Abstract
Background
Pitx2 (paired-like homeodomain 2 transcription factor) is crucial for heart development, but its role in heart failure (HF) remains uncertain. The present study lays the groundwork implicating Pitx2 signalling in different modalities of HF.
Methodology/Principal Findings
A variety of molecular, cell-based, biochemical, and immunochemical assays were used to evaluate: (1) Pitx2c expression in the porcine model of diastolic HF (DHF) and in patients with systolic HF (SHF) due to dilated and ischemic cardiomyopathy, and (2) molecular consequences of Pitx2c expression manipulation in cardiomyocytes in vitro. In pigs, the expression of Pitx2c, physiologically downregulated in the postnatal heart, is significantly re-activated in left ventricular (LV) failing myocardium which, in turn, is associated with increased expression of a restrictive set of Pitx2 target genes. Among these, Myf5 was identified as the top upregulated gene. In vitro, forced expression of Pitx2c in cardiomyocytes, but not in skeletal myoblasts, activates Myf5 in dose-dependent manner. In addition, we demonstrate that the level of Pitx2c is upregulated in the LV-myocardium of SHF patients.
Conclusions/Significance
The results provide previously unrecognized evidence that Pitx2c is similarly reactivated in postnatal/adult heart at distinct HF phenotypes and suggest that Pitx2c is involved, directly or indirectly, in the regulation of Myf5 expression in cardiomyocytes.
Introduction
The Pitx2 homeobox transcription factor gene was originally identified as the candidate gene for the human Axenfeld-Rieger's syndrome [1], which is characterized by severe eye, teeth, craniofacial and umbilical abnormalities; less common features include heart defects [2], [3]. Shortly after its identification, Pitx2 was found to play an important role in early development, as revealed by the generation of constitutive knockout mouse models. Consistent with Pitx2 expression patterns, homozygous disruption of the mouse Pitx2 gene led to mid-embryonic lethality due to defects in cardiac morphogenesis, in addition to severe abdominal wall and other tissue malformations [4]–[7]. Subsequently, the study of the Pitx2 gene has become the object of continued research efforts aimed at identifying its role in the fetal, adult and diseased myocardium (reviewed in [8]–[11]).
Selective Pitx2 deletion in the developing myocardium resulted in delayed differentiation of ventricular (but not atrial) cardiomyocytes, as development proceeded from embryonic to prenatal stages. During postnatal development, these mutants displayed dilatation and enlargement of right heart chambers and asymmetric hypertrophy of the interventricular septum associated with severely impaired ventricular systolic function [12]. Chamber-specific inactivation of Pitx2 within atrial myocardium led to dilatation of both the left (LA) and right atrium (RA), in the absence of significant defects in ventricular chambers of mutant foetuses. However, after birth the mutants displayed a maladaptive remodelling of both atria and ventricles, associated with electrophysiological dysfunction, preferentially in the LA [13]. The atrial conduction system is particularly sensitive to Pitx2 gene dose because mice heterozygous for Pitx2 deficiency did not display altered cardiac morphology and contractile function in any of the heart chambers, but under electrical stimulation showed atrial arrhythmias [14], [15].
Three Pitx2 transcript variants (Pitx2a, Pitx2b, and Pitx2c) have been identified in mammals, Pitx2c being the predominant or the only transcript detected in the adult mouse and human heart [13]–[17]. Of note, Pitx2c is differentially expressed across the cardiac chambers with maximal expression in the LA [15]. Pitx2c expression in the mouse LA is downregulated from fetal to postnatal stages [14]. The roles played by Pitx2 within the four-chambered postnatal heart are poorly understood, even though its requirement is unquestioned. Conditional mouse mutants, in which Pitx2 expression in atrial myocardium was turned off at birth, developed atrial ultrastructural remodelling and sinus node dysfunction [18]. Pitx2 plays a pivotal role restricting pacemaker activity in the developing myocardium by repressing a nodal gene program and activating the working myocardium gene program [14], [19], [20]. Two groups independently found that Pitx2c expression is downregulated both in LA and in RA of patients with atrial fibrillation (AF), suggesting that Pitx2c dysfunction could be causatively linked to AF pathophysiology [13], [15]. Although using different in-vivo and in-vitro approaches, both groups provided congruent evidence that the reduction of Pitx2c expression in the adult atrial myocardium promotes its susceptibility to AF. These results are well in line with recent findings which suggest that Pitx2c loss-of-function mutations play a role in the genesis of the so-called “lone” AF in patients with structurally normal atria [21].
This study uncovers a novel, previously unrecognized link between heart failure (HF) and activation of the Pitx2 gene in failing myocardium. To address this issue, we examined Pitx2c expression in left ventricular (LV) myocardium in the porcine model of diastolic HF (DHF) with preserved LV-ejection fraction. The expression of Pitx2c, physiologically downregulated in the postnatal heart, is significantly re-activated in failing myocardium which, in turn, is associated with increased expression of a restrictive set of Pitx2 target genes. Among these, Myf5 was identified as the top upregulated gene. In vitro, forced expression of Pitx2c in cardiomyocytes, but not in skeletal myoblasts, activates Myf5 in dose-dependent manner. In addition, we demonstrate that the level of Pitx2c is upregulated in the LV-myocardium at systolic HF with severe reduced LV-ejection fraction in patients. Our present study provides the initial framework for analyzing putative contribution of Pitx2c re-activation in failing myocardium to the overall progression of HF, possibly through the involvement of Pitx2c in regulation of myogenic regulatory factor genes.
Materials and Methods
Heart failure porcine samples
Experimental procedures were carried out in accordance with the European Commission Directive 86/609/EEC on the protection of animals used for experimental and other scientific purposes, and all protocols were approved by the Animal Care and Use Ethical Committee of the University of La Coruña, Spain (approval N°:CE 012/2012). Newborn and early neonatal “Large White” piglets were obtained from a local commercial breeder (La Coruña, Spain), maintained in a conventional Nürtinger nursery system for days 6 after birth, randomized in two groups, and assigned to receive a single intravenous injection of isotonic PBS (phosphate-buffered saline) or 1.5–2.0 mg/kg of doxorubicin (Dox; Sigma, Madrid, Spain) [22], [23]. Untreated control group consisted of six non-injected piglets. On day 24 after injection (i.e., on day 30 after birth), hemodynamic parameters (surface ECG, blood pressure, heart rhythm, cardiac output, and extravascular lung water) were monitored in closed-chest piglets, whereas the measurements of LV end-systolic and end-diastolic pressure were performed in open-chest piglets as described [24], [25]. Dox-injected piglets displayed diastolic dysfunction (see Table S1). The piglets were euthanized and the entire heart was rapidly removed, weighed, and photographed while still beating. Then the isolated heart was placed on an ice-cold petri dish, partially sectioned at the midpoint of the LV length and photographs of the open ventricular chambers were taken. Immediately after this step, the LV free wall (LVFW) and left atrium (LA) were dissected into several samples (100–200 mg each). Samples were frozen in liquid nitrogen immediately after isolation and then stored until use at −80°C. The thickness of the LVFW was measured using digitized photoimages of the ventricular-chamber cross-sections as described [22], [26].
Heart failure patient samples
Human heart samples were obtained from the heart tissue bank set up by MGC-L at the University Hospital Center of La Coruña (La Coruña, Spain). Written informed consent was obtained from patients or their relatives, and the use of human cardiac tissues was approved by the Ethical Committee of Clinical Investigation of Galicia, Spain (approval N°: 2007/239). The investigation conforms to the principles outlined in the Declaration of Helsinki. RNA and protein expression were assayed in explanted hearts from end-stage heart failure (HF) transplantation patients with ischemic (ICM) or idiopathic dilated cardiomyopathy (DCM) and compared with samples obtained from non-failing donor hearts that did not meet criteria for transplantation. All HF patients (functionally classified according to the New York Heart Association criteria) manifested a severely reduced systolic function; the donor group was not known to have any history of overt cardiovascular disease (see Table S2). Transmural samples biopsied from the anterolateral wall of the left ventricle (LV) of failing and non-failing human hearts were stored at −80°C until assayed for RT-PCR and Western blotting.
RNA extraction and reverse transcription
Deep-frozen samples of piglet and human myocardium were directly disrupted in RLT buffer (Qiagen, Madrid, Spain) using a high-speed rotor-stator homogenizer (Ultra-Turrax T8, Germany), digested with Proteinase K (Qiagen), loaded onto RNeasy Mini/Midi columns (Qiagen), subjected to on-column digestion of DNA with RNase-free DNase (Qiagen), and processed in accordance with the manufacturer's recommendations. Total RNA from cultured HL-1 and Sol8 cells was extracted with the TriPure isolation reagent (Roche Diagnostics, Barcelona, Spain), treated with RNase-free DNase (Roche) for 1 h at 37°C and purified using a standard phenol–chloroform protocol. RNA yield and purity was determined spectrophotometrically, and RNA integrity was verified by running samples on 1.2% agarose gels and staining with ethidium bromide. Resulting RNA preparations were resolved in nuclease-free water (Ambion, Madrid, Spain) and kept at −80°C. Two microgram aliquots of individual total RNA were reverse transcribed using SuperScript III or SuperScript RNase H-minus (Invitrogen, Barcelona, Spain) reverse transcriptase and oligo-dT primers according to the manufacturer's instructions. Negative control reactions were performed in the same conditions without reverse transcriptase.
Microarray
Total RNAs isolated from LV biopsies of three failing (i.e., Dox-injected) and three non-failing (i.e., PBS-injected) piglets were transported on dry ice to the “KFB – Center of Excellence for Fluorescent Bioanalytics” (University of Regensburg, Regensburg, Germany) and the quality of RNAs was assessed by using gel electrophoresis and spectrophotometry running eukaryotic total RNA Nano Series II program. The qualified RNA samples were independently hybridized on the Affymetrix GeneChip Porcine Genome Array (Affymetrix, Santa Clara, USA). Sample processing, array hybridization, scanning, and quantification were performed at the Affymetrix Service Provider and Core Facility, “KFB – Center of Excellence for Fluorescent Bioanalytics” (University of Regensburg, Regensburg, Germany; www.kfb-regensburg.de) as described in the Affymetrix GeneChip Expression Analysis Technical Manual. Briefly, the Ambion MessageAmp Premier kit, starting with 150 ng total RNA, was used for labeling. Hybridization was followed standard Affymetrix protocol in Affymetrix GeneChip Hybridization Oven 640. All chips passed standard quality controls to eliminate scans with abnormal characteristics (high versus low affinity binding). The scanning and image acquisition were performed by using the Affymetrix GeneChip Scanner 3000 7G and core software GCOS v.1.4. Expression data generation was performed by using the Affymetrix GeneChip Operating Software (GCOS v. 1–4; MAS5 algorithm with default settings). A set of the algorithms implemented in the Affymetrix Microarray Suite Version 5.0 was used for statistical treatment of the data. Genes were considered as altered if the folds-change was at least 2.0 and adjusted p value ≤0.05. The complete microarray data is available at NCBI through GEO (Gene Expression Omnibus) accession number GSE30110 (http://www.ncbi.nlm.nih.gov/geo).
Conventional PCR
Conventional PCR was performed in a Biometra II system (Gottingen, Germany) as described [24]. The piglet LV/LA cDNAs were used as templates to detect different Pitx2 transcript variants using the primers indicated in Table S3. The plasmid DNAs encoding pig full-length Pitx2a, Pitx2b and Pitx2c were used as a positive reference control. All PCR setups, including no-RT and no template (NT) controls, were performed at least in duplicate. Each PCR sample was resolved on a 2% agarose gel, and PCR products were visualized with ethidium bromide staining and UV illumination. Band intensity was estimated by densitometry (VersaDoc 1000) and Quantity One software (Bio-Rad, Madrid, Spain). The PCR products were cloned and sequenced (Secugen, Madrid, Spain) to confirm their identity.
Real-time quantitative PCR (qRT-PCR)
qRT-PCR was performed on Bio-Rad IQ5 instrument (Bio-Rad, Madrid, Spain) and MxPro Mx3005p PCR thermal cycler (Stratagene, Madrid, Spain) using, respectively, SYBR Green (Bio-Rad, Madrid, Spain) and DyNAmo HS SYBR Green (Thermo Scientific, Madrid, Spain) master mix as described previously [24], [27]. The primer pairs were located in different exons to rule out genomic DNA amplification. Each primer pair used yielded a single peak of dissociation on the melting curve and a single band with the expected size on PAGE gels. Identity of the PCR products was confirmed by sequencing. NT and non-RT RNA template reactions were used as negative controls. All PCR setups were performed at least in duplicate. Relative quantifications were calculated with the comparative ΔCt cycle method with normalization to the expression of housekeeping genes coding for ribosomal protein L19 (Rpl19), glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and β-D-glucuronidase (Gusb). The efficiency of target and reference amplifications was tested to be approximately equal. Primer sequences and additional data are given in Table S3.
Plasmid constructs
Pig Pitx2a (sequence deduced from the swine genomic sequence NW_003610943) and Pitx2c (NM_001206435) were amplified from oligo-dT-primed cDNA from left atrium of newborn piglets; pig Pitx2b (sequence deduced from the swine genomic sequence NW_003610943) was amplified from skeletal muscle cDNA of 20-day-old neonatal animals. Each of the full-length Pitx2 constructs was directionally cloned into p3xFLAG-CMV-14 expression vector (Sigma, Madrid, Spain) at the EcoRI (5′) and BamHI (3′) restriction sites and verified by sequencing [28]. Mouse full-length Pitx2c (NM_001042502.1) was amplified from Sol8 myoblast cDNA by PCR with specific primers containing HindIII and XbaI restriction sequences [27]. Subsequently, the PCR product was inserted into the pcDNA3.1/Zeo(-) plasmid (Invitrogen, Barcelona, Spain), which was modified to generate the V5-tagged Pitx2c as reported [29]. The plasmids were purified by using a PureLink HiPure plasmid filter purification kit (Invitrogen, Barcelona, Spain) according to the manufacturer's protocol. Purified plasmids were formulated in nuclease-free water (Ambion, Madrid, Spain).
Cell culture and transfection in vitro
The following cell lines were used: COS-7 (a fibroblast-like cell line derived from monkey kidney tissue was purchased from the European Collection of Cell Cultures, Salisbury, Wiltshire, UK), HL-1 (a mouse atrial cardiac myocyte cell line was provided by Prof. William C. Claycomb [30]), and Sol8 (a mouse skeletal myogenic cell line was purchased from the American Type Culture Collection, Barcelona, Spain). COS-7 and Sol8 cells were cultured in Dulbecco's modified Eagle's Medium (Gibco, Barcelona, Spain) supplemented with 10% fetal bovine serum and penicillin–streptomycin–glutamine (Gibco); HL-1 cells were cultured in similarly supplemented Claycomb's growth medium [30]. Cells were trypsinized at 70–80% confluence and cell numbers were determined using an automated cell counter (Countess, Invitrogen, Barcelona, Spain). Cells were plated at a density of 105 cells per 35 mm well, allowed to attach overnight and transfected with plasmids expressing: (1) FLAG-tagged pig Pitx2a, Pitx2b and Pitx2c (COS-7 cells/500 and 1000 ng of plasmid DNA), and (2) V5-tagged mouse Pitx2c (HL-1 and Sol8 cells/100–400 ng of plasmid DNA). All transfections were carried out with Lipofectamine LTX and PLUS Reagents (Invitrogen, Barcelona, Spain) following the manufacturer's instructions. For each plasmid, from three to six separate transfection assays were employed, and in each assay, transfections were performed in duplicate. The transfection protocol typically yielded 60–70% transfection efficiency, as revealed by transfection of CMV-EGFP (enhanced green fluorescent protein) vector [27]. Controls consisted of mock- and empty vector–transfected cells. For Pitx2c-silencing experiments, HL-1 and Sol8 cells were seeded in twenty four-well plates (30000 cells per well) and co-transfected with V5-tagged Pitx2c (at 400 ng) and Pitx2c-specific siRNA heteroduplex [13] (at 80 nM) using the Lipofectamine 2000 Transfection Reagent (Invitrogen, Barcelona, Spain), following the manufacturer's protocol. The cells transfected with V5-tagged Pitx2c, under the same conditions, were used as a reference. In each assay, transfections were performed in triplicate. The cells were harvested at 24–48 hours after transfection and processed for RNA and protein extraction as described [24], [27].
Antibodies
The following primary antibodies were used: (1) rabbit polyclonal antibodies to PITX2A,B,C (Capra Science, Ängelholm, Sweden; at 1∶2000 dilution), (2) rabbit polyclonal antibodies to PITX2B (Capra Science, Ängelholm, Sweden; at 1∶1000 dilution).The specificity of the anti-PITX2 antibodies was independently validated in this study by Western blot analysis of COS-7 cells expressing each PITX2 isoform (see Fig 1 A, B), (3) rabbit monoclonal antibodies to MYF5 (Abcam, Cambridge, UK; at 1∶10000 dilution), (4) rabbit polyclonal antibodies to cardiac troponin I (Abcam, Cambridge, UK; at 1∶40000 dilution), (5) rabbit polyclonal antibodies to cardiac calsequestrin-2 (Abcam, Cambridge, UK; at 1∶10000 dilution), (6) rabbit polyclonal antibodies to ANKRD1 (at 1∶1000 dilution) were generated by Davids Biotechnologie (Regenburg, Germany) using the N-terminal region of pig ANKRD1 as immunogen [22], (7) mouse monoclonal anti-FLAG M2 antibody (Sigma, Madrid, Spain; at 1∶5000 dilution), (8) mouse monoclonal anti-V5 antibody (Sigma, Madrid, Spain, at 1∶5000 dilution), and (9) mouse monoclonal anti-GAPDH antibody (Sigma, Madrid, Spain, at 1∶10000 dilution). Secondary peroxidase conjugated anti-rabbit and anti-mouse IgG (Fab-specific) antibodies were purchased from Sigma (Madrid, Spain).
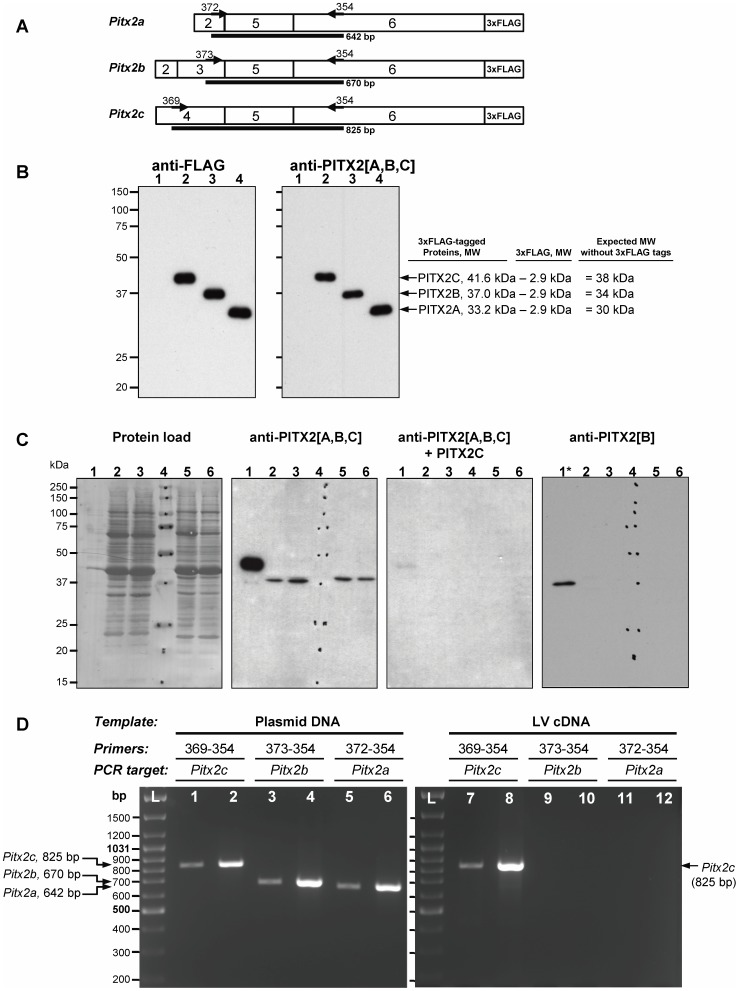
Figure 1. PITXC, but not PITX2A and PITX2B, is detected in the normal porcine and human left ventricular myocardium.
A - Schematic representation of FLAG-tagged Pitx2a, Pitx2b and Pitx2c constructs. 2–6 – exons. The approximate location of the primers for downstream PCR analyses (D) is indicated. The expected size of the corresponding amplicons is shown in black lines. B - Lysates of COS-7 cells transfected with FLAG-tagged Pitx2 constructs were electrophoresed and immunoblotted with anti-FLAG or anti-PITX2A,B,C antibodies. Cells transfected with: vector only (Lane 1), Pitx2c (Lane 2), Pitx2b (Lane 3) and Pitx2a (Lane 4) constructs. C - Lysates of COS-7 cells transfected with FLAG-tagged Pitx2c (Lane 1) or Pitx2b (Lane 1*) and left ventricle (LV) samples from normal 30-day-old porcine (Lane 2 and 3) and non-failing human (Lane 5, 6) hearts were blotted and probed with anti-PITX2A,B,C or anti-PITX2B antibodies. Lane 4 – Precision Plus WesternC Standards, kDa. Protein load – membrane stained with Amido Black 10B. Anti-PITX2A,B,C+PITX2C – antibodies neutralized by the extract of COS-7 cells transfected with FLAG-tagged Pitx2c construct. D - PCR amplifications of Pitx2 transcript variants using the Pitx2-encoding plasmid DNAs (left panel, lane 1–6) or porcine LV cDNA (right panel, lane 7–12) as templates. Reactions were run in duplicate using 2-fold dilutions of both templates with each primer pairs. L – DNA size standards (GeneRuler DNA ladder mix; Fermentas).
SDS-PAGE and Western blotting
Tissue/cell samples were homogenized in standard 2x Laemmli buffer (Invitrogen, Barcelona, Spain) supplemented with complete protease inhibitor cocktail (Roche, Madrid, Spain) as previously described [24], [25]. Following centrifugation at 20000 g for 30 minutes, the concentration of supernatant proteins was analyzed using the Bio-Rad DC Protein Assay Kit (Bio-Rad, Hercules, USA) according to the manufacturer's protocol. The protein extracts were normalized to total protein concentration; the results of normalization were confirmed by SDS-PAGE and Coomassie staining before Western blotting analysis [25], [31]. Protein supernatants (loading range of 5–15 µg/run) were resolved on a 12% SDS-PAGE (Mini-Protean-III, Bio-Rad, Hercules, USA) and blotted onto PVDF-membranes (Hybond-P, Amersham Biosciences, Barcelona, Spain). Molecular weight (MW) standards (Precision Plus Protein WesternC Standards from Bio-Rad and SeeBlue Plus2 Pre-Stained Standard from Invitrogen) were included on each gel. Blots were probed with the antibodies indicated above and visualized by the Super-Signal West Pico chemiluminescent substrate (Pierce Biotechnology, Madrid, Spain). Equivalence of protein loading was confirmed by Amido-Black 10B (Merck, Barcelona, Spain) staining of blots after immunodetection. The blots were re-probed with anti-cardiac calsequestrin 2 and anti-GAPDH antibodies as additional control for loading. Quantification of Western blot signals was obtained by using a Bio-Rad GS800 calibrated densitometer with Quantity One software.
Statistical Analysis
Values presented are expressed as mean ± S.E.M. All comparisons between groups were performed using an unpaired Student's t test. Differences were considered statistically significant for p value ≤0.05.
Results
PITX2C is the predominant splice-variant isoform in porcine and human left ventricular myocardium
PITX2 protein expression was studied by Western blot in left ventricular (LV) samples of normal porcine and non-failing human hearts. The FLAG-tagged constructs containing each of the three different PITX2 isoforms (Fig. 1 A) were used to evaluate the specificity of antibodies against PITX2A,B,C. Lysates from COS-7 cells transiently transfected with constructs were probed by Western blotting with anti-FLAG and anti-PITX2A,B,C antibodies (Fig. 1 B). Anti-FLAG detection revealed the expression of three PITX2 isoform-specific products, each migrating in SDS-PAGE as one band. The same protein products were also detected by anti-PITX2A,B,C antibodies indicating that these antibodies specifically recognize all PITX2 isoforms equally well (Fig. 1 B, right panel). However, only one band, with MW around 38–39 kDa corresponding to the expected size of PITX2C, could be seen in both porcine and human LV samples when probed with anti-PITX2A,B,C antibodies (Fig. 1 C). After neutralization of these antibodies with FLAG-tagged PITX2C protein, neither band was detected on the LV-derived blots. In addition, the antibodies against PITX2B isoform did not reveal any band in LV-extracts assayed under the same experimental conditions.
The results of Western blot assays were consistent with the expression levels of Pitx2 variants determined by PCR analysis (Fig. 1 D). In control experiments, with the use of Pitx2-encoding plasmid DNAs as templates, each of three Pitx2 transcript variants was amplified with comparable efficiency (Fig 1 D, left panel). However, only the largest Pitx2c variant was detected in LV samples using the same primer sets (Fig. 1 D, right panel). Taken together, the results strongly evidenced that PITX2C is the most abundant and predominant isoform in the LV of pigs and humans.
Ventricular Pitx2c expression is downregulated after birth but markedly reactivated in piglet diastolic heart failure
In neonatal piglets, the LV undergoes rapid hypertrophic growth as development proceeds [22], [32]. Likewise, in this work, we found that the average LVFW thickness was approximately 2-fold higher in normal 30-day-old piglets than in newborns (Fig. 2 A, B). Comparative qRT-PCR analysis of the LV-samples from these two groups of normal animals revealed that Pitx2c mRNA levels at birth are 16-fold higher than are those detected in 30-day-old animals (Fig. 2 C). These results suggest that a rapid progression of LV-concentric remodelling is associated with downregulation of Pitx2c expression in healthy neonatal piglets.
Figure 2. In the pig left ventricular myocardium, Pitx2c expression is downregulated after birth but re-activated in DHF.
A, B - Cross-sections through left ventricular (LV) chambers of newborn (NB) and 30-day-old (30 d) piglets (Bar – 1 cm) and LVFW thickness values, respectively. C - Overall relative levels of Pitx2c transcript in LV-samples of newborn versus 30-day-old animals, *p≤0.05. D - Overall relative levels of Pitx2c transcript in LV-samples of PBS-injected versus Dox-injected 30-day-old piglets, *p≤0.05. E - Representative Western blots of PITX2C levels in the LV myocardium of PBS-injected (Lane 1, 2, 3) and Dox-injected (Lane 4, 5, 6) 30-day-old piglets. Each LV-sample (1–6) was derived from an individual animal. Western blot replicates were probed with antibodies against: (F) - cardiac calsequestrin 2 (CASQ2) and (G) - cardiac troponin I (TNNI3). MW values (kDa) of the bands detected are shown. H – Membrane stained with Amido Black 10B. I - Overall relative levels of PITX2C protein in LV-samples as based on average values from each group studied. *p≤0.05.
By utilizing our protocol [24], [25], diastolic heart failure (DHF) syndrome was induced in 6-day-old piglets by a single Dox injection. At day 30, neonatal piglets developed a diastolic dysfunction resulting in pulmonary congestion, but with nearly-normal (“preserved”) systolic function in terms of LV-end systolic pressure and global cardiac output values. The average fold-increase of both the normally spliced Nppb (natriuretic peptide precursor B) and ΔE2-Nppb (exon 2-skipped) mRNA [33] in the failing versus control LV myocardium was 5.8 and 9.4, respectively (see Table S1).
We quantified the relative amounts of the Pitx2c splice variant in the LV myocardium of control (i.e., PBS-injected) and experimental (i.e., Dox-injected) piglets (Fig. 2 D) by qRT-PCR. Expression of Pitx2c transcript was enhanced 6.7-fold in the failing versus control myocardium. The specificity of the unique Pitx2c amplification product was determined by melting curve and PAGE analyses (Figure S1 A–D). A similar 6.5 fold-increase was observed for the PITX2C protein in failing as compared to non-failing piglet LV myocardium (Fig. 2 E–I).
Thus, these results demonstrated that expression of Pitx2c, which is physiologically downregulated in the early postnatal heart, becomes significantly re-activated in an experimental DHF setting.
Myf5 is identified as the most upregulated gene among Pitx2 targets in piglet failing myocardium
Marked reactivation of PITX2C protein expression suggested that downstream targets could also be activated in DHF myocardium. A variety of putative targets of Pitx2 have been suggested over the years, many in associative gene expression studies of non-cardiac tissues by using microarray-based methods [34]–[38]. In adult ventricular myocardium, the exact Pitx2 signalling pathway is not defined, nor the Pitx2 targets involved. Our microarray expression profiling of LV-samples from non-failed and failed porcine hearts revealed that Pitx2 upregulation is associated with increased expression of a restrictive set of known Pitx2 target genes (Table 1). It is noteworthy that previous studies have demonstrated that these genes have PITX2 binding site(s) in their promoter region, as demonstrated by chromatin immunoprecipitation and functional promoter-reporter assays (see references in Table 1). Changes in the expression of three transcription factors, Myf5, Pax3, and FoxJ1, as well as Pitx2 itself were further verified by real time qRT-PCR in the same (three failing versus three non-failing) samples used in microarray assays (Table 1). Among Pitx2 targets, most notable was the marked upregulation of the myogenic factor MYF5 in DHF myocardium. mRNA levels for the Pax3 transcription factor, which can activate Myf5 in embryonic muscle progenitor cells [39], were also elevated in failing LV-myocardium, although to a lesser extent as compared to Pitx2c and Myf5 (see Table 1). Pax3, Pitx2, and Myf5 are upstream genes which differentially regulate the kernel myogenic program in cell- and time-dependent manner [40], [41]. However, the expression of downstream myogenic factors - MYOG (myogenin; see Fig. 3 A, right panel), MYOD, and MYF6 (also known as MRF4) - was not significantly altered in failing compared to non-failing LV myocardium. In addition, there were subtle differences in the expression levels of skeletal muscle-specific genes (Tnnt1, Tnnt3, Myh4, and Tpm2), with exception of the Myl1 gene which expression was markedly downregulated in failing myocardium (see Table S4 and complete microarray data set at GEO accession number GSE30110). LV expression of the FoxJ1 (forkhead box J1) transcription factor, which is involved in cardiac morphogenesis and remodelling [42], was moderately upregulated in Dox-injected versus that in control PBS-injected animals (see Table 1).
Table 1. Upregulation of genes sensitive to Pitx2 dosage in the piglet DHF model.
| Gene coding for | Gene symbol | Fold change | PITX2 binding to the promoter | Reference | |
| microarray | qRT-PCR | ||||
| Paired-like homeodomain 2 | Pitx2 | 2.14 | 6.60±1.6* | ||
| Natriuretic peptide precursor A | Nppa | 4.42 | HV | + | [73] |
| Cyclin D2 | Ccnd2 | 2.38 | HV | + | [51] |
| Forkhead box protein J1 | Foxj1 | 2.66 | 3.20±0.4 | + | [74] |
| Myogenic factor 5 | Myf5 | 5.15 | 4.80±0.5 | + | [57] |
| Paired box 3 | Pax3 | 3.39 | 1.80±0.3 | + | [45] |
Three pairs of failing-non failing samples were assayed by both microarray and qRT-PCR. *Quantitative RT-PCR analysis with the use of the primer pair (350–353, see Table S3) annealing to the sequence common to the different Pitx2 variants. HV – highly variable expression within each sample sets.
Figure 3. Expression of Myf5, but not Myog, is upregulated in the failing pig left heart as determined by qRT-PCR and Western blot analyses.
A – Relative levels for Myf5 (left) and Myog (right) transcripts in the left ventricular (LV) samples from PBS- and Dox-injected piglets. B - Representative Western blots of MYF5 levels in the LV samples from PBS- (Lane 1, 2, 3) and Dox-injected (Lane 4, 5, 6) piglets (blots probed with anti-PITX2A,B,C antibodies). Western blot replicates were probed with antibodies against cardiac calsequestrin 2 (CASQ2) and cardiac troponin I (TNNI3). MW values (kDa) of the bands detected are shown. C – Control of protein loading: Western blot membrane probed with anti-MYF5-antibodies was then stained with Amido Black 10B to detect blotted proteins. D – Overall relative levels of MYF5 protein in LV-samples as based on average values from each group studied. *p≤0.05. E – Representative Western blots of PITX2C and MYF5 levels in the left atrium (LA) from PBS- (Lane 1, 2, 3) and Dox-injected (Lane 4, 5,6) piglets. MW values (kDa) of the bands detected are shown. F – Membrane stained with Amido Black 10B. G – Overall relative levels of PITX2C and MYF5 in LA-samples as based on average values from each group studied. *p≤0.05. H – RT-PCR amplifications (Lane 1–6) of Pitx2 transcript variants using the porcine LA cDNA as template. Reactions were run in duplicate using 2-fold template dilutions with each primer pairs. L – DNA size standards (GeneRuler DNA ladder mix; Fermentas).
Recently, a search for cardiac Pitx2 targets by ChIP-Seq and microarray assays revealed several novel putative targets negatively regulated by Pitx2 in transfection assays using the 293FT cell line derived from human embryonic kidney [18]. The expression of these genes (coding of calcium voltage-gated channel subunit alpha-1d; potassium voltage-gated channel, KQT-like subfamily, member 1; caveolin 1; emerin) was not altered in LV myocardium of Dox-injected versus PBS-injected piglets (see complete microarray data set at GEO accession number GSE30110).
We further validated the upregulation of Myf5 in DHF LV-myocardium by performing quantitative RT-PCR and Western blot analyses in a larger cohort of Dox- versus PBS-injected animals. On average, the level of Myf5 transcript was enhanced 4.8-fold in the failing (n = 12) versus control (n = 8) LV-myocardium (Fig. 3 A, left panel). The specificity of the unique Myf5 amplification product was determined by melting curve and PAGE analyses (see Figure S2). A seemingly similar 3.5 fold-increase was observed for the MYF5 protein in failing as compared to non-failing piglet LV myocardium (Fig. 3 B–D).
Recent clinical evaluations indicated that LV diastolic dysfunction can result in functional and structural remodelling of the left atrium (LA) in patients [43], [44]. In line with these data, we studied the expression of Pitx2c and Myf5 in LA samples from Dox- versus PBS-injected piglets. As revealed by Western blot, the levels of both PITX2C and MYF5 were about 10-fold higher in the LA of Dox-treated as compared to those in the LA of PBS-treated piglets (Fig. 3 E–G). Only the 38-kDa band, corresponding to the porcine PITX2C isoform, was detected on LA-derived blots probed with anti-PITX2A,B,C antibodies (Fig. 3 E), although these antibodies, as we demonstrated, recognize the all PITX2 isoforms (see Fig. 1 B, right panel). The RT-PCR analysis for LA-samples (Fig. 3 H) revealed the predominant expression of the Pitx2c variant, while other Pitx2 alternative transcripts were either not detected (Pitx2b) or detected in traces (Pitx2a).
Collectively, the findings from the porcine model of DHF revealed a strong upregulation of Pitx2c and Myf5 under stress conditions, suggesting that Pitx2c could be involved in transcriptional regulation of Myf5 expression, and probably to a lesser extent of FoxJ1 too, in postnatal cardiomyocytes.
Forced expression of Pitx2c increases the expression of Myf5 in cultured cardiomyocytes
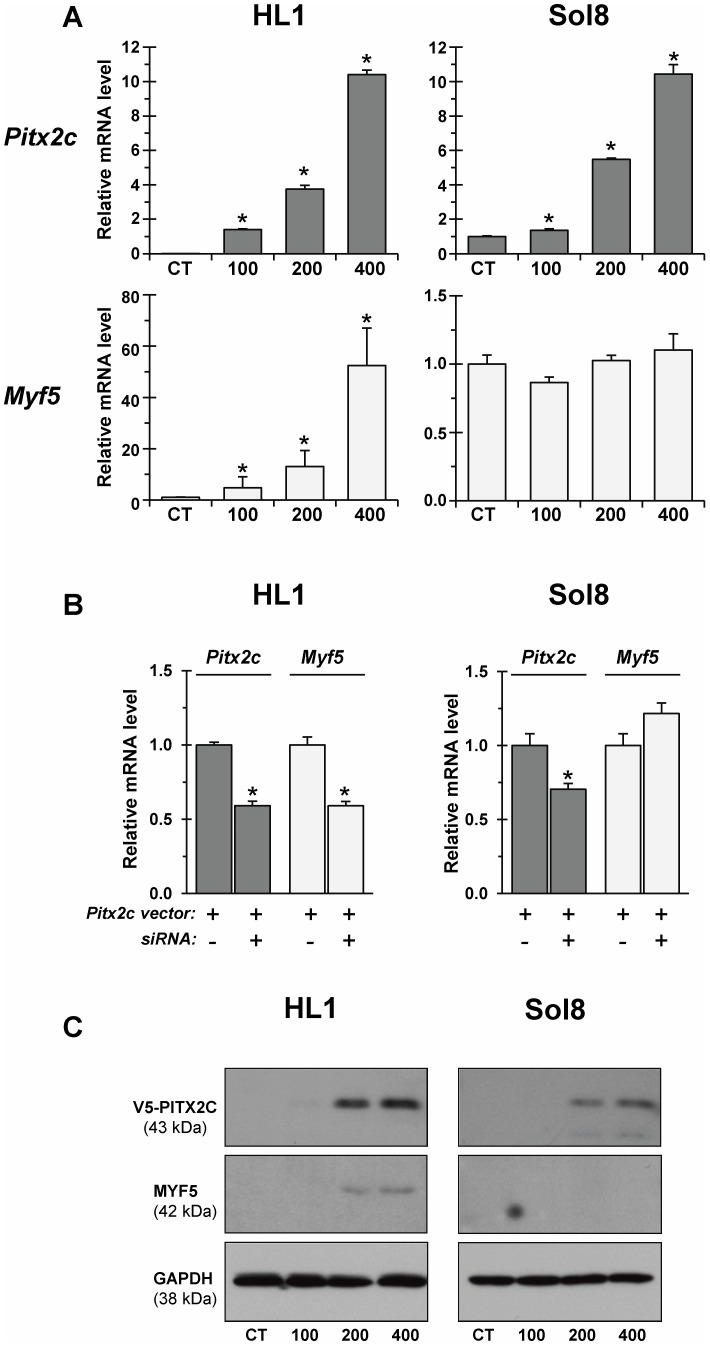
To explore the possibility that Pitx2c may activate Myf5 and/or FoxJ1 expression specifically in cardiomyocytes, we overexpressed the full-length Pitx2c variant in HL-1 cardiomyocyte as well as undifferentiated Sol8 skeletal muscle cell lines. The Sol8 cell line was used as a control since, as previously demonstrated, Pitx2c overexpression promotes proliferation and arrests muscle differentiation of Sol8 cells [45].
Transient transfections of HL-1 cardiomyocytes strongly enhanced, in a dose-dependent manner, the expression of the Myf5 gene compared to that in mock- and empty vector-transfected cells. This effect appears to be cell-type dependent, as no stimulation of Myf5 transcript expression was observed in Pitx2c-transfected Sol8 myoblasts (Fig. 4 A). In this direction, the inhibition of Pitx2c overexpression, using Pitx2-siRNAs, led to a decrease of Pitx2c-induced activation of the Myf5 gene in HL-1 cardiomyoblasts, whereas co-transfection with Pitx2c siRNA did not affect the basal level of Myf5 expression in Pitx2c-transfected Sol8 myoblasts (Fig. 4 B). The ability of Pitx2c to activate Myf5 expression in HL-1 cardiomyoblasts was further confirmed by Western blot analysis. Ectopic PITX2C upregulation resulted in activation of MYF5 protein expression in HL-1, but not in Sol8 cells (Fig. 4 C). Under our experimental conditions, we could not detect any MYF5 protein in either control or Pitx2c-transfected Sol8 myoblasts.
Figure 4. In vitro forced expression of Pitx2c results in increased Myf5 expression in cultured cardiomyocytes but not skeletal myoblasts.
A - Overall relative levels of Pitx2c (black) and Myf5 (grey) transcripts in HL-1 and Sol8 cells transfected with V5-tagged Pitx2c vector at different doses (100-400 ng). CT – empty-vector transfected cells. Shown are results of qRT-PCR analysis. Data from six replicates of each transfection were pooled and averaged. In controls, the relative levels of transcripts for Myf5 were assigned to a value of 1, whereas those for Pitx2c were assigned to a value of 10−4. *p≤0.05. B – Overall relative levels of Pitx2c and Myf5 transcripts in HL-1 and Sol8 cells co-transfected with V5-tagged Pitx2c (at 400 ng) and Pitx2c-specific siRNA heteroduplex (at 80 nM). Data from three replicates were pooled and averaged. C - HL-1 and Sol8 cells transfected with V5-tagged Pitx2c (at 100–400 ng) were pooled (from triplicate wells in each transfections), lysed, electrophoresed, and immunoblotted with antibodies against PITX2A,B,C, MYF5 and GAPDH. MW values (kDa) of the bands detected are shown.
With respect to expression of FoxJ1, the results of Pitx2c transfections of HL-1 cells were not very conclusive but there appeared to be a statistically significant and slightly dose-dependent increase of FoxJ1 expression as compared to control cells (see Figure S3). By contrast, in transfected Sol8 myoblasts FoxJ1 was downregulated at doses of 100 and 200 ng of Pitx2c plasmid DNA followed by a return to basal levels at a highest dose used (400 ng). The cause of this down-regulation of FoxJ1 expression is not known at present.
We next asked whether forced expression of Pitx2c is able to modulate the expression of PAX3 and MYOG myogenic factors in HL-1 versus Sol8 cells. We found that forced Pitx2c expression does not affect the levels of Pax3 expression in either HL-1 or Sol8 transfected cells, but results in decreased expression of the Myog gene selectively in Sol8 undifferentiated myoblasts (see Figure S3). The latter is in accordance with previous experimental data reporting Myog downregulation in Sol8 myoblasts transfected with Pitx2c expression vectors [27], [45].
Essentially, data from transfection assays showed that Pitx2c upregulation does result in strong and dose-dependent activation of Myf5 expression in cardiomyocytes and to a minor extend of FoxJ1.
Ventricular Pitx2c expression is reactivated in human systolic heart failure
Controversy about whether diastolic (DHF) and systolic HF (SHF) represent overlapping [46] or distinct [47] phenotypes of severely impaired cardiac function remains unsolved. We aimed to investigate whether re-activation of Pitx2c expression in DHF, as revealed in the porcine model, is also related to SHF in patients undergoing heart transplantation due to dilated (DCM) or ischemic cardiomyopathy (ICM). On average, the level of expression of the Pitx2c transcript was nearly 6-fold (DCM) and 8-fold (ICM) higher in LV tissue samples from explanted hearts as compared with those from donor human hearts (Fig. 5 A). However, Western blotting analysis showed that PITX2C protein upregulation in falling human myocardium was significantly less (2–3-fold increase) than that observed at the RNA level (see Fig. 5 B, C). In the studied failing myocardium, the expression of ANKRD1 (ankyrin repeat domain 1 protein), the surrogate marker of HF [48], was significantly upregulated (Fig. 5 B, E), while the levels of TNNI3 (cardiac troponin I; the marker of acute myocardial ischemia) were unchanged or slightly augmented as compared to non-failing samples (see Fig. 5 B).
Figure 5. Pitx2c expression is reactivated, while expression of Myf5 is downregulated in human systolic heart failure as determined by qRT-PCR and Western blot analyses.
A - Overall relative levels of Pitx2c, Myf5, and Myog transcripts in the left ventricular (LV) samples from non-falling (NF) and failing human hearts due to idiopathic dilated (DCM) and ischemic (ICM) cardiomyopathy. *p≤0.05. B - Representative Western blots of PITX2C levels (top) in the LV samples from NF (Lane 1–3), DCM (lane 4–6) and ICM (lane 7–9) failing hearts. Western blot replicates were probed with antibodies against MYF5, cardiac calsequestrin 2 (CASQ2), cardiac troponin I (TNNI3) and ankyrin repeat domain 1 protein (ANKRD1). MW values (kDa) of the bands detected are shown. Protein load - Membrane stained with Amido Black 10B. Overall relative levels of PITX2C (C), MYF5 (D) and ANKRD1 (E) protein in LV-samples as based on average values from each group studied. *p≤0.05.
In addition, we studied the expression of Myf5 and Myog genes in human failing hearts. Expression of Myf5 was found to be nearly 2-fold decreased at both transcript (Fig. 5 A) and protein (Fig. 5 B, D) levels in falling as compared to non-failing human myocardium, whereas the expression of Myog was unchanged (Fig. 5 A). Although not statistically significant, there was a trend toward less decrease of MYF5 protein in ICM than in DCM samples (Fig. 5 D).
Collectively, the results suggest that Pitx2c re-activation in ventricular myocardium is a common molecular signature of HF with preserved (DHF) as well as reduced systolic (SHF) function, while the expression of Myf5 is distinct between the two HF phenotypes studied.
Discussion
In this paper we provide what are, to our knowledge, the first reported results, demonstrating: (1) the re-activation of Pitx2c expression in failing myocardium in different HF settings, (2) the marked upregulation of Myf5 myogenic factor expression in failing ventricular and atrial myocardium as well as in cultured cardiomyocytes in response to forced expression of Pitx2c, and (3) the expression of the large repertoire of myogenic regulatory factors (MRFs) in normal and failing myocardium. Each of these data sets is discussed in the context of the underlying molecular mechanisms of HF.
Activation of myocardial PITX2c expression as a hallmark of heart failure
A possible involvement of PITX2C in ventricular HF has not yet been explored. We demonstrate herein a similar upregulation of Pitx2c expression in the LV myocardium, at transcript and protein levels, in a porcine model of DHF as well as in patients with end-stage SHF. In patients, LV diastolic dysfunction can lead to elevation of LA pressure, resulting in increased LA wall tension and progressive LA dilatation, thereby increasing the risk and frequency of atrial fibrillation development [44], [49]. By using the porcine DHF model, we evidence that Pitx2c is upregulated in the LA myocardium. This new observation adds an additional layer of complexity to the challenge linking perturbations of Pitx2c expression in the LA to the development of atrial fibrillation [11]. Mechanistically, the results of this study strongly suggest that Pitx2c activation is a common feature of the failing heart, in line with recent reports of myocardial Pitx2c upregulation in HF due to ventricular septal defects in human foetuses [50]. The question is whether Pitx2c upregulation deteriorates the failing myocardium as one of the factors for HF progression or its activation is merely a secondary manifestation of impaired LV function that might even have a compensatory effect in HF? Certainly, any answer to such a question is still speculative as current knowledge on Pitx2c functions in postnatal ventricular myocardium is incomplete and largely uncertain. In addition, the role of Pitx2c in failing myocardium may be multifaceted depending on the cause, type, stage, and severity of HF. Our results in fact suggest that an excessive re-activation of Pitx2c expression could contribute to an overall disturbance of the cardiac gene regulatory network in failing myocardium.
Pitx2 expression is upregulated by a canonical Wnt/β-catenin signalling pathway involved in activation of cell proliferation of embryonic cardiac precursors [51]. However, the canonical Wnt signalling negatively regulates the proliferation of adult cardiac progenitor cells, contributing to negative LV-remodelling [52]. There is substantial evidence that inhibition of the canonical Wnt/catenin pathway can be beneficial for HF [53]. Recently, Pitx2 is recognized for being a downstream transcription factor in the TGF-β (transforming growth factor beta) signalling pathway in non-muscle cells [36]. Myocardial TGF-β signalling is activated in animal models of myocardial infarction and cardiac hypertrophy, and patients with dilated and hypertrophic cardiomyopathy. Reducing of TGF-β signalling is seen as a promising therapeutic approach for cardiovascular diseases (reviewed in [54], [55]). Notably, our microarray analysis revealed that the components of the TGF-β signalling pathway, such as TGFβR3 (TGFβ receptor 3; 6.2-fold increase) and SMAD3 (2.0-fold increase), are upregulated in the failing porcine LV-myocardium (see Table S4). Hence, it is likely that upregulation of the canonical TGF-β signalling pathway, that is believed to represent a maladaptive response of diseased heart to stress, may contribute to Pitx2c re-expression in porcine and human myocardium in advanced and late stages of HF. The endogenous targets of Pitx2 in the adult heart remain largely enigmatic. Many genes involved in cell-junction assembly, ion transport and cell proliferation/migration were found to be upregulated in mouse mutants with conditionally inactivated Pitx2 in the postnatal atrial myocardium, but no supporting molecular evidence of Pitx2-dependent downregulation of these genes in cardiomyocytes has been reported [18].
Although the data mentioned above do not directly involve Pitx2c signalling in pathogenesis of HF, our preliminary assumption is that Pitx2c re-activation in the failing heart might be maladaptive. The finding that forced expression of Pitx2c dramatically upregulates the myogenic transcription factor MYF5 in cultured cardiomyocytes, lends some credence to this suggestion.
Myf5 upregulation as a consequence of Pitx2c activation
Given that gene regulatory networks for cardiac and skeletal muscle are different in many ways, the enhanced expression of two key regulators of skeletal myogenesis - Myf5 and Pax3 - in failing porcine myocardium has attracted our special attention. To establish whether the upregulation of these genes is mediated via Pitx2c, HL-1 cardiomyocytes (with barely detectable Myf5 transcript levels) and Sol8 skeletal myocytes (with low detectable Myf5 transcript levels) were transiently transfected with Pitx2c expression vector. Forced expression of Pitx2c dramatically stimulated the expression of Myf5 (at both transcript and protein levels) exclusively in HL-1 cardiomyocytes, suggesting such regulation is cell-content dependent. The observation that the expression of Myf5 is decreased following Pitx2c silencing in only HL-1 cardiomyoblasts, overexpressing Pitx2c, does also argue in this direction. In Pitx2c-transfected HL-1 cardiomyocytes, augmented expression of both Pitx2c and Myf5 did not affect Myog levels, contrary to Myog downregulation in Sol8 skeletal myoblasts overexpressing Pitx2c. The Pax3 expression was not significantly altered in HL-1 cardiomyocytes that transiently overexpressed Pitx2c. The latter suggests that Pax3 does not contribute to activation of Myf5 expression in HL-1 cardiomyocytes, similarly to that occurring in the progenitor cells of postnatal skeletal muscle [56].
The Pitx2-binding sites in the Myf5 promoter region are conserved among species, and two potential Pitx2-binding sites exist in the mouse Myf5 promoter. It has been demonstrated that PITX2 physically associates with these binding sites and activates the Myf5 promoter in limb- and extra-ocular muscle-derived precursor cell lines [57]. In addition, Pitx2 potently stimulates the myogenic differentiation program in adult skeletal muscle satellite cells [58]. The results of our transfection assays raise the intriguing possibility that Pitx2-Myf5 expression crosstalk can also be operative in cardiomyocytes. In this regard, expression of both Pitx2 and Myf5 was detected in cardiomyocyte-like (CML) cells generated from pluripotent stem cells of patients with DCM due to mutations in the cardiac troponin T gene [59]. In-vitro forced expression of sarcoplasmic reticulum Ca2+ ATPase (Serca2a) improved contractility function of CML cells that, in turn, was associated with downregulation of Pitx2 and Myf5 mRNA levels in Serca2a-transduced cells (see the data set available at NCBI through accession number GSE35108). On the other hand, and complementary to these results, ectopic overexpression of Myf5 in the heart activates a skeletal muscle gene expression that results in progressive cardiomyopathy [60], [61]. The latter suggests that aberrant Pitx2-Myf5 co-activation seen in our model of DHF may negatively impact on diastolic cardiac function.
Formally viewed, our findings suggest the involvement of Pitx2c in regulation of Myf5 expression in the stressed myocardium. However, the expression of Myf5 is modulated by a large number of endogenous signalling factors, each of which can have either positive or negative effects on Myf5 expression, depending on cell type and physiological/pathological context [40], [62]. An inverse correlation in expression of Pitx2c (i.e., upregulation) and Myf5 (i.e., downregulation) was detected in human samples of DCM and ICM (see Fig. 5). The discordance in Myf5 expression in porcine versus human failing myocardium could be attributable to multiple factors, including differences in HF dysfunction phenotypes (diastolic versus systolic HF), stage and severity of HF (mid-advanced versus end-stage HF), age at which HF occurs (neonatal versus adult/aged heart), and medication (non-drug for DHF versus multidrug therapy for SHF). In addition, we observed a certain variability in Pitx2c and Myf5 expression between DCM and ICM samples (see Fig. 5). At the molecular level, the difference in cardiac expression of inhibitors of skeletal myogenesis might also play a role. Myostatin, a negative regulator of myogenesis and Myf5/MyoD expression [63], is downregulated in our model of porcine DHF [64], while significantly upregulated in human HF due to DCM and ICM [65], supporting thus this hypothesis.
Expression of the MRF transcriptional cassette in the heart
In the present study we demonstrated the expression of Myf5 in porcine and human LV-myocardium at both transcript and protein levels via qRT-PCR and Western blotting, respectively (see Fig. 3 and 5). Surprisingly, our gene expression studies revealed that, in addition to the Myf5 gene, a number of genes involved in regulation of skeletal myogenesis are also expressed in postnatal pig (MyoD, Myog, Myf6/Mrf4, Pax3) and adult human (Myog) myocardium. These data are consistent with detection of MyoD expression in postnatal mouse heart [66] as well as MyoD, Myog [67] and Myf5 [68] transcripts in postnatal/adult rat ventricular myocardium.
Forced expression of MRFs, especially MyoD and Myf5, can covert different non-muscle cell types into skeletal myoblasts in vitro (reviewed in [69]. However, the ectopic misexpression of either Myf5 or MyoD in the heart in-vivo, triggering skeletal muscle-specific gene expression, does not lead to terminal skeletal muscle differentiation [70], [71]. It is tempting to speculate that cardio-enriched miRs might suppress the expression level of MRFs, thus preventing skeletal myoblast differentiation in postnatal myocardium. In this sense, it was found that two miRs of miR669 family (miR669a and miR669q) inhibit skeletal myogenesis in postnatal cardiac progenitors by suppressing MyoD expression. Notably, miR669a expression is reduced in cardiac progenitors isolated from patients with cardiomyopathies due to myocardial infarction or pathological LV-hypertrophy [72]. In this context, it appears to be important to determinate miR networks regulated by Pitx2c in adult myocardium and their putative actions on MRF expression in the failing heart.
In the classical muscle molecular context, expression of the MRFs is sensitive to Pitx2 gene dose. It could be foreseeable, for this reason, that Pitx2c upregulation in the failing myocardium in response to different pathological stimuli could contribute to MRF expression and thus impaired cardiac function. Our results provide previously unrecognized evidence that Pitx2c is reactivated in postnatal/adult heart at HF that in turn is associated with modulation of Myf5 expression in failing myocardium. These findings emphasize the particular importance for future studies to define the Pitx2 signalling pathways in principal cell types (cardiomyocytes, fibroblasts, smooth muscle and vascular endothelium cells) of postnatal myocardium.
Supporting Information
Representative qRT-PCR amplification plots for Pitx2c transcripts in the piglet left ventricle. A - Newborn (NB, green) versus 30-day-old (30 d, blue) normal piglets. B - PBS-injected (PBS, blue) versus Dox-injected (Dox, red) 30-day-old animals. Rpl19 - internal standard levels. Arrows - threshold cycle (CT). FT - fluorescent threshold. RFU - relative fluorescent units. Under experimental conditions used, each primer pair yielded a single peak of dissociation on the melting curve (C) and a single band with the expected size on PAGE gel post-stained with SYBR Green I (D).
(TIF)
Representative qRT-PCR amplification plots for Myf5 and Myog transcripts in the left ventricle of PBS- versus Dox-injected piglets. A - PBS-injected (PBS, blue) versus Dox-injected (Dox, red) 30-day-old animals. Rpl19 - internal standard levels. Arrows - threshold cycle (CT). FT - fluorescent threshold. RFU - relative fluorescent units. Under experimental conditions used, each primer pair yielded a single band with the expected size on PAGE gel post-stained with SYBR Green I (B).
(TIF)
Expression of Myog, Foxj1 and Pax3 genes in Pitx2c-transfected HL-1 and Sol8 cells. Overall relative levels of Myog, Foxj1 and Pax3 transcripts in HL-1 and Sol8 cells transfected with V5-tagged Pitx2c vector at different doses (100-400 ng). CT – empty-vector transfected cells. Shown are results of qRT-PCR analysis. Data from six replicates of each transfection were pooled and averaged. *p≤0.05.
(TIF)
Baseline characteristics of neonatal piglets injected with Dox or PBS three weeks after injections.
(DOCX)
Patient characteristics for samples employed qRT-PCR and Western blot analyses.
(DOCX)
Primers used in this study.
(DOCX)
Selective data set derived from the microarray database.
(DOCX)
Acknowledgments
This work was carried out within the framework agreement between the Institute of Health Sciences/University of La Coruña and Department of Experimental Biology/University of Jaen, Spain. The HL-1 cell line was kindly provided by Dr. W. C. Claycomb (Louisiana State University Medical Center, USA). Assistance of Alberto Centeno and Eduardo López (Experimental Surgical Unit/University Hospital Center of La Coruña) in establishment of DHF model was greatly appreciated. We also thank Dr. N. Domenech García for allowing us to use the Cryobank facility at the University Hospital Center of La Coruña.
Funding Statement
ATM, MT and AC-B were supported by funds from the Institute of Health Sciences and by a grant (GRC 2013/061) from the Autonomic Government of Galicia, Spain. DF and FH-T were supported by Excellence Project of the Consejeria de Innovacion, Ciencia y Empresa of the Junta de Andalucia (grant # CTS1416) and a CNIC-T translactional grant (CNIC-T 2009/08). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, et al. (1996) Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet 14: 392–399. [DOI] [PubMed] [Google Scholar]
- 2. Hjalt TA, Semina EV (2005) Current molecular understanding of Axenfeld-Rieger syndrome. Expert Rev Mol Med 7: 1–17. [DOI] [PubMed] [Google Scholar]
- 3. Tumer Z, Bach-Holm D (2009) Axenfeld-Rieger syndrome and spectrum of PITX2 and FOXC1 mutations. Eur J Hum Genet 17: 1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gage PJ, Suh H, Camper SA (1999) Dosage requirement of Pitx2 for development of multiple organs. Development 126: 4643–4651. [DOI] [PubMed] [Google Scholar]
- 5. Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, et al. (1999) Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development 126: 5749–5758. [DOI] [PubMed] [Google Scholar]
- 6. Lin CR, Kioussi C, O'Connell S, Briata P, Szeto D, et al. (1999) Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401: 279–282. [DOI] [PubMed] [Google Scholar]
- 7. Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF (1999) Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature 401: 276–278. [DOI] [PubMed] [Google Scholar]
- 8. Franco D, Campione M (2003) The role of Pitx2 during cardiac development. Linking left-right signaling and congenital heart diseases. Trends Cardiovasc Med 13: 157–163. [DOI] [PubMed] [Google Scholar]
- 9. Franco D, Chinchilla A, Aranega AE (2012) Transgenic insights linking pitx2 and atrial arrhythmias. Front Physiol 3: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Franco D, Chinchilla A, Daimi H, Dominguez JN, Aranega A (2011) Modulation of conductive elements by Pitx2 and their impact on atrial arrhythmogenesis. Cardiovasc Res 91: 223–231. [DOI] [PubMed] [Google Scholar]
- 11. Franco D, Christoffels VM, Campione M (2014) Homeobox transcription factor Pitx2: The rise of an asymmetry gene in cardiogenesis and arrhythmogenesis. Trends Cardiovasc Med 24: 23–31. [DOI] [PubMed] [Google Scholar]
- 12. Tessari A, Pietrobon M, Notte A, Cifelli G, Gage PJ, et al. (2008) Myocardial Pitx2 differentially regulates the left atrial identity and ventricular asymmetric remodeling programs. Circ Res 102: 813–822. [DOI] [PubMed] [Google Scholar]
- 13. Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, et al. (2011) PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet 4: 269–279. [DOI] [PubMed] [Google Scholar]
- 14. Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, et al. (2010) Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci USA 107: 9753–9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, et al. (2011) PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet 4: 123–133. [DOI] [PubMed] [Google Scholar]
- 16. Kahr PC, Piccini I, Fabritz L, Greber B, Scholer H, et al. (2011) Systematic analysis of gene expression differences between left and right atria in different mouse strains and in human atrial tissue. PLoS ONE 6: e26389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hsu J, Hanna P, Van Wagoner DR, Barnard J, Serre D, et al. (2012) Whole genome expression differences in human left and right atria ascertained by RNA sequencing. Circ Cardiovasc Genet 5: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao Y, Zhang M, Li L, Bai Y, Zhou Y, et al.. (2014) Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ Cardiovasc Genet: doi 10.1161/CIRCGENETICS.1113.000259. [DOI] [PMC free article] [PubMed]
- 19. Christoffels VM, Smits GJ, Kispert A, Moorman AF (2010) Development of the pacemaker tissues of the heart. Circ Res 106: 240–254. [DOI] [PubMed] [Google Scholar]
- 20. Ammirabile G, Tessari A, Pignataro V, Szumska D, Sutera Sardo F, et al. (2012) Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc Res 93: 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou YM, Zheng PX, Yang YQ, Ge ZM, Kang WQ (2013) A novel PITX2c loss-of-function mutation underlies lone atrial fibrillation. Int J Mol Med 32: 827–834. [DOI] [PubMed] [Google Scholar]
- 22. Torrado M, Lopez E, Centeno A, Castro-Beiras A, Mikhailov AT (2004) Left-right asymmetric ventricular expression of CARP in the piglet heart: regional response to experimental heart failure. Eur J Heart Fail 6: 161–172. [DOI] [PubMed] [Google Scholar]
- 23. Torrado M, Lopez E, Centeno A, Medrano C, Castro-Beiras A, et al. (2003) Myocardin mRNA is augmented in the failing myocardium: expression profiling in the porcine model and human dilated cardiomyopathy. J Mol Med 81: 566–577. [DOI] [PubMed] [Google Scholar]
- 24. Torrado M, Iglesias R, Centeno A, Lopez E, Mikhailov AT (2011) Targeted gene-silencing reveals the functional significance of myocardin signaling in the failing heart. PLoS ONE 6: e26392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Torrado M, Nespereira B, Bouzamayor Y, Centeno A, Lopez E, et al. (2006) Differential atrial versus ventricular ANKRD1 gene expression is oppositely regulated at diastolic heart failure. FEBS Lett 580: 4182–4187. [DOI] [PubMed] [Google Scholar]
- 26. Torrado M, Iglesias R, Nespereira B, Mikhailov AT (2010) Identification of candidate genes potentially relevant to chamber-specific remodeling in postnatal ventricular myocardium. J Biomed Biotechnol 2010: 603159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lozano-Velasco E, Contreras A, Crist C, Hernandez-Torres F, Franco D, et al. (2011) Pitx2c modulates Pax3+/Pax7+ cell populations and regulates Pax3 expression by repressing miR27 expression during myogenesis. Dev Biol 357: 165–178. [DOI] [PubMed] [Google Scholar]
- 28. Torrado M, Iglesias R, Nespereira B, Centeno A, Lopez E, et al. (2009) Intron retention generates ANKRD1 splice variants that are co-regulated with the main transcript in normal and failing myocardium. Gene 440: 28–41. [DOI] [PubMed] [Google Scholar]
- 29. Hernandez-Torres F, Rastrojo A, Aguado B (2013) Intron retention and transcript chimerism conserved across mammals: Ly6g5b and Csnk2b-Ly6g5b as examples. BMC Genomics 14: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Claycomb WC, Lanson NA, Stallworth BS, Egeland DB, Delcarpio JB, et al. (1998) HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA 95: 2979–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eaton SL, Roche SL, Llavero Hurtado M, Oldknow KJ, Farquharson C, et al. (2013) Total protein analysis as a reliable loading control for quantitative fluorescent Western blotting. PLoS ONE 8: e72457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beinlich CJ, Rissinger CJ, Morgan HE (1995) Mechanisms of rapid growth in the neonatal pig heart. J Mol Cell Cardiol 27: 273–281. [DOI] [PubMed] [Google Scholar]
- 33. Torrado M, Iglesias R, Centeno A, Lopez E, Mikhailov AT (2010) Exon-skipping brain natriuretic peptide variant is overexpressed in failing myocardium and attenuates brain natriuretic peptide production in vitro. Exp Biol Med (Maywood) 235: 941–951. [DOI] [PubMed] [Google Scholar]
- 34. Diehl AG, Zareparsi S, Qian M, Khanna R, Angeles R, et al. (2006) Extraocular muscle morphogenesis and gene expression are regulated by Pitx2 gene dose. Invest Ophthalmol Vis Sci 47: 1785–1793. [DOI] [PubMed] [Google Scholar]
- 35. Huang Y, Huang K, Boskovic G, Dementieva Y, Denvir J, et al. (2009) Proteomic and genomic analysis of PITX2 interacting and regulating networks. FEBS Lett 583: 638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iwata J, Tung L, Urata M, Hacia JG, Pelikan R, et al. (2012) Fibroblast growth factor 9 (FGF9)-pituitary homeobox 2 (PITX2) pathway mediates transforming growth factor beta (TGFbeta) signaling to regulate cell proliferation in palatal mesenchyme during mouse palatogenesis. J Biol Chem 287: 2353–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paylakhi SH, Fan JB, Mehrabian M, Sadeghizadeh M, Yazdani S, et al. (2011) Effect of PITX2 knockdown on transcriptome of primary human trabecular meshwork cell cultures. Mol Vis 17: 1209–1221. [PMC free article] [PubMed] [Google Scholar]
- 38. Campbell AL, Shih HP, Xu J, Gross MK, Kioussi C (2012) Regulation of motility of myogenic cells in filling limb muscle anlagen by Pitx2. PLoS ONE 7: e35822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bajard L, Relaix F, Lagha M, Rocancourt D, Daubas P, et al. (2006) A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev 20: 2450–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Francetic T, Li Q (2011) Skeletal myogenesis and Myf5 activation. Transcription 2: 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mok GF, Sweetman D (2011) Many routes to the same destination: lessons from skeletal muscle development. Reproduction 141: 301–312. [DOI] [PubMed] [Google Scholar]
- 42. Van der Heiden K, Egorova AD, Poelmann RE, Wentzel JJ, Hierck BP (2011) Role for primary cilia as flow detectors in the cardiovascular system. Int Rev Cell Mol Biol 290: 87–119. [DOI] [PubMed] [Google Scholar]
- 43. Rossi A, Cicoira M, Florea VG, Golia G, Florea ND, et al. (2006) Chronic heart failure with preserved left ventricular ejection fraction: diagnostic and prognostic value of left atrial size. Int J Cardiol 110: 386–392. [DOI] [PubMed] [Google Scholar]
- 44. Teo SG, Yang H, Chai P, Yeo TC (2010) Impact of left ventricular diastolic dysfunction on left atrial volume and function: a volumetric analysis. Eur J Echocardiogr 11: 38–43. [DOI] [PubMed] [Google Scholar]
- 45. Martinez-Fernandez S, Hernandez-Torres F, Franco D, Lyons GE, Navarro F, et al. (2006) Pitx2c overexpression promotes cell proliferation and arrests differentiation in myoblasts. Dev Dyn 235: 2930–2939. [DOI] [PubMed] [Google Scholar]
- 46. De Keulenaer GW, Brutsaert DL (2011) Systolic and diastolic heart failure are overlapping phenotypes within the heart failure spectrum. Circulation 123: 1996–2005. [DOI] [PubMed] [Google Scholar]
- 47. Borlaug BA, Redfield MM (2011) Diastolic and systolic heart failure are distinct phenotypes within the heart failure spectrum. Circulation 123: 2006–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mikhailov AT, Torrado M (2008) The enigmatic role of the ankyrin repeat domain 1 gene in heart development and disease. Int J Dev Biol 52: 811–821. [DOI] [PubMed] [Google Scholar]
- 49. Zakeri R, Chamberlain AM, Roger VL, Redfield MM (2013) Temporal relationship and prognostic significance of atrial fibrillation in heart failure patients with preserved ejection fraction: a community-based study. Circulation 128: 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Su D, Li Q, Guan L, Gao X, Zhang H, et al. (2013) Down-regulation of EBAF in the heart with ventricular septal defects and its regulation by histone acetyltransferase p300 and transcription factors smad2 and cited2. Biochim Biophys Acta 1832: 2145–2152. [DOI] [PubMed] [Google Scholar]
- 51. Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, et al. (2002) Identification of a Wnt/Dvl/beta-Catenin –> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell 111: 673–685. [DOI] [PubMed] [Google Scholar]
- 52. Oikonomopoulos A, Sereti KI, Conyers F, Bauer M, Liao A, et al. (2011) Wnt signaling exerts an antiproliferative effect on adult cardiac progenitor cells through IGFBP3. Circ Res 109: 1363–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bergmann MW (2010) WNT signaling in adult cardiac hypertrophy and remodeling: lessons learned from cardiac development. Circ Res 107: 1198–1208. [DOI] [PubMed] [Google Scholar]
- 54. Dobaczewski M, Chen W, Frangogiannis NG (2011) Transforming growth factor (TGF)-beta signaling in cardiac remodeling. J Mol Cell Cardiol 51: 600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Doetschman T, Barnett JV, Runyan RB, Camenisch TD, Heimark RL, et al. (2012) Transforming growth factor beta signaling in adult cardiovascular diseases and repair. Cell Tissue Res 347: 203–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, et al. (2006) Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol 172: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zacharias AL, Lewandoski M, Rudnicki MA, Gage PJ (2011) Pitx2 is an upstream activator of extraocular myogenesis and survival. Dev Biol 349: 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Knopp P, Figeac N, Fortier M, Moyle L, Zammit PS (2013) Pitx genes are redeployed in adult myogenesis where they can act to promote myogenic differentiation in muscle satellite cells. Dev Biol 377: 293–304. [DOI] [PubMed] [Google Scholar]
- 59. Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, et al. (2012) Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med 4: 130ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Santerre RF, Bales KR, Janney MJ, Hannon K, Fisher LF, et al. (1993) Expression of bovine myf5 induces ectopic skeletal muscle formation in transgenic mice. Mol Cell Biol 13: 6044–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Edwards JG, Lyons GE, Micales BK, Malhotra A, Factor S, et al. (1996) Cardiomyopathy in transgenic myf5 mice. Circ Res 78: 379–387. [DOI] [PubMed] [Google Scholar]
- 62.Sweetman D (2012) The myogenic regulatory factors: critical determinants of muscle identity in development, growth and regeneration. In: Cseri J, editor. Skeletal muscle - from myogenesis to clinical relations. Rijeka: InTech. pp. 31–48.
- 63. Rios R, Carneiro I, Arce VM, Devesa J (2002) Myostatin is an inhibitor of myogenic differentiation. Am J Physiol Cell Physiol 282: C993–999. [DOI] [PubMed] [Google Scholar]
- 64.Torrado M, Iglesias R, Centeno A, Mikhailov AT (2011) Molecular signature analysis reveals a set of genes that can be involved in balance between concentric and eccentric cardiac growth. Eur J Heart Fail Suppl 10: S215–S217.
- 65. George I, Bish LT, Kamalakkannan G, Petrilli CM, Oz MC, et al. (2010) Myostatin activation in patients with advanced heart failure and after mechanical unloading. Eur J Heart Fail 12: 444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zu L, Bedja D, Fox-Talbot K, Gabrielson KL, Van Kaer L, et al. (2010) Evidence for a role of immunoproteasomes in regulating cardiac muscle mass in diabetic mice. J Mol Cell Cardiol 49: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Clause KC, Tchao J, Powell MC, Liu LJ, Huard J, et al. (2012) Developing cardiac and skeletal muscle share fast-skeletal myosin heavy chain and cardiac troponin-I expression. PLoS ONE 7: e40725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yang CF, Chou KY, Weng ZC, Hung SC, Lai ST, et al. (2008) Cardiac myocyte progenitors from adult hearts for myocardial regenerative therapy. J Chin Med Assoc 71: 79–85. [DOI] [PubMed] [Google Scholar]
- 69. Berkes CA, Tapscott SJ (2005) MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 16: 585–595. [DOI] [PubMed] [Google Scholar]
- 70. Miner JH, Miller JB, Wold BJ (1992) Skeletal muscle phenotypes initiated by ectopic MyoD in transgenic mouse heart. Development 114: 853–860. [DOI] [PubMed] [Google Scholar]
- 71. Murry CE, Kay MA, Bartosek T, Hauschka SD, Schwartz SM (1996) Muscle differentiation during repair of myocardial necrosis in rats via gene transfer with MyoD. J Clin Invest 98: 2209–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Crippa S, Cassano M, Messina G, Galli D, Galvez BG, et al. (2011) miR669a and miR669q prevent skeletal muscle differentiation in postnatal cardiac progenitors. J Cell Biol 193: 1197–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ganga M, Espinoza HM, Cox CJ, Morton L, Hjalt TA, et al. (2003) PITX2 isoform-specific regulation of atrial natriuretic factor expression: synergism and repression with Nkx2.5. J Biol Chem 278: 22437–22445. [DOI] [PubMed] [Google Scholar]
- 74. Venugopalan SR, Amen MA, Wang J, Wong L, Cavender AC, et al. (2008) Novel expression and transcriptional regulation of FoxJ1 during oro-facial morphogenesis. Hum Mol Genet 17: 3643–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative qRT-PCR amplification plots for Pitx2c transcripts in the piglet left ventricle. A - Newborn (NB, green) versus 30-day-old (30 d, blue) normal piglets. B - PBS-injected (PBS, blue) versus Dox-injected (Dox, red) 30-day-old animals. Rpl19 - internal standard levels. Arrows - threshold cycle (CT). FT - fluorescent threshold. RFU - relative fluorescent units. Under experimental conditions used, each primer pair yielded a single peak of dissociation on the melting curve (C) and a single band with the expected size on PAGE gel post-stained with SYBR Green I (D).
(TIF)
Representative qRT-PCR amplification plots for Myf5 and Myog transcripts in the left ventricle of PBS- versus Dox-injected piglets. A - PBS-injected (PBS, blue) versus Dox-injected (Dox, red) 30-day-old animals. Rpl19 - internal standard levels. Arrows - threshold cycle (CT). FT - fluorescent threshold. RFU - relative fluorescent units. Under experimental conditions used, each primer pair yielded a single band with the expected size on PAGE gel post-stained with SYBR Green I (B).
(TIF)
Expression of Myog, Foxj1 and Pax3 genes in Pitx2c-transfected HL-1 and Sol8 cells. Overall relative levels of Myog, Foxj1 and Pax3 transcripts in HL-1 and Sol8 cells transfected with V5-tagged Pitx2c vector at different doses (100-400 ng). CT – empty-vector transfected cells. Shown are results of qRT-PCR analysis. Data from six replicates of each transfection were pooled and averaged. *p≤0.05.
(TIF)
Baseline characteristics of neonatal piglets injected with Dox or PBS three weeks after injections.
(DOCX)
Patient characteristics for samples employed qRT-PCR and Western blot analyses.
(DOCX)
Primers used in this study.
(DOCX)
Selective data set derived from the microarray database.
(DOCX)