Abstract
Cellular adhesion by classical cadherins depends critically on the exact proteolytic removal of their N-terminal prosequences. In this combined solution NMR and X-ray crystallographic study, the consequences of propeptide cleavage of an epithelial cadherin construct (domains 1 and 2) were followed at atomic level. At low protein concentration, the N-terminal processing induces docking of the tryptophan-2 side-chain into a binding pocket on the same molecule. At high concentration, cleavage induces dimerization (KD=0.72 mM, koff=0.7 s−1) and concomitant intermolecular exchange of the βA-strands and the tryptophan-2 side-chains. Thus, the cleavage represents the switch from a nonadhesive to the functional form of cadherin.
Keywords: cellular adhesion, prodomain, protein dynamics, protein structure, proteolytic activation
Introduction
Cell–cell adhesion in vertebrates as well as in many invertebrates is largely based on the specificity and activity of cell-surface receptors of the cadherin superfamily. Cadherins are calcium-dependent, homophilic cell adhesion molecules, which exert important functions in tissue morphogenesis, neuronal development, and signal transduction (Takeichi, 1995; Gumbiner, 1996). The adhesive activity of mature cadherins is regulated by several mechanisms including protein recruitment to regions of cell contacts, calcium binding, internalization and modulation of cadherin–catenin interactions (Gumbiner, 2000). The generation of mature, functional active cadherins, however, occurs via a proteolytic event. Many cadherins are synthesized as inactive precursor proteins containing a prosequence followed by five Ig-like cadherin domains (CAD1–5) (Nollet et al, 2000). Domains CAD1 and CAD2 are functional in cell adhesion (Takeichi, 1995; Tomschy et al, 1996; Boggon et al, 2002; He et al, 2003). The length of cadherin prosequences varies from 105 to 120 residues in type I cadherins and desmocollins and is shorter (20–35 residues) in type II cadherins and desmogleins. E- and N-cadherins (ECAD and NCAD) belong to the first type (for definitions of type I and II, see Nollet et al, 2000). The N-terminal prosequence is proteolytically cleaved off in the late Golgi at the conserved consensus site R-X1-K-R-X2-W, and the mature cadherin (X2-W- - -) is then transported to the plasma membrane (Ozawa and Kemler, 1990). Strikingly, cadherins are completely nonadhesive when only few amino-acid residues remain attached to the CAD1 domain by incorrect cleavage (Ozawa and Kemler, 1990; Ozawa, 2002).
Structural details regarding how propeptide processing renders cadherins functionally active have not been reported. Only limited and controversial information was obtained regarding the active, adhesive state of cadherins from structural studies of fragments containing CAD1 domains with additional N-terminal residues (Overduin et al, 1995; Nagar et al, 1996; Tamura et al, 1998; Pertz et al, 1999; Häussinger et al, 2002). The data suggest that the functionally essential and conserved tryptophan residue in position 2 (W2) is strongly influenced by the state of the closely adjacent N-terminus (Koch et al, 1999). W2 point mutants are inactive (Tamura et al, 1998; Pertz et al, 1999), and crystallographic data suggest that formation of an N-terminal β-strand exchange dimer with the W2 side-chain docked into a hydrophobic pocket of the partner molecule is essential for activity (Shapiro et al, 1995; Boggon et al, 2002).
