were used for the search: range of m/z values (extraction window), 15 ppm m/z; minimum intensity of peak, 1000 a.u.; minimum value of baseline signal intensity ratio, 10. If an identified metabolite was detected in at least five MS runs out of seven in specie one, while in specie two it was not detected, or it was detected only once, we attributed this metabolite to be present in the first species and to be absent in the second one.
Comparative analysis of the S. melliferum, M. gallisepticum, and A. laidlawii metabolomes
A pairwise sample comparison of metabolomic data for three species was performed using the XCMS online service, providing a direct comparison of two sample groups [21]. From the resulting features of every compared pair of oganisms, we selected only those with a p value≤0.05 in order to state that compound concentration is different in two species. Additionally these compounds were analyzed manually by counting the metabolite identifications among the seven repetitions of every species for both acquisition modes. The quantification of an identified metabolite is possible only if it was detected at least three times in each species. In the case a compound was detected less than three times of seven, only a qualitative comparison in the pair was possible, based on the detection of MS counts for this compound in the pair.
Results
MS spectra acquisition
Bacteria were cultivated in liquid media to more easily determine the growth phase. Bacteria were harvested in the logarithmic growth phase, which was determined based on the culture duplication time (see “Bacteria strain and growth conditions”). Cold methanol extraction was used to instantly quench the metabolism [16]. The high effectiveness of this method and its low loss of compounds has been demonstrated previously [22]. Metabolite detection was carried out using HPLC analysis in combination with electrospray ionization quadrupole time-of-flight mass spectrometry (HPLC-MS Q-TOF). The parameters of the chromatographic method, as well as the mass spectrometric peak recording, were optimized using standards to obtain the most reliable data (see “Materials and Methods” section). Metabolites were initially identified using the Metabolomic Analysis and Visualization Engine (MAVEN) software [19]. The metabolite identity was then confirmed by analysis of the detected parent ions fragmentation spectra using targeted mass spectrometry [23].
There were seven independent repeats for metabolite detection for each of the three studied bacterial species. Usually one LC-MS analysis run produces ∼120 peaks in which the m/z values are close to the exact metabolite m/z values (deviating by less than 10 ppm). However, in MS acquisition, some compounds show unstable signals and display low reproducibility; their peak intensities are comparable to the noise level. Each compound was considered to be present in bacteria if it was observed at least three times (in seven runs) in any of the ionization modes (positive or negative) and the intensity of the corresponding peak was significantly higher than that of the background. List of identified metabolites is shown in Table 1.
Table 1. Identified metabolites of S. melliferum, M. gallisepticum and A. laidlawii.
| Compound name | Compound ID | Detection mode | ||
| A. laidlawii | S. melliferum | M. gallisepticum | ||
| L-Tyrosine | C00082 | pos+neg | pos+neg | pos+neg |
| L-Tryptophan | C00078 | pos+neg | pos+neg | pos+neg |
| L-Aspartate | C00049 | neg | pos+neg | pos+neg |
| L-Serine | C00065 | neg | pos+neg | pos+neg |
| L-Citrulline | C00327 | pos+neg | ||
| L-Threonine | C00188 | pos+neg | pos+neg | pos+neg |
| L-Proline | C00148 | pos+neg | pos+neg | pos+neg |
| L-Isoleucine/L-Leucine | C00407/C00123 | pos+neg | pos+neg | pos+neg |
| L-Lysine | C00047 | pos+neg | pos+neg | |
| L-Glutamine | C00064 | pos | ||
| L-Phenylalanine | C00079 | pos+neg | pos+neg | pos+neg |
| L-Methionine | C00073 | pos+neg | pos+neg | pos+neg |
| L-Arginine | C00062 | pos | pos+neg | |
| L-Histidine | C00135 | pos | pos | pos |
| L-Glutamate | C00025 | pos+neg | pos+neg | |
| 4-Methyl-2-oxopentanoate | C00233 | pos | pos | pos+neg |
| 2–3-Dihydroxy-3-methylpentanoate | C06007 | pos | pos | pos |
| L-Cysteine | C00097 | pos | ||
| L-Arogenate | C00826 | pos | ||
| L-Glycine | C00037 | neg | neg | |
| Mevalonate-5-phosphate | C01107 | pos | ||
| S-Ribosyl-L-homocysteine | C03539 | pos | pos | |
| S-Adenosyl-L-methionine | C00019 | pos | pos | |
| N-Formyl-L-methionine | C03145 | neg | neg | |
| 5-Oxopentanoate | C03273 | pos | pos | |
| Thymine | C00178 | pos | pos | pos |
| Adenine | C00147 | pos+neg | pos+neg | pos+neg |
| Xanthine | C00385 | pos+neg | pos+neg | pos+neg |
| Hypoxanthine | C00262 | pos | pos+neg | pos+neg |
| Guanine | C00242 | pos+neg | pos+neg | pos+neg |
| Uracil | C00106 | pos+neg | pos+neg | pos+neg |
| Thymidine | C00214 | pos+neg | neg | pos+neg |
| Deoxycytidine | C00881 | pos+neg | pos | pos |
| Cytidine | C00475 | pos+neg | pos+neg | pos+neg |
| Cytosine | C00380 | pos | pos | pos |
| Uridine | C00299 | pos+neg | pos+neg | pos+neg |
| Guanosine | C00387 | pos+neg | pos | pos+neg |
| Adenosine/Deoxyguanosine | C00212/C00330 | pos | pos | pos+neg |
| Deoxyadenosine | C00559 | pos | pos | pos |
| CMP | C00055 | pos+neg | pos+neg | pos+neg |
| dUMP | C00365 | pos+neg | pos+neg | neg |
| UDP | C00015 | pos+neg | pos | pos+neg |
| GMP | C00144 | pos+neg | pos+neg | pos+neg |
| dGMP/AMP | C00362/C00020 | pos+neg | pos+neg | pos+neg |
| dAMP | C00360 | pos+neg | pos+neg | neg |
| UMP | C00105 | pos+neg | pos+neg | pos+neg |
| dGDP/ADP | C00361/C00008 | pos+neg | pos+neg | pos+neg |
| dTMP | C00364 | neg | neg | neg |
| ITP | C00081 | pos | pos | pos |
| dUTP | C00460 | pos | pos | pos |
| 2-Deoxyinosine 5-phosphate | C06196 | pos | pos | |
| Xanthosine | C01762 | pos | neg | |
| Inosine | C00294 | neg | neg | |
| XTP | C00700 | neg | neg | |
| dTDP | C00363 | neg | neg | |
| dUDP | C01346 | neg | ||
| Deoxyuridine | C00526 | pos+neg | neg | |
| dTTP | C00459 | neg | neg | |
| dCMP | C00239 | neg | ||
| S-Adenosyl-L-homocysteine | C00021 | pos | ||
| a-a-Trehalose/Maltose/Sucrose/Lactose | C01083/C00208/C00185/C00089/C00243 | pos+neg | ||
| D-Mannitol/D-Sorbitol | C00392/C00794/C01697 | pos+neg | pos+neg | pos+neg |
| Sedoheptulose 7-phosphate | C05382 | neg | neg | neg |
| D-Ribose 5-phosphate | C00117 | neg | neg | |
| GDP-6-deoxy-D-mannose | C03117 | neg | pos | |
| D-Mannitol 1-phosphate/D-Sorbitol-1-phosphate | C00644/C01096 | pos+neg | ||
| 3-Phospho-D-glycerate | C00197 | pos | ||
| Phosphoenolpyruvate | C00074 | neg | neg | |
| N-Acetyl-D-galactosamine 6-phosphate/n-acetyl-D-glucosamine-1-phosphate/N-Acetyl-D-mannosamine 6-phosphat | C06376/C04501/C04257 | neg | ||
| Glycerone phosphate/glyceraldehydes-3-phosphate | C00111/C00118 | neg | neg | |
| beta-D-Fructose 1–6-bisphosphate | C05378 | pos | ||
| Glycerone | C00184 | neg | neg | |
| 5-Phospho-alpha-D-ribose 1-diphosphate | C00119 | pos | pos | pos |
| Thiamin diphosphate | C00068 | pos | pos+neg | pos+neg |
| Formate | C00058 | neg | neg | neg |
| Geranylgeranyl diphosphate | C00353 | neg | neg | |
| 2-C-Methyl-D-erythritol 2–4-cyclodiphosphate | C11453 | neg | neg | neg |
| 5–10-Methenyltetrahydrofolate | C00445 | pos | pos | pos |
| 5-Amino-6-(5-phospho-ribosylamino)uracil | C01268 | neg | neg | neg |
| 5-Amino-6-(5-phospho-D-ribitylamino)uracil | C04454 | neg | neg | |
| sn-Glycerol 3-phosphate | C00093 | pos | neg | pos+neg |
| 2-C-Methyl-D-erythritol 4-phosphate | C11434 | neg | ||
| 5–10-Methylenetetrahydrofolate | C00143 | pos+neg | neg | |
| NADH | C00004 | neg | pos | |
| FMN | C00061 | pos+neg | pos+neg | |
| FAD | C00016 | neg | ||
| Nicotinate D-ribonucleoside | C05841 | pos+neg | pos+neg | pos+neg |
| NAD | C00003 | pos+neg | pos+neg | |
| Deamino-NAD+ | C00857 | pos+neg | pos+neg | pos+neg |
| Nicotinurate | C05380 | pos | pos | pos |
| 2-Acetolactate | C00900 | pos | pos | |
| 4-Methyl-3-oxoadipate | C18312 | pos | pos | |
| Nitrobenzene | C06813 | pos | ||
| UDP-N-acetyl-D-galactosaminuronic acid | C13952 | neg | ||
| UDP-N-acetyl-D-mannosamine | C01170 | neg | pos+neg | pos |
Fragmentation spectra were obtained for most of the detected metabolites using targeted mass spectrometry. The fragmentation spectra were compared with standard fragmentation spectra, which were obtained under similar ionization conditions (polarity of ionization and collision energy values were taken into account) and provided by the Metlin MS/MS Spectrum Match service [24], [25], [26]. Figure S1 shows an example of the metabolite identification as provided by the MS/MS. Interestingly, all fragmentation spectra confirmed the primary identification of the compounds based on the m/z values alone. There were no compounds whose fragmentation spectra contradicted the identification based on the m/z values. The provided m/z values usually did not deviate from the theoretical value by more than 10 ppm (see Table 2). Among the detected metabolites, there were several compounds that presented a relatively large deviation from their theoretical ion m/z values (up to 15 ppm m/z). Nevertheless, MS/MS fragmentation spectra render the unambiguous identification of such compounds possible.
Table 2. Fragmentation spectra results for the detected metabolites of S. melliferum, M. gallisepticum and A. laidlawii, and fragmentation spectra of the analyzed ion standards from the Metlin database [23] in the same experimental conditions (fixed collision energy of 20 eV).
| compound | Ionization Mode | dppm | Parent Mass (m/z) | Product Mass (m/z) from database Metlin | Found Product Mass (m/z) | Retention Time (minutes) |
| Adenine | + | 13 | 136.060 | 136,062;119,035;94,040 | 136,0575;119,030;94,034 | 28,5 |
| Thymine | + | 12 | 127.048 | 127.0502;110.0229;109.0382;84.0439;82.0286;56.0501;54.0340 | 127.0502;110.0229;109.0382;84.0439;82.0286;56.0501;54.0340 | 16–17 |
| Hypoxanthine | − | 12 | 135.030 | 92,025;65,015;66,0097 | 92,0305;65,0187;66,015 | 28,3 |
| Uracil | − | 8 | 111.021 | 41.998 | 41.999 | 9 |
| Guanine | + | 4 | 152.056 | 152,056;135,030;110,035;82,041 | 152,056;135,030;110,0361;82,038 | 23,3 |
| Deoxyadenosine monophosphate | + | 10 | 332.072 | 332.313;136.061;81.0333 | 332.313;136.061;81.0333 | 26.3 |
| Guanosine | + | 5 | 284.099 | 152.0559;135.0286;110.0347 | 284.0989;152.0559 | 25 |
| Cytidine | + | 7 | 244.093 | 112.0492 | 244.0928;112.0492 | 25 |
| CMP | + | 5 | 324.059 | 112.0502 | 324.0591;112.0502 | 27 |
| GMP | − | 0 | 362.051 | 78,959;96,968;150,042;210,999 | 78,959;96,968;150,042;211,001 | 38,9 |
| AMP | − | 0 | 346.056 | 134,047;96,969;78,959 | 134,0476;96,9698;78,9598 | 39,2 |
| ADP | − | 0 | 426.022 | 328,045;276,952;158,924;134,046;78,959 | 328,0505;276,952;158,9235;134,0475;78,9549 | 39,5 |
| UDP | − | 12 | 403.000 | 323,028;111,020;96,969;78,959; | 323,028;111,020;96,969;78,959; | 24,9 |
| CMP | − | 0 | 322.045 | 110,033;96,969;78,958 | 110,025;96,960;78,950 | 48,4 |
| dTDP | − | 14 | 401.010 | − | 195,007;78,964;176,997;96,969 | 39,3 |
| UMP | − | 1 | 323.029 | 211,001;150,980;138,980;111,0200;96,969;78,959;192,990 | 211,0014;193,000;150,913;138,980;111,027;96,972;78,959 | 45,4 |
| dTMP | − | 0 | 321.050 | 195,005;125,034;78,959;176,995;96,967 | 195,0089;125,034;78,9584;176,995;96,972 | 38,8 |
| L-Glutamate | + | 10 | 147.074 | 102.0549;84,045;56,0502;41,039;85.0291 | 148.058;102.0549;84,0445;56,0488 | 5,3 |
| L-Tryptophan | − | 0 | 203.082 | 142,071;116,055;74,0307 | 34,5 | |
| L-Leucine | + | 0 | 132.102 | 86,097;44,050;43,055;30,034 | 86,09499;44,050;43,055;30,034 | 35,1 |
| L-Isoleucine | + | 0 | 132.102 | 86,097;57,058;44,050;43,055;30,034 | 86,0965;57,059;44,051;43,056;30,0345 | 36,4 |
| L-Cysteine | + | 11 | 122.026 | 58,996; | 58,996; | 38,67 |
| L-Threonine | − | 2 | 118.051 | 74,059;57,035;56,050 | 74,0601;56,0498 | 39,0 |
| L-Proline | + | 3 | 116.071 | 70.066 | 70.064 | 34 |
| L-Phenylalanine | + | 2 | 166.086 | 120,081;103,054 | 120,0801;103,0539 | 35 |
| L-Histidine | + | 7 | 156.076 | 110,071;95,061;93,045;83,061 | 110,0668;95,0575;81,046 | 36,2 |
| L-Tyrosine | − | 0 | 180.067 | 72,0093;93,0343;119,0500;163,038;74,0253 | 72,010;119,0516;163,0389 | 47,2 |
| L-Lysine | + | 4 | 147.112 | 84,081; | 84,0766; | 36,5 |
| L-Aspartate | − | 0 | 132.030 | 115,003;88,0401;72,008;71,014;59,0133;42,0328 | 115,011;88,0454;72,032;71,014;59,0187;42,0384 | 38,3 |
| L-Serine | − | 3 | 104.036 | 74.025 | 74.0272 | 39,7 |
| L-Arginine | + | 8 | 175.1157 | 116.0707;70.0658;60.0653 | 175.1157;70.0658;60.0653 | 29.2 |
| L-Methionine | + | 4 | 150.057 | 56.0501;61.0114;104.0524;133.0313;102.0544;74.0244;87.0263 | 56.0501;61.0114;104.0524 | 27 |
| D-Glyceraldehyde 3-phosphate/Dihydroxyacetone phosphate | − | 3 | 168.990 | 96,969;78,959 | 96,974;78,964 | 39,4 |
| Sucrose | + | 9 | 365.1022 | 365.1052;203.0518;185.0415 | 365.1052;203.0518;185.0415 | 25.2 |
| 2-Acetolactate | − | 2 | 131.035 | − | 85,8767;45,0019;41,993 | 32,3 |
| N-Acetyl-D-glucosamine 6-phosphate | − | 3 | 300.050 | − | 260,387;78,961;32,675;92,923;94,920;96,964 | 44,8 |
| Sorbitol/Mannitol | − | 12 | 181.075 | 119,035;101,024;89,024;89,025;73,029;71,0145;59,0144;44,999;43,019 | 119,050;101,0188;89,018573,025;71,009;59,0140;44,996;43,016 | 43,05 |
| NADH | + | 3–21 | 665.115 | 542,062;524,060;428,035;323,084;136,061 | 542,061;524,059;428,0389;232,084;136,061 | 38,9 |
| Betaine | 118.086 | 118.086;59.0739;58.0660 | 118.086;59.0739;58.0660 | 28–30.7 | ||
| Cytosine | + | 11 | 112.0489 | 112.0505;95.0242;94.0401;69.0452;68.0136;67.0296;52.0189 | 112.0505;95.0242 | 26.3 |
| NAD | + | 3 | 664.1164 | 542.0622;524.060;428.035;232.0836;136.0603 | 664.1164;542.0622;524.060;428.035;232.0836;136.0603 | 27 |
| dGMP | + | 9 | 386.0227 | 152.0544;81.033 | 386.0227;152.0544 | 24.5 |
Metabolite content of S. melliferum, M. gallisepticum, and A. laidlawii
The strategy for metabolome analysis includes the following steps: 1) Metabolic map reconstruction; 2) Prediction of all possible metabolism intermediates; 3) Comparison of metabolic enzymes of Mollicute species; 4) Experimental MS detection and mapping of the detected compounds on the metabolic map for each specie; 5) Juxtaposition of the metabolome data to enzyme differences between three Mollicute species.
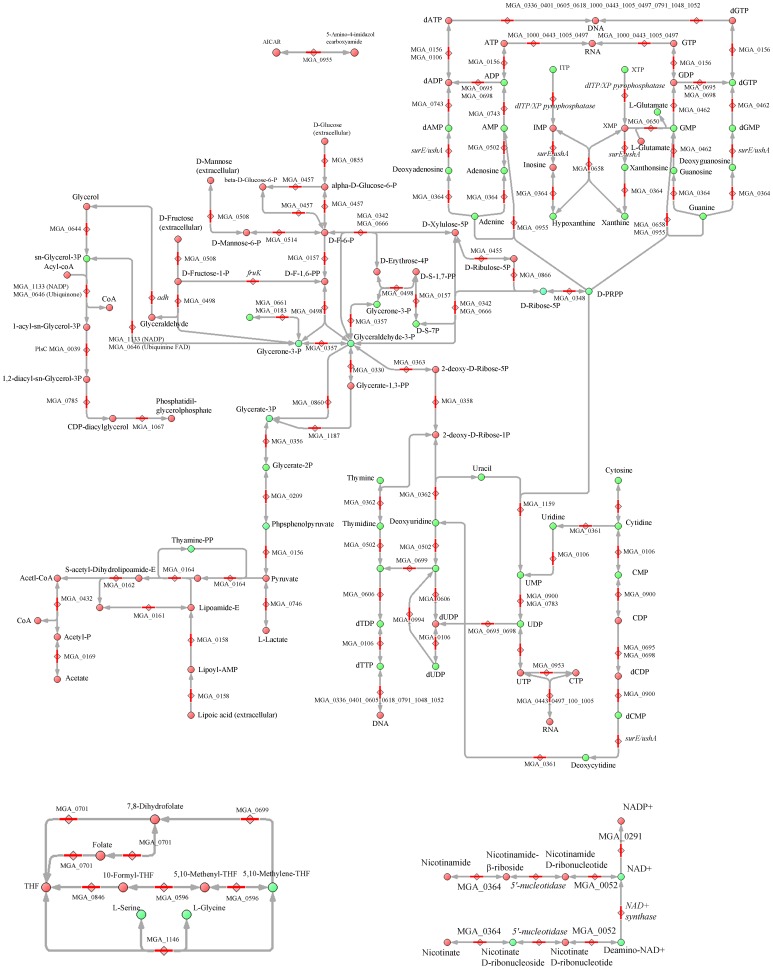
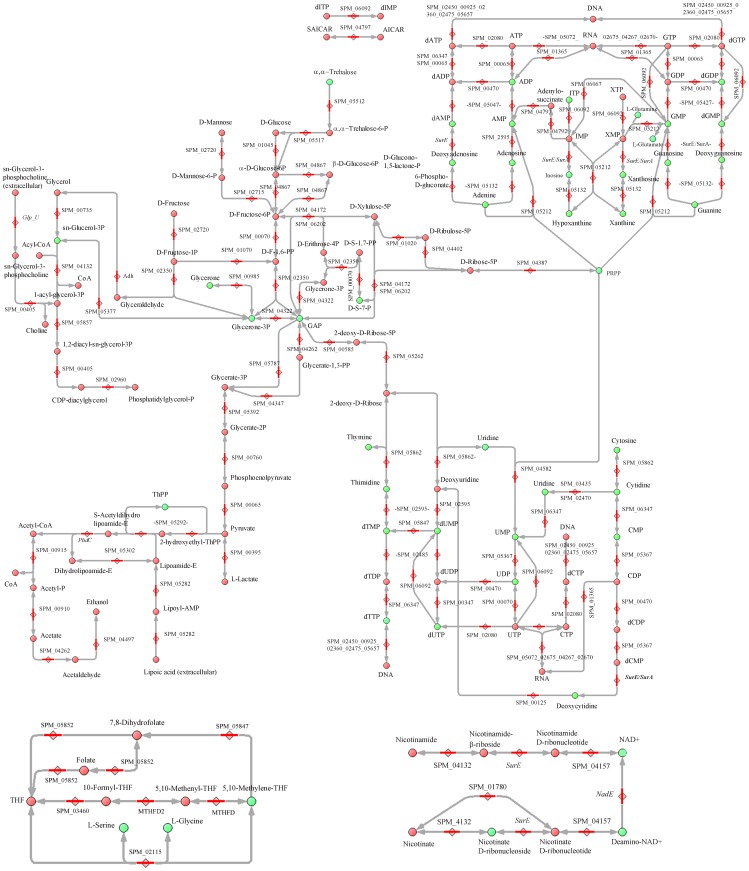
We manually reconstructed the metabolic network of M. gallisepticum, S. melliferum, and A. laidlawii in order to fill the network with the detected intermediates. To build a comprehensive metabolic network for each species, we used the Kyoto Encyclopedia of Genes and Genomes database and UniprotKB proteogenomic annotation data provided earlier by our lab [3], [11], [27]. For enzymes that could carry out more than one reaction, we removed the reactions that were decoupled from pathways and those for which the substrate was unavailable (i.e. there is no way for substrate synthesis and transport into the cell according to the proteogenomic annotation). The final result is a map without gaps, isolated reactions, or open metabolic loops for each species (Figures 1–3, see text S1 and Figures S2, S3).
Figure 1. Reconstructed metabolic map of M. gallisepticum.
The pathways common for three Mollicute species are represented. Metabolites are shown as circles; compounds identified by LC-MS are marked in green, other predicted compounds are marked in red. Proteins that catalyze metabolic reactions are shown as diamonds, and their ID numbers are indicated. Enzymatic activities, which are not associated with annotated proteins, are indicated in italics. Abbreviations: D-F-6-P - D-fructose-6-phosphate; D-F-1,6-PP - D-fructose-1,6-bisphosphate; D-S-1,7-PP - D-sedoheptulose-1,7-bisphosphate; D-S-7-P - D-sedoheptulose-7-phosphate; GAP - glyceraldehyde-3-P).
Figure 3. Reconstructed metabolism of S. melliferum.
The pathways common for three Mollicute species are represented. Abbreviations and symbols are given in figure 1.
Figure 2. Reconstructed metabolic map of A. laidlawii.
The pathways common for three Mollicute species are represented. Abbreviations and symbols are given in figure 1.
For each species a list of predicted metabolism intermediates was formed. The map includes all compounds, which serve as substrates and products for all enzymes and transporters predicted by the proteogenomic annotation. The metabolic database for raw MS data analysis by MAVEN includes all metabolism intermediates predicted for the three species. This database contains 500 items and is suitable for the analysis of each species. The database used is wider than the list of possible metablolites for each species. It therefore permits detection of metabolites produced by putative uncharacterized enzymes.
Metabolic pathways were reconstructed with all the identified metabolites of S. melliferum, M. gallisepticum and A. laidlawii positioned in the maps (Figures 1–3). The metabolome predicted by proteogenomic annotation consists of 323 metabolites for A. laidlawii, 225 for M. gallisepticum, and 361 for S. melliferum. We have identified 75, 74 and 74 metabolites for each species, respectively. Accordingly, we were able to detect 20 to 33% of the predicted metabolites by LC-MS using HILIC chromatography (Figure 4 and Table 1).
Figure 4. Overlap of the detected metabolites for three Mollicute species.

Yellow circle represents metabolites of M. gallisepticum, violet circle represents metabolites of S. melliferum, and green circle represents metabolites of A. laidlawii. The list of identified metabolites of S. melliferum, M. gallisepticum and A. laidlawii is presented in Table 1.
Reconstruction of the three species' metabolic maps reveals many pathways with enzymatic gaps in the middle. For pathways in which only one enzyme is missing, the gap can be filled by adding an unassigned reaction as performed earlier for another Mycoplasma species, M. pneumoniae [23]. Alternatively, for a missing enzymatic reaction, a bypass can be found by the manual analysis of the known enzyme's side activities.
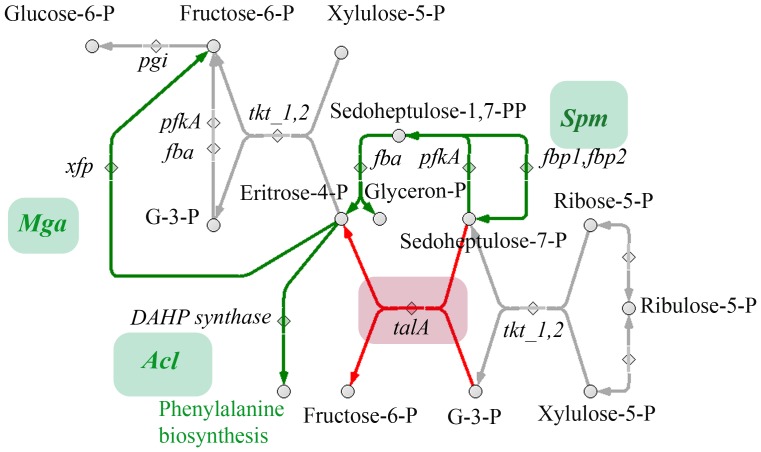
One example is the link between pentoses and hexoses (the pentose phosphate pathway) in Mollicutes. This link is omnipresent in living cells, although in many organisms, including mycoplasmas, only its non-oxidative branch is annotated. Two enzymes, transketolase (tktA) and transaldolase (tal), act in this branch of the pentose phosphate pathway (Figure 5). Transketolase transforms two pentoses, xylulose-5-phosphate and ribose-5-phosphate, into the seven-carbon product sedoheptulose-7-phosphate and the three-carbon glyceraldehyde-3-phosphate. To produce the hexose, fructose 6-phosphate, and glyceraldehyde-3-phosphate and to finish the pentose phosphate pathway, the same enzyme uses the four-carbon erythrose-4-phosphate and the pentose xylulose-5-phosphate. A key enzyme, transaldolase, acts to link these two transketolase reactions; it removes three-carbon fragment from sedoheptulose-7-phosphate and condenses it with glyceraldehyde-3-phosphate, forming the fructose 6-phosphate and the erythrose-4-phosphate [28].
Figure 5. Reactions of the pentose phosphate pathway.
Metabolites involved in the pathway are shown as circles; enzymes are shown as diamonds; and the path that we propose to convert sedoheptulose-7-phosphate is marked in green.
Annotation of genome sequence shows that Mollicutes lack transaldolase, and this obviously creates a gap in the metabolic branch. Nevertheless, we detected a unique intermediate of transketolase reaction, sedoheptulose-7-phosphate, in all three species studied (along with less specific intermediates). It suggests that the pentose phosphate pathway is functional. The connection of the glycolytic pathway with a functional pentose phosphate pathway was earlier demonstrated for M.pneumoniae as 13C-6 glucose carbons were detected in ribose 5-phosphate using mass spectrometry [23].
Database search for transaldolase-like proteins in all other Mollicute genomes produces a negative result. Therefore we believe that transaldolase activity is absent in this genus. To fill the gap, it is possible to postulate the presence of aldolase activity by an unknown mycoplasma protein. The other way is to find a bypass of this reaction performed by other known mycoplasma enzymes. The literature shows that sedoheptulose-7-phosphate can act as acceptor for 6-phosphofructokinase; fructose-bisphosphate aldolase can bind with sedoheptulose-1,7-bisphosphate and decompose it to glycerone phosphate and erythrose-4-phosphate (see Figure 5) [29]. Glycerone phosphate can be isomerized to glyceraldehyde-3-phosphate by triosephosphate isomerase [28].
The proposed modifications in the pentose phosphate pathway make it possible to utilize ribose as an energy source despite the lack of transaldolases. The described pathway is reversible in S. melliferum, and A. laidlawii. The first reaction, phosphorylation of sedoheptulose-7-phosphate, catalyzed by 6-phosphofructokinase, is irreversible; however, dephosphorylation of sedoheptulose-1,7-bisphosphate can be carried out by fructose -1,6-bisphosphatase [30], annotated in S. melliferum [11].
Fructose 1,6-bisphosphatase is not annotated in A. laidlawii, and eritrose-4-phosphate, formed as a result of the transketolase reaction between fructose-6- phosphate with glyceraldehyde-3- phosphate, must be utilized in some other way. Eritrose-4-phosphate may be utilized in phenylalanine synthesis, as all enzymes of phenylalanine biosynthesis are annotated in A. laidlawii [3]. A lack of fructose-1,6-bisphosphatase, as well as the absence of transaldolase activity in M. gallisepticum, was experimentally shown by J.D. Pollack and M.V. Williams [31]. We assume that the resultant eritrose-4-phosphate in M. gallisepticum is utilized by D-fructose-6-phosphate D-erythrose-4-phosphate-lyase; this enzyme is not annotated for M. gallisepticum, but it is annotated for another Mycoplasma species (M. fermentans). We believe that this activity should be assigned for M. gallisepticum. This is a way to implement the nonoxidative reactions of the pentose phosphate pathway for M. gallisepticum.
Among the detected metabolites we found compounds that participate in the main metabolic pathways, such as glycolysis, amino acid, sugar, and amino sugar metabolism, synthesis of terpenoids, riboflavines, purine and pyrimidine metabolism, etc. We used the LC-MS/MS method to analyze 95 compounds (see Table 1), representing a substantial fraction of the >500 compounds of the total metabolome of three Mollicute species, M. gallisepticum, S. melliferum, and A. laidlawii. These compound were predicted by protein annotation, and performed by our lab in previous studies [3], [11]. In particular, the purine and pyrimidine metabolism maps were the most uniformly and densely filled maps with the identified metabolites (see Figures 1–3). We detected a variety of purine and pyrimidine metabolism products: nitrogenous bases- adenine, thymine, guanine, xanthine, hypoxanthine, uracil, cytosine; their nucleosides and deoxynucleosides and some of their mono-, di-, triphosphate nucleotides and deoxyribonucleotides. It should be noted that we did not consider inosine mono- and dinucleotide, as according to V.M. Boer et al., this compound is not detectable, as it can be confused with peak interference of ADP and AMP [32]. Expression of the annotated proteins that are functionally associated with the metabolism of purine and pyrimidine was demonstrated in our previous studies [3], [11]. In the S. melliferum metabolome, we did not detect deoxyuridine, dTDP, XTP, xanthosine, 2-deoxyinosine 5-phosphate; however, these compounds have been detected in M. gallisepticum and A. laidlawii. Among M. gallisepticum and A. laidlawii metabolites, we did not identify inosine and dTTP, respectively.
There are many nutrient carriers in the Mycoplasma proteome, so the description of membrane fluxes and the discovery of exogenous metabolites acquired by the membrane transport is an important issue in metabolome research. Carbohydrates and their derivatives (such as glycolysis metabolites; see Figures 1–3) including sedoheptulose-7-phosphate, D-ribose-5-phosphate, D-mannitol 1-phosphate/D-sorbitol 1-phosphate, fructose-1,6-bisphosphate, deoxy-D-ribose-1-phosphate, 5-phospho-alpha-D-ribose-1-diphosphate, 3-phospho-D-glycerate, phosphoenolpyruvate, and n-acetyl-D-galactosamine-6-phosphate/n-acetyl-D-glucosamine-6-phosphate/N-Acetyl-D-mannosamine 6-phosphate were detected in all three species. We detected deoxy-D-ribose-1-phosphate in M. gallisepticum and A. laidlawii metabolome, while the peak of this compound was considered to be peak interference according to V.M. Boer et al. [32]. We detected n-acetyl-D-galactosamine-6-phosphate and beta-D-fructose-1–6-bisphosphate in the A. laidlawii metabolome, while 3-phospho-D-glycerate is a characteristic feature of the M. gallisepticum metabolome. The latter compound was also detected in M. pneumoniae [18]. Detection of phosphorylated sugar products (mannitol 1-phosphate) is in good agreement with proteogenomic annotation [3], [11] (PTS system mannitol-specific (MtlA)-like IIB domain protein MGA_1283) and with the identification of phosphorylated sugars in M. pneumoniae [18].
In all three bacteria, using both positive and negative ionization modes, we detected non-phosphorylated forms of mannitol/sorbitol/galactitol (sugars and some amino acids like leucine/isoleucine isomers cannot be differentiated using the HPLC-MS method, since they have particularly identical fragmentation spectra and retention times) and alpha-alpha-trehalose/maltose/cellobiose/sucrose/lactose.
Sugars are the most important energy source for the Mollicutes; their import in the cytoplasm is usually coupled with phosphorylation [11]. Here we directly demonstrate that mycoplasmas contain non-phosphorylated sugars. It was assumed that non-phosphorylated forms of carbohydrates can be accumulated from media or adsorbed to the cell surface [1], [33], [34], [35], [36]. Sugar ABC transporter systems which are not associated with phosphorylation were also annotated (MGA_1076, MGA_1077, MGA_1078 for M. gallisepticum, SPM_2645 S. melliferum). It cannot be ruled out, but it does seem unlikely that mycoplasmas store sugars to consume them during possible carbohydrate starvation.
We detected 17 of the 20 amino acids in the three Mollicute species. Their presence in the metabolome can be attributed to their exogenous nature and the fact that they are actively imported into the cells through specific transporters (permeases), since no amino acid synthesis enzymes have been annotated in these organisms [11]. Peaks in the MS spectra that corresponded to amino acids have the highest intensity. Only asparagine, alanine, and valine were not detected. In experiments employing the same equipment and another bacteria (not shown), we were able to detect these three amino acids. This suggests that the concentration of the three amino acids, which were not found in S. melliferum, M. gallisepticum and A. laidlawii total metabolome, was significantly lower than the concentration of the seventeen detected amino acids. Of note, in M. pneumoniae these three amino acids were detected [18]. Alanine is the most abundant amino acid in proteins and its absence in the metabolome could be explained by its rapid involvement into protein synthesis through aminoacyl-tRNA synthetase.
In M. pneumoniae riboflavin was detected by LC-MS [18]. We have detected intermediates of riboflavin biosynthesis, 5-amino-6-(5-phospho-D-ribitylamino)uracil, 5-amino-6-(5-phosphoribosylamino)uracil, FMN and FAD. Among the explored three species, only A. laidlawii proteogenomic annotation contains proteins responsible for the diaminohydroxyphosphoribosylaminopyrimidines conversions in addition to the bifunctional FAD synthetase/riboflavin kinase. However, in the S. melliferum and M. gallisepticum metabolomes 5-amino-6-(5-phosphoribosylamino) uracil mass-spectral peaks were reliably detected with a high intensity. This finding can indicate activity of a metabolic branch responsible for FMN biosynthesis from GTP in S. melliferum and M. gallisepticum.
Discussion
The obtained list of metabolism intermediates taken together with our previous studies on the protein annotation of the same cells makes it possible to understand the exact mechanisms of the biochemical reactions in three Mollicute species (M. gallisepticum, S. melliferum, and A. laidlawii). Metabolome studies of the simplest bacteria reveal compounds and pathways that are important for the basic functions of bacteria maintenance. There were some metabolites that were not detected in our experiment but were present in the list of metabolites based on the bacterial proteome [3], [11]. Some metabolites cannot be detected by LC-MS, because, for example, they are unstable under ionization conditions. Metabolites that are non-soluble in methanol cannot be detected either. Another reason why some metabolite intermediates were not detected is their possible consumption during cell wash in poor medium or even due to the residual enzymatic activity during the methanol extraction [22]. We detected several glycolysis intermediates (3-phospho-D-glycerate, phosphoenolpyruvate, glyceraldehyde-3-phosphate, beta-D-fructose 1–6-bisphosphate), so we conclude that the glycolysis rate is significantly decreased under the quenching conditions that we use. Similar sample preparation procedures revealed a similar glycolysis intermediate content in M. pneumoniae [18]. Several items that we did not expect to be revealed as metabolism components were detected. For example, in S. melliferum, S-ribosyl-L-homocysteine and 5-oxopentanoate were detected. As no enzymes responsible for their utilization or synthesis were annotated, we believe that such compounds are the products of amino acid degradation.
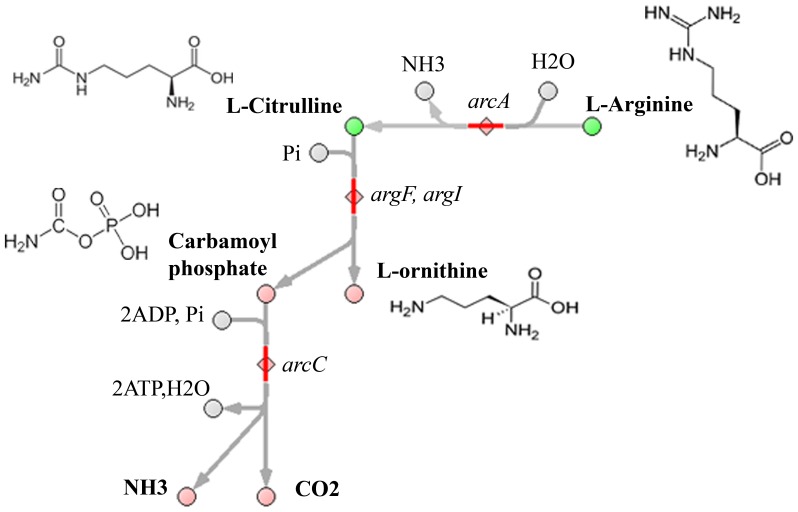
The differences that we identified primarily concerned amino acids and lipid biosynthesis and mechanism of carbohydrate assimilation (owing to Mollicutes' parasitic nature, many nutrients are received from the host), and the biosynthesis of terpenoids and riboflavins. We found that citrulline concentration is higher in M. gallisepticum compared to A. laidlawii and S. melliferum while arginine concentration is lower in S. melliferum as compared to A. laidlawii (Table 3). Citrulline is an intermediate in the arginine deiminase (ADI) pathway. The ADI path converts arginine to citrulline and ammonia, ornithine transcarbamylase subsequently converts citrulline to ornithine and carbamoyl phosphate, and finally carbamate kinase cleaves carbamyl phosphate to ammonia and CO2, generating an ATP in that process (see Figure 6). The system also requires an arginine/ornithine antiporter to rid the cell of the end-product, ornithine, while importing a new arginine substrate [27]. Ammonia that is generated by arginine deamination can alkalinize the environment [37], and helps the survival of cultures during acid stress. Some Mycoplasmas (M. hominis, M. arthritidis, M. gallinarum and others) use the arginine pathway only as an alternative energy source [38].The central enzyme of this pathway, arginine deiminase, was identified in M. gallisepticum and S. melliferum genome and proteome in our previous studies, while it is absent from A. laidlawii [3], [11]. Only the S. melliferum genome annotation has proteins responsible for providing a complete ADI cycle, including arginine/ornithine antiporter [11], while M. gallisepticum lacks ornithine carbamoyltransferase [39].
Table 3. Metabolite peaks counting and pairwise comparison of three Mollicute species.
| Compound | Compound ID | Spectral counts | Normalized peak intensities ratio | Conclusion | ||
| M.gal. | A.laid. | S.melli. | ||||
| L-Lysine | C00047 | - | 6 | 10 | S.mellif./A.laid. = 3.0 | S.mellif.>A.laid.>M.gal. = 0 |
| L-Glutamate | C00025 | 5 | - | 4 | S.mellif./M.gal. = 1.2 | S.mellif. = M.gal.>A.laid. = 0 |
| L-Glutamine | C00064 | - | - | 4 | - | In S.mellif. only |
| CMP | C00055 | 8 | 7 | 12 | S.mellif./M.gal. = 3.7 S.mellif./A.laid. = 1.6 A.laid./M.gal. = 1.8 | S.mellif.>A.laid.>M.gal. |
| sn-Glycerol 3-phosphate | C00093 | 11 | 7 | 4 | A.laid./S.mellif./ = 1.4 M.gal./A.laid. = 24 M.gal./S.mellif. = 53 | M.gal.>A.laid.≥S.mellif. |
| 5-Amino-6-(5-phospho-D-ribitylamino)uracil | C04454 | - | 6 | - | - | In A.laid. only |
| 5-Amino-6-(5-phosphoribosylamino)uracil | C01268 | 3 | 7 | 7 | A.laid./M.gal. = 5.4 S.mellif/A.laid = 2.6 S.mellif/M.gal. = 30 | S.mellif.>A.laid.>M.gal. |
| 2-C-Methyl-D-erythritol 4-phosphate | C11434 | - | 6 | - | - | In A.laid. only |
| 2-C-Methyl-D-erythritol 2–4-cyclodiphosphate | C11453 | 7 | 6 | 4 | M.gal./A.laid. = 9.1 M.gal./S.mellif. = 20 A.laid./S.mellif = 1.3 | M.gal.>A.laid.≥S.mellif. |
| L-Arginine | C00062 | - | 5 | 4 | A.laid./S.mellif. = 2.2 | A.laid.>S.mellif.>M.gal = 0 |
| L-Citrulline | C00327 | 11 | - | - | - | In M.gal.only |
| Geranylgeranyl diphosphate | C00353 | 0 | 7 | 5 | A.laid./S.mellif. = 3.3 | A.laid.>S.mellif.>M.gal = 0 |
| a-a-Trehalose/Maltose/Sucrose/Lactose | /C00208/C00185/C00089/C00243 | - | - | 5 | - | In S.mellif.only |
Results are summarized for the positive and negative acquisition modes.
Figure 6. Arginine deiminase (ADI) pathway.
In this pathway, metabolites are shown as circles; enzymes are shown as diamonds; and compounds that we detected are marked in green.
Another way that microbes use to respond to acidification is by producing enzymes that can convert acidic metabolites to neutral products or neutral metabolites to alkaline products. Good examples of this type of enzymes are glutamate decarboxylase, lysine decarboxylase, and arginine decarboxylase of Escherichia coli, all of which exhibit increased expression at external acidic pH [40]. Ammonia can also be produced by the degradation of L-histidine, such as by the activity of L-histidine ammonialyase detected in ureaplasmas [41]. Glutamate decarboxylase is absent from all Mycoplasma species. The A. laidlawii annotation lacks arginine deiminase, glutamate decarboxylase or arginine decarboxylase, while the lysine decarboxylase annotated for A. laidlawii is unique among all Mollicutes. Lysine is a basic amino acid and possesses two amino groups. We found that lysine concentration is decreased in A. laidlawii compared to S. melliferum. Lysine decarboxylation can possibly serve as a mechanism to aid the survival of cultures in acid extracellular conditions.
Host infection caused by bacteria of both species, S. melliferum and M. gallisepticum, necessitates acid adaptation of the pathogens. When infected, chickens and turkeys mycoplasmosis bacteria M. gallisepticum should adapt to acidic environment conditions. The clinical manifestation following the infection is called chronic respiratory disease in chickens and infectious sinusitis in turkeys [26]. As the infection begins with the colonization of the respiratory tract, tracheitis and airsacculitis are the predominant symptoms of a localized infection in chickens [42]. In turn, bees are the main reservoir for spiroplasmas, where they primarily invade the gut lumen. Some species expanded their habitat range to include hemolymph, ovaries, fat bodies, hypodermis, and salivary glands. Spiroplasma infection of bees occurs with nectar consumption. The invertase of the pharyngeal gland secretions inverts sucrose into glucose and fructose, and then glucose oxidase converts glucose into gluconic acid to acidify honey (pH 4–5). To survive, spiroplasmas need to overcome this pH-barrier. The balance of citrulline/arginine during the growth of Mycoplasmas will be investigated in future research.
We suggest that A. laidlawii has adaptation mechanisms different from M. gallisepticum and S. melliferum and uses decarboxylation of lysine since A. laidlawii is widely distributed in nature and has infects a wide range of hosts [43]. Members of the Acholeplasma genus do not have specific host associations and have been found in almost every type of living organism. They are one of the five common reasons of cell culture contamination, and a causative agent of some plant diseases [44], [45], [46], [47].
To check whether M. gallisepticum, A. laidlawii and S. melliferum are able to survive in acidic environment, we used the “color test” method to estimate a fraction of the surviving cells and found that M. gallisepticum, A. laidlawii and S. melliferum can survive at low values of pH (∼5.0–5.3); this type of adaptation can be necessary during an infection.
We detected intermediates of purine metabolism (Figures 1–3). It should be noted that a direct transition between purine nitrogen base and monophosphate nucleotide is catalyzed by AMP pyrophosphorylase, which is found in all three species. Sequential conversion of nucleotides- to nucleosides and then to nitrogen bases is performed by the protein 5′-nucleotidase. This protein is not annotated for S. melliferum (in A. laidlawii, this protein is annotated, while M. gallisepticum contains the protein thymidine kinase, which is capable to catalyzing this reaction). Purine nucleoside phosphorylase is available in all three species. 5′-nucleotidase makes it possible to 1) consume AMP and use it as an energy source; 2) to use ribose from AMP as a building block for other nucleotide synthesis. However, we do not believe that 5′-nucleotidase should be assigned for S. melliferum, as was suggested for M. pneumoniae [23]. We believe that the purine metabolism pathway in S. melliferum is directed from phosphoribosyl-pyrophosphate to DNA and RNA synthesis while the reverse direction is absent (S. melliferum does not utilize nucleotides).
We detected a high level of glycerol-3-phosphate in A. laidlawii and a decreased CMP level compared to S. melliferum and M. gallisepticum (see Table 3). The composition of the A. laidlawii membrane differs from that of other Mollicutes, since the main lipids of its cytoplasmic membrane are the neutral glycolipids monoglucosyl diacylglycerol (MGDG) and diglucosyl diacylglycerol (DGDG), the phosphoglycolipid glycerophosphoryl diglucosyl diacylglycerol (GPDGDG), and the phospholipid phosphatidylglycerol (PG), whereas cholesterol is a major membrane component of other mycoplasmas [8]. An important reaction in phospholipid synthesis is the replacement of the cytosine unit by glycerol-3-phosphate to form phosphatidylglycerol (PG) and CMP. PG is a precursor of lipoglycans, the main component of the Acholeplasma membrane. Glycerol kinase is absent from the A. laidlawii genome annotation, while glycerol-3-phosphate (G3P) dehydrogenase is present [3]. G-3-P dehydrogenase catalyzes the reversible redox conversion of dihydroxyacetone phosphate to sn-glycerol 3-phosphate and serves as a major link between carbohydrate metabolism and lipid metabolism. A decreased level of glycerol-3-phosphate and a high content of CMP indicate an active synthesis of the main structural component of biological membranes - glycerophospholipids in A. laidlawii.
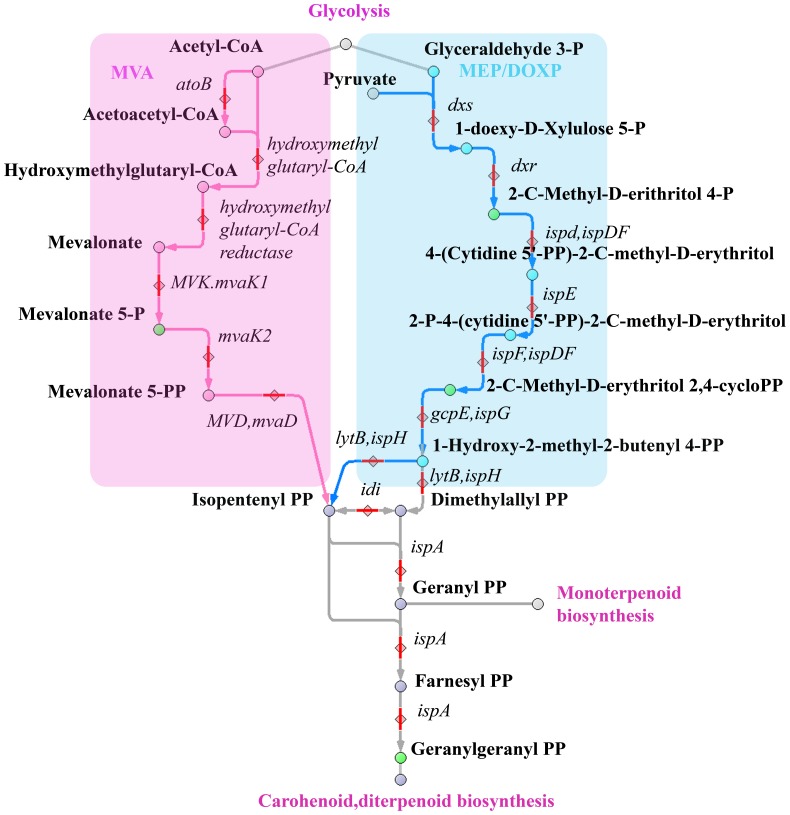
There are two metabolic pathways of terpenoid synthesis: the mevalonate acid pathway (MVA) and nonmevalonate pathway (2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose-5-phosphate pathway (MEP/DOXP pathway)) (Figure 7). Although both pathways, MVA and MEP/DOXP, are mutually exclusive in most organisms, interactions between these pathways have been reported in plants and several bacterial species [48], [49]. Many organisms manufacture terpenoids through the mevalonate acid pathway (MVA), that also produces cholesterol. These reactions, directed to carotenoid biosynthesis, take place in the A. laidlawii [3]. We have detected a high level of 2-C-methyl-D-erythritol 4-phosphate in A. laidlawii, we have not detected any metabolites involved the MVA path.
Figure 7. Terpenoid backbone biosynthesis pathway.
Metabolites involved in this pathway are shown as circles; enzymes are shown as diamonds; and compounds that we detected are marked in green.
The alternative nonmevalonate pathway or 2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose-5-phosphate pathway (MEP/DOXP pathway) of isoprenoid biosynthesis is an alternative metabolic pathway leading to the formation of isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). For numerous microbial pathogens such as Enterobacteria, Mycobacterium tuberculosis, M. gallisepticum and S. melliferum, the nonmevalonate pathway is the exclusive source of terpenoids [49]. Because the enzymes of the nonmevalonate pathway have no orthologs in mammalian hosts, they are attractive targets for the development of novel antibiotic or antiprotozoal agents. In M. gallisepticum and S. melliferum metabolome, we detected the presence of substantial amounts of 2-C-methyl-D-erythritol-2,4-cyclodiphosphate (component of the MEP/DOXP branches of metabolism), and found geranylgeranyl diphosphate (an important precursor in the biosynthesis of all higher terpenoids and carotenoid biosynthesis) only in S. melliferum. In M. gallisepticum and S. melliferum metabolisms, the presence of MEP/DOXP pathway proteins can be found, but the path terminated at the 1-hydroxy-2-methyl-2-butenyl 4-diphosphate [11] (Whole Genome Shotgun ID: AFFR00000000). Nevertheless, the identification of MEP/DOXP pathway components in the metabolome suggests active expression of annotated proteins and terpenoid backbone biosynthesis in M. gallisepticum and S. melliferum. Interestingly, we have also detected mevalonate-5-phosphate in S. melliferum metabolome; however, no proteins of the mevalonate acid pathway are noted in these bacteria.
We have systematically studied three bacterial species that belong to the class Mollicutes: the smallest and simplest bacteria, Spiroplasma melliferum, Mycoplasma gallisepticum, and Acholeplasma laidlawii. We described the basic difference in the principles of three species' metabolic adaptation to acidic environmental conditions and analyzed three species' their metabolomes to confirm our assumptions. The metabolic pathways of three species were reconstructed and analyzed using the proteogenomic annotation data provided by our lab in previous studies [3], [11].
Supporting Information
Metabolome reconstruction.
(DOCX)
MS/MS Spectrum match of Uracil fragmentation spectrum obtained for the S. melliferum sample. Uracil fragmentation spectrum obtained for the S. melliferum sample is above the OX axis and standard Uracil fragmentation spectrum in positive ionization mode collision energy 20 eV from Metlin Metabolites database is under the OX axis [30]; Uracil M+H: m/z = 113.03344, Δ = 10 ppm.
(TIF)
Reconstructed pathways characteristic for A. laidlawii which are absent in M. gallisepticum . For description see legend to Figure 1.
(TIF)
Reconstructed pathways characteristic for S. melliferum which are absent in M. gallisepticum . For description see legend to Figure 1.
(TIF)
Acknowledgments
We thank G.B. Smirnov for the critical review of the manuscript and A.A.Vassilevski for his help during the preparation of the manuscript.
Funding Statement
This work was supported by a Grant (12-04-31696) from Russian Foundation for Basic Research (RFBR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Razin S (1973) Physiology of mycoplasmas. Adv Microb Physiol 10: 1–80. [DOI] [PubMed] [Google Scholar]
- 2. Razin S, Michmann J, Shimshoni Z (1964) The Occurrence of Mycoplasma (Pleuropneumonia-Like Organisms, Pplo) in the Oral Cavity of Dentulous and Edentulous Subjects. J Dent Res 43: 402–405. [DOI] [PubMed] [Google Scholar]
- 3. Lazarev VN, Levitskii SA, Basovskii YI, Chukin MM, Akopian TA, et al. (2011) Complete genome and proteome of Acholeplasma laidlawii. J Bacteriol 193: 4943–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Laidlaw PP, Elford WJ (1936) A new group of filterable organisms. B Biol Sci 292–303. [Google Scholar]
- 5. Leach RH (1967) Comparative studies of mycoplasma of bovine origin. Ann N Y Acad Sci 143: 305–316. [DOI] [PubMed] [Google Scholar]
- 6. Stipkovits L (1979) The pathogenicity of avian mycoplasmas. Zentralbl Bakteriol Orig A 245: 171–183. [PubMed] [Google Scholar]
- 7. Pollack JD, Tryon VV, Beaman KD (1983) The metabolic pathways of Acholeplasma and Mycoplasma: an overview. Yale J Biol Med 56: 709–716. [PMC free article] [PubMed] [Google Scholar]
- 8. Cocaign-Bousquet M, Even S, Lindley ND, Loubiere P (2002) Anaerobic sugar catabolism in Lactococcus lactis: genetic regulation and enzyme control over pathway flux. Appl Microbiol Biotechnol 60: 24–32. [DOI] [PubMed] [Google Scholar]
- 9. van der Greef J, Stroobant P, van der Heijden R (2004) The role of analytical sciences in medical systems biology. Curr Opin Chem Biol 8: 559–565. [DOI] [PubMed] [Google Scholar]
- 10. Levitskiy SA, Sycheva AM, Kharlampieva DD, Oberto J, Kamashev DE, et al. (2011) Purification and functional analysis of recombinant Acholeplasma laidlawii histone-like HU protein. Biochimie 93: 1102–1109. [DOI] [PubMed] [Google Scholar]
- 11. Alexeev D, Kostrjukova E, Aliper A, Popenko A, Bazaleev N, et al. (2012) Application of Spiroplasma melliferum proteogenomic profiling for the discovery of virulence factors and pathogenicity mechanisms in host-associated spiroplasmas. J Proteome Res 11: 224–236. [DOI] [PubMed] [Google Scholar]
- 12. Kamashev D, Oberto J, Serebryakova M, Gorbachev A, Zhukova Y, et al. (2011) Mycoplasma gallisepticum produces a histone-like protein that recognizes base mismatches in DNA. Biochemistry 50: 8692–8702. [DOI] [PubMed] [Google Scholar]
- 13. Urano K, Kurihara Y, Seki M, Shinozaki K (2010) ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr Opin Plant Biol 13: 132–138. [DOI] [PubMed] [Google Scholar]
- 14. Tully JG, Whitcomb RF, Clark HF, Williamson DL (1977) Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science 195: 892–894. [DOI] [PubMed] [Google Scholar]
- 15. Gorbachev AY, Fisunov GY, Izraelson M, Evsyutina DV, Mazin PV, et al. (2013) DNA repair in Mycoplasma gallisepticum. BMC Genomics 14: 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maharjan RP, Ferenci T (2003) Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal Biochem 313: 145–154. [DOI] [PubMed] [Google Scholar]
- 17. Bolten CJ, Kiefer P, Letisse F, Portais JC, Wittmann C (2007) Sampling for metabolome analysis of microorganisms. Anal Chem 79: 3843–3849. [DOI] [PubMed] [Google Scholar]
- 18. Maier T, Marcos J, Wodke JA, Paetzold B, Liebeke M, et al. (2013) Large-scale metabolome analysis and quantitative integration with genomics and proteomics data in Mycoplasma pneumoniae. Mol Biosyst 9: 1743–1755. [DOI] [PubMed] [Google Scholar]
- 19. Melamud E, Vastag L, Rabinowitz JD (2010) Metabolomic analysis and visualization engine for LC-MS data. Anal Chem 82: 9818–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanehisa M (1997) A database for post-genome analysis. Trends Genet 13: 375–376. [DOI] [PubMed] [Google Scholar]
- 21. Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78: 779–787. [DOI] [PubMed] [Google Scholar]
- 22. Villas-Boas SG, Hojer-Pedersen J, Akesson M, Smedsgaard J, Nielsen J (2005) Global metabolite analysis of yeast: evaluation of sample preparation methods. Yeast 22: 1155–1169. [DOI] [PubMed] [Google Scholar]
- 23. Yus E, Maier T, Michalodimitrakis K, van Noort V, Yamada T, et al. (2009) Impact of genome reduction on bacterial metabolism and its regulation. Science 326: 1263–1268. [DOI] [PubMed] [Google Scholar]
- 24. Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, et al. (2005) METLIN: a metabolite mass spectral database. Ther Drug Monit 27: 747–751. [DOI] [PubMed] [Google Scholar]
- 25. Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, et al. (2012) An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol 30: 826–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vogl G, Plaickner A, Szathmary S, Stipkovits L, Rosengarten R, et al. (2008) Mycoplasma gallisepticum invades chicken erythrocytes during infection. Infect Immun 76: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moat A, Foster J, Spector M (2002) Microbial Physiology.: Wiley-Liss,Inc. [Google Scholar]
- 28.Lehninger A (2008) Princioals of biology.
- 29. Karadsheh NS, Tejwani GA, Ramaiah A (1973) Sedoheptulose-7-phosphate kinase activity of phosphofructokinase from the different tissues of rabbit. Biochim Biophys Acta 327: 66–81. [DOI] [PubMed] [Google Scholar]
- 30. Pontremoli S, Traniello S, Luppis B, Wood WA (1965) Fructose diphosphatase from rabbit liver. I. Purification and properties. J Biol Chem 240: 3459–3463. [PubMed] [Google Scholar]
- 31. Pollack JD, Williams MV (1986) PPi-dependent phosphofructotransferase (phosphofructokinase) activity in the mollicutes (mycoplasma) Acholeplasma laidlawii. J Bacteriol 165: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD (2010) Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol Biol Cell 21: 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Terry TM, Zupnik JS (1973) Weak association of glucosamine-containing polymer with the Acholeplasma laidlawii membrane. Biochim Biophys Acta 291: 144–148. [DOI] [PubMed] [Google Scholar]
- 34. Cole RM, Tully JG, Popkin TJ, Bove JM (1973) Morphology, ultrastructure, and bacteriophage infection of the helical mycoplasma-like organism (Spiroplasma citri gen. nov., sp. nov.) cultured from “stubborn” disease of citrus. J Bacteriol 115: 367–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robertson J, Smook E (1976) Cytochemical evidence of extramembranous carbohydrates on Ureaplasma urealyticum (T-strain Mycoplasma). J Bacteriol 128: 658–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schiefer HG, Gerhardt U, Brunner H, Krupe M (1974) Studies with lectins on the surface carbohydrate structures of mycoplasma membranes. J Bacteriol 120: 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pereyre S, Sirand-Pugnet P, Beven L, Charron A, Renaudin H, et al. (2009) Life on arginine for Mycoplasma hominis: clues from its minimal genome and comparison with other human urogenital mycoplasmas. PLoS Genet 5: e1000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hahn RG, Kenny GE (1974) Differences in arginine requirement for growth among arginine-utilizing Mycoplasma species. J Bacteriol 117: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Papazisi L, Gorton TS, Kutish G, Markham PF, Browning GF, et al. (2003) The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low). Microbiology 149: 2307–2316. [DOI] [PubMed] [Google Scholar]
- 40. Castanie-Cornet MP, Foster JW (2001) Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147: 709–715. [DOI] [PubMed] [Google Scholar]
- 41. Ajello F, Romano N, Massenti MF (1977) L-histidine ammonia-lyase from a T-strain Mycoplasma (Ureaplasma urealyticum). Boll Ist Sieroter Milan 56: 343–350. [PubMed] [Google Scholar]
- 42.Ley D (2003) Mycoplasma gallisepticum infection; M. SY, editor. Ames.: Iowa State Press. [Google Scholar]
- 43. Lee IM, Davis RE, Gundersen-Rindal DE (2000) Phytoplasma: phytopathogenic mollicutes. Annu Rev Microbiol 54: 221–255. [DOI] [PubMed] [Google Scholar]
- 44. Scripal IG, Bilay VI (1988) Microorganisms - agents of the plant diseases. Naukova Dumka [Google Scholar]
- 45. Chernov VM, Mukhametshina NE, Gogolev YV, Nesterova TN, Chernova OA (2007) Mycoplasma adaptation to adverse growth conditions: nanotransformation and phytopathogenicity of Acholeplasma laidlawii PG8. Dokl Biochem Biophys 413: 57–60. [DOI] [PubMed] [Google Scholar]
- 46. David SA, Volokhov DV, Ye Z, Chizhikov V (2010) Evaluation of Mycoplasma inactivation during production of biologics: egg-based viral vaccines as a model. Appl Environ Microbiol 76: 2718–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Folmsbee M, Howard G, McAlister M (2010) Nutritional effects of culture media on mycoplasma cell size and removal by filtration. Biologicals 38: 214–217. [DOI] [PubMed] [Google Scholar]
- 48. Lichtenthaler HK (1999) The 1-Deoxy-D-Xylulose-5-Phosphate Pathway of Isoprenoid Biosynthesis in Plants. Annu Rev Plant Physiol Plant Mol Biol 50: 47–65. [DOI] [PubMed] [Google Scholar]
- 49. Rohdich F, Hecht S, Gartner K, Adam P, Krieger C, et al. (2002) Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc Natl Acad Sci U S A 99: 1158–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolome reconstruction.
(DOCX)
MS/MS Spectrum match of Uracil fragmentation spectrum obtained for the S. melliferum sample. Uracil fragmentation spectrum obtained for the S. melliferum sample is above the OX axis and standard Uracil fragmentation spectrum in positive ionization mode collision energy 20 eV from Metlin Metabolites database is under the OX axis [30]; Uracil M+H: m/z = 113.03344, Δ = 10 ppm.
(TIF)
Reconstructed pathways characteristic for A. laidlawii which are absent in M. gallisepticum . For description see legend to Figure 1.
(TIF)
Reconstructed pathways characteristic for S. melliferum which are absent in M. gallisepticum . For description see legend to Figure 1.
(TIF)