In the present combined solution NMR and X-ray crystallographic study, we have followed the structural rearrangements of an epithelial cadherin construct as a consequence of the cleavage of an artificial N-terminal peptide, which should serve as a model for propeptide processing. Note that it would be desirable to follow this process on the native E-cadherin prosequence. However, such an approach is currently not possible, because the specific E-cadherin endoprotease has not been identified (Ozawa and Kemler, 1990). The artificial precursor cadherin (HisXa-ECAD12) was generated from epithelial cadherin ectodomains CAD1 and CAD2 preceded by a 28-amino-acid-long prosequence including a His10-tag and a factor Xa cleavage site (Figure 1). Proteolytic cleavage of this construct generates a protein (ECAD12) with the mature N-terminus of E-cadherin. This N-terminal processing leads to substantial structural rearrangements that are observable by solution NMR spectroscopy. At low protein concentrations, the cleavage induces tight, intramolecular binding of W2 to the tryptophan binding pocket of the same cadherin molecule. Additional strong chemical shift changes in the vicinity of W2 and for residues that line the tryptophan binding pocket at higher protein concentrations indicate an associated form of the protein and the onset of strand exchange dimerization. The strand exchange was confirmed by an X-ray crystallographic study of mature ECAD12 in the presence of calcium. These findings present a dynamical view of the strand exchange mechanism (Shapiro et al, 1995; Boggon et al, 2002) and explain conflicting results on the absence of strand exchange in other studies of cadherins (Overduin et al, 1995; Nagar et al, 1996; Shapiro and Colman, 1998; Pertz et al, 1999; Häussinger et al, 2002).
Figure 1.
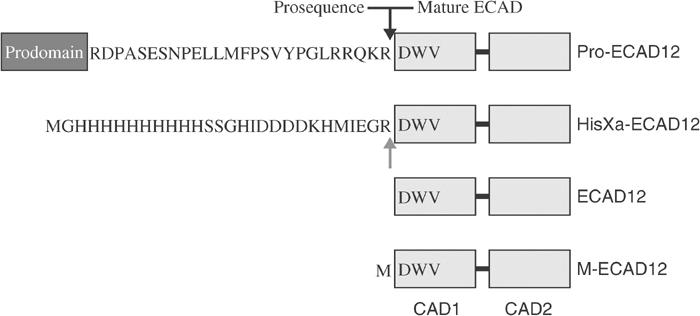
Schematic representation of prodomain–CAD1 domain boundaries of wild-type murine E-cadherin and E-cadherin constructs used in this study. The cleavage site of the prodomain in the wild-type protein is indicated by a black arrow. In HisXa-ECAD12, this cleavage site is mimicked by the factor Xa cleavage sequence IEGR (gray arrow).
Results
Domain boundary between prodomain and CAD1 and assignment of constructs
Figure 1 shows the N-terminal sequence differences between murine wild-type epithelial cadherin with its prosequence (Pro-ECAD12) and the ECAD12 constructs used in this investigation. The prodomain of classical type I cadherins such as E-cadherin is connected to the first cadherin domain via a flexible linker (AW Koch, personal communication) that contains a cleavage site for an endogenous protease. Our construct HisXa-ECAD12 mimics this arrangement with a His10-tag followed by a spacer and a factor Xa cleavage site.
Backbone and side-chain chemical shifts of HisXa-ECAD12 and ECAD12 in the presence of 1 mM CaCl2 or 1 mM EDTA were assigned by standard three- and four-dimensional NMR experiments. The assignments for M-ECAD12 have been reported before (Häussinger et al, 2002).
Activation by proteolytic cleavage
The 1H, 15N, 13C′, 13Cα, and 13Cβ chemical shifts of HisXa-ECAD12 are very similar (∣Δδ∣<∼0.5 ppm; Figure 2A, Supplementary Figure S1A) to M-ECAD12 (Häussinger et al, 2002) in which an additional methionine precedes the native N-terminus of ECAD12.
Figure 2.
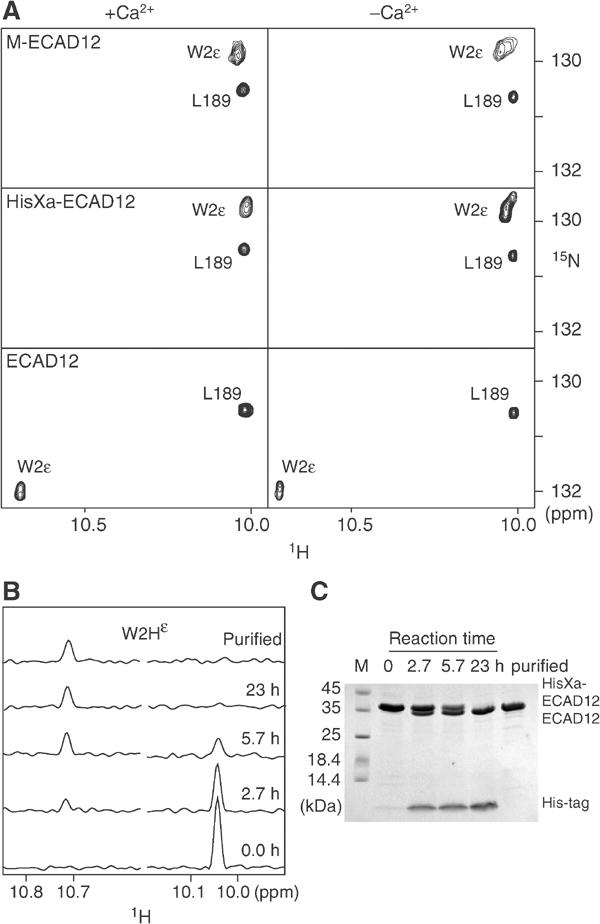
Changes in solution NMR spectra induced by N-terminal processing of ECAD12 in the presence or absence of calcium. (A) Small regions of the 1H-15N TROSY spectra of M-ECAD12 (0.6 mM), HisXa-ECAD12 (1.1 mM), and ECAD12 (0.6 mM) released from HisXa-ECAD12 after protease processing. Spectra are labelled with assignment information. (B) Real-time observation of the effect of HisXa-ECAD12 N-terminal processing on the W2Hɛ resonance. One-dimensional 1H traces were taken from 1H-15N TROSY spectra at the W2Nɛ position of ECAD12 (left) and HisXa-ECAD12 (right) at the indicated times after cleavage start by factor Xa and after the removal of the cleaved-off His-tag. (C) SDS–PAGE observation of HisXa-ECAD12 N-terminal cleavage by factor Xa in the same samples as in (B).
In contrast, when HisXa-ECAD12 is subjected to proteolysis by factor Xa, thereby releasing the mature ECAD12 molecule, strong chemical shift changes (up to 5 ppm) occur across the entire CAD1 domain of ECAD12 whereas the CAD2 domain remains virtually unchanged (Supplementary Figure S1B). These chemical shift changes indicate substantial structural rearrangements and are most pronounced in the vicinity of W2 and its binding pocket, for example, for residues V3, K25, I24, H79, and T97. Particularly striking is the prominent, calcium-independent downfield shift of the W2 indole 1Hɛ and 15Nɛ resonances by 0.67 and 2.1 ppm, respectively (Figure 2A). Such downfield shifts are indicative of the formation of a strong hydrogen bond (H-bond), in this case to the carbonyl oxygen of residue D90 (see below). The time course of the cleavage reaction was monitored by NMR (Figure 2B) and SDS–PAGE (Figure 2C) and, upon completion after 23 h, the cleaved-off His-tag was removed by Ni-affinity chromatography.
The addition of calcium to either ECAD12, HisXa-ECAD12, or M-ECAD12 does not induce strong changes in the W2ɛ resonances (Figure 2A). However, as described earlier (Häussinger et al, 2002), calcium induces changes for residues that are close to the calcium-binding sites, for example, R68-A70, D100-E107, T133-I146, D195, G200, and L201, and similar changes in chemical shift are observed for HisXa-ECAD12 and ECAD12 (Supplementary Figure S1C). In addition to this calcium effect, there is a pronounced dependence of the spectra on the protein concentration for the mature ECAD12 molecule, which is not observed for the HisXa-ECAD12 or M-ECAD12 construct. With increasing protein concentration, a second, strongly shifted spectral species appears (see below, Figure 6) that is in slow chemical exchange with the first species. Analysis of 15N relaxation data reveals that the first species is a monomeric form of ECAD12 whereas the second species must be an associated form of ECAD12. In the following, we analyze first the behavior of the monomeric species, which is dominant at low protein concentration, and discuss later the associated species, which is dominant at high concentrations.
Figure 6.
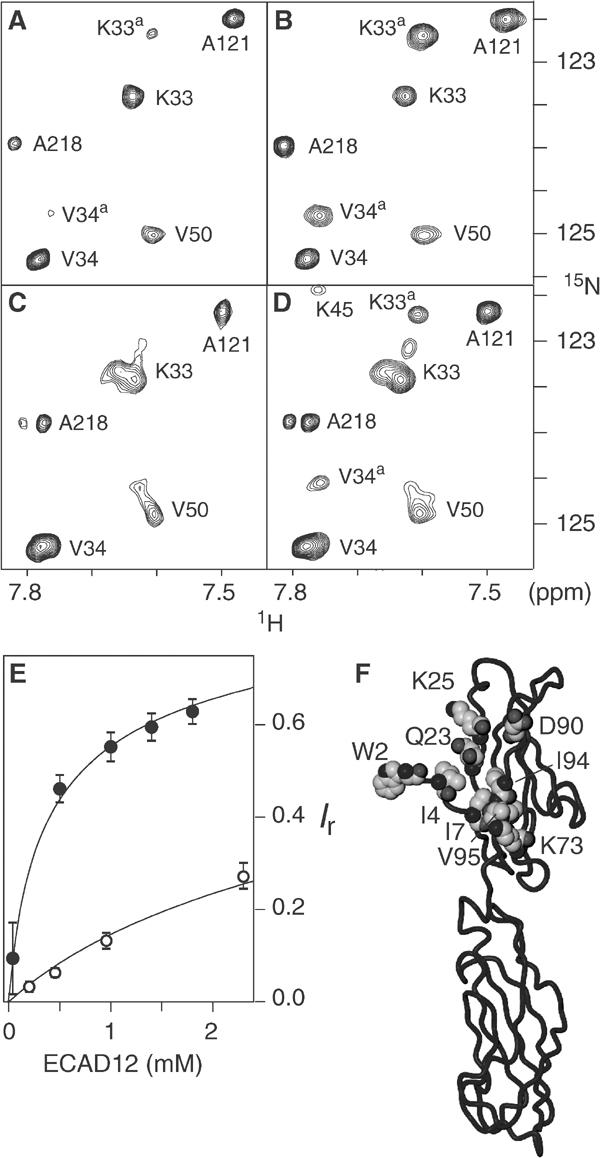
NMR spectroscopic evidence of ECAD12 homoassociation in the presence and absence of calcium. Small regions of 1H-15N TROSY spectra are shown for total ECAD12 concentrations of 40 μM (A) and 0.5 mM (B) in the presence of calcium, and for 60 μM (C) and 1.1 mM (D) in the absence of calcium. Resonances are labelled with assignment information. The associated species is indicated by a superscript ‘a'. (E) Relative intensity Ir=Ia/(Im+Ia) of the associated species as a function of total ECAD12 concentration. Intensities of the monomeric (Im) and associated (Ia) species were obtained as an average of peak volumes for residues, which showed well-separated resonances. Filled and open circles correspond to calcium-bound and calcium-free conditions, respectively. The solid lines present fits to the data assuming a bimolecular reaction with a KD of 0.72 mM for the calcium-bound and of 10 mM for the calcium-free protein, respectively. (F) Residues that show the strongest observable 1H-15N chemical shift differences between the monomer and associated species in the calcium-bound form are shown in space-fill representation in the crystal structure of ECAD12.
Apparently, the strong chemical shift changes induced by the removal of the N-terminal prosequence are the result of a structural rearrangement within the CAD1 domain. This rearrangement was followed by 15N-edited NOE spectra and is most evident for the W2 side-chain and for the D90, M92, V3, and K25 backbone 1H-15N moieties (Figure 3). NOEs between W2Hɛ and either D90HN or M92HN are expected when the tryptophan side-chain is inserted into its binding pocket of the CAD1 domain with its side-chain ɛ-nitrogen forming an H-bond to the carbonyl oxygen of D90 (see also Figure 4). For HisXa-ECAD12 (Figure 3A) and M-ECAD12 (Figure 3B), such NOEs are below the detection limit, regardless of protein and calcium concentration. Exchange peaks for W2Hɛ, D90HN, and M92HN with water indicate that these protons are not involved in strong H-bonds and are readily accessible to solvent. This is supported by a heteronuclear {1H}-15N NOE of 0.44 for the W2ɛ group in HisXa-ECAD12 and 0.30 in M-ECAD12 (data not shown), which gives direct evidence for considerable flexibility of the tryptophan side-chain on the subnanosecond time scale.
Figure 3.
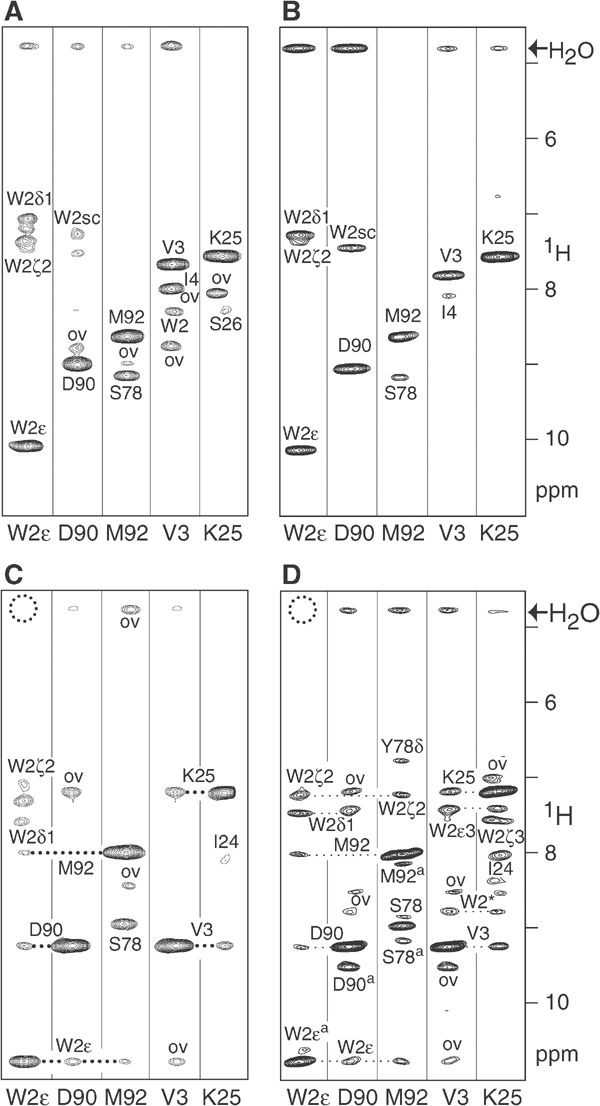
NOE evidence for changes in the conformation of the ECAD12 N-terminus upon N-terminal processing in the presence and absence of calcium. Small regions of the 15N-edited NOESY spectra for several forms of ECAD12 were extracted at the 1H-15N resonance positions of the W2 side-chain and backbone amides of D90, M92, V3, and K25. In all cases, the resonance frequencies of the monomeric species were used. (A) Calcium-free HisXa-ECAD12 (total concentration 1.1 mM). (B) Calcium-free M-ECAD12 (3.5 mM). (C) Calcium-free ECAD12 (1.1 mM). (D) Calcium-bound ECAD12 (1.0 mM). (A–D) Resonances are labelled with assignment information. Peaks resulting from overlap with unrelated resonances are marked by ‘ov'. Owing to incomplete deuteration, several NOEs to W2 side-chain protons are visible. W2sc indicates an unassigned proton resonance of the W2 side-chain. A dotted circle indicates the absence of an exchange peak with water for W2Hɛ. In (D), several peaks show correlations to residues of the associated form due to chemical exchange and are marked by a superscript ‘a'.
Figure 4.
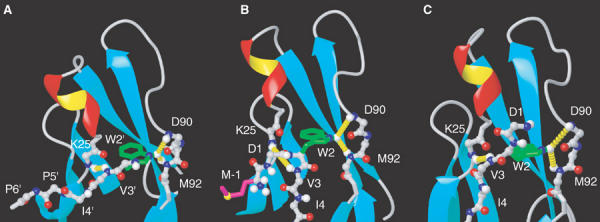
Conformation of the N-terminal region of ECAD12 in different crystal forms. (A) Details of the crystal structure of dimeric ECAD12 (1q1p) showing the docking of W2 into the pocket of the partner molecule. Residues that stem from the symmetry equivalent molecule are marked by an apostrophe ('). The same region is shown for the crystal structures of M-ECAD12 (B, 1ff5) (Pertz et al, 1999) and human ECAD1 in complex with internalin (C, 1o6s) (Schubert et al, 2002). Long-range NOEs observed between W2Hɛ↔D90HN/M92HN and V3HN↔K25HN for the monomeric form of ECAD12 (see text and Figure 3C and D) are highlighted by dashed yellow cylinders in all crystal forms (A–C).
In contrast, after removal of the prosequence, NOE correlations from W2Hɛ to D90HN and M92HN are readily observed for ECAD12 in the absence (Figure 3C) as well as in the presence of calcium (Figure 3D). In addition, the exchange with water is either absent or very strongly reduced for these protons, and the heteronuclear {1H}-15N NOE for the W2ɛ group is increased to 0.86. Therefore, these data indicate that the N-terminal cleavage induces a switch for W2 from a highly flexible to a rigid state, where the side-chain is tightly bound within the specific binding pocket of the CAD1 domain.
Intramolecular docking of tryptophan 2
A particularly striking difference between the low concentration form of ECAD12 and HisXa-ECAD12 is the NOE pattern for V3HN. In ECAD12 (Figure 3C and D), V3HN shows a correlation to K25HN, which is not present in HisXa-ECAD12 or M-ECAD12 (Figure 3A and B). In the crystal structures that show intermolecular W2 strand exchange, that is, NCAD1 (Shapiro et al, 1995), CCAD1–5 (Boggon et al, 2002), and ECAD12 (Figure 4A), the amide nitrogen of V3 forms an intermolecular H-bond to the carbonyl oxygen of residue K25′ of the neighboring molecule such that V3HN and K25′HN are in close contact. No such intermolecular H-bond exists in the other cadherin structures that do not show strand exchange.
It was therefore of importance to determine whether the observed NOEs for W2Hɛ and V3HN in the mature form of ECAD12 correspond to intermolecular strand exchange or whether they are caused by intramolecular contacts. For this purpose, 15N-selected/15N-edited NOE experiments were carried out for 1:1 mixtures of uniformly 15N-labelled and natural abundance 14N-ECAD12 in the absence of calcium (Figure 5). The spectra clearly show that for the spectral species that dominates at low concentrations, the NOEs between W2Hɛ and D90HN/M92HN as well as between V3HN and K25HN are only observed between protons that are both bonded to 15N nuclei (Figure 5A). No such NOEs are visible in a variant of this experiment where only NOEs between 15N-bonded and not 15N-bonded protons are detected (Figure 5B). Therefore the contacts must be of intramolecular nature. This result was obtained irrespective of the presence of calcium or whether the mixture of molecules had been subjected to an unfolding/refolding procedure in 7 M urea. Hence under these solution conditions, W2 binds tightly to the binding pocket on the same molecule.
Figure 5.
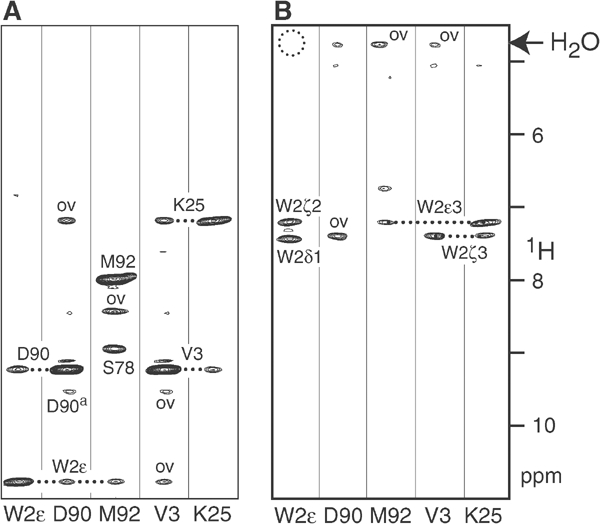
15N-selected/15N-edited NOESY experiments to distinguish between intra- and intermolecular interactions. Experiments were carried out on a sample consisting of a 1:1 mixture of uniformly 15N-labelled and natural abundance 14N-ECAD12 (total concentration 1.3 mM, calcium-free conditions) (A) Results of a 15N-selected/15N-edited 3D NOESY experiment. NOE connectivities are only present between protons that are both bonded to 15N nuclei. Associated forms are marked by ‘a'. (B) Results of a (not-15N)-selected/15N-edited NOESY experiment. NOE connectivities are only present between protons that are not bonded to 15N nuclei (vertical axis) and protons bonded to 15N nuclei (horizontal axis).
Apparently, this conformation is similar, but not identical, to the 1ff5 crystal structure of M-ECAD12 (Figure 4B) where the side-chain indole of W2 forms an intramolecular H-bond to the carbonyl of D90 and the amide protons of D90 and M92 are in close contact with W2Hɛ. An important difference exists between these two conformations: in M-ECAD12, a sharp turn in the protein backbone at residue W2 puts residues M-1 and D1 between residues V3 and K25 such that the H-bond between V3HN and K25O cannot be closed. In this conformation, the distance between the V3 and K25 amide protons amounts to 5.8 Å, which would hardly be detectable by our NOE spectra. Apparently, after the removal of the additional N-terminal residues, the structure around W2 can snap into a conformation where this distance is much shorter, such that an NOE can be detected. This coincides with a strong (1.7 ppm) downfield shift of the V3 amide proton (Supplementary Figure S1B), which is characteristic of H-bond formation. We interpret this behavior as the closing of the intramolecular V3HN/K25O H-bond in the monomeric state of mature ECAD12. Thus, the NMR data provide evidence that the structure of the monomeric, mature ECAD12 N-terminus including the H-bond pattern for W2Hɛ and V3HN is almost identical to the conformation in the dimeric strand exchange form (Figure 4A), with the sole difference that the contacts between residues W2/V3 and the rest of the protein are of intramolecular instead of intermolecular nature.
It is interesting to observe that this conformation of the monomeric, mature ECAD12 N-terminus is realized in the structure of monomeric, human ECAD1 bound to Listeria internalin (Schubert et al, 2002) (PDB code 1o6s; Figure 4C). Although the N-terminus of this structure contained the foreign sequence GPLG before residue 1 and the incorrect residue S1 instead of the conserved D1, its conformation for W2 and V3 corresponds exactly to the structure expected from the observed intramolecular NOE connectivities for V3HN and W2Hɛ in mature, monomeric ECAD12. Apparently in this case, the perturbation by the nonproper N-terminus is not strong enough to cause deviations from the structure of mature, monomeric cadherin.
Transition from monomeric to associated form of ECAD12
As mentioned before, there is a pronounced dependence of the spectra on the protein concentration for the mature ECAD12 molecule. A comparable dependence is not observed for the HisXa-ECAD12 or M-ECAD12 construct. With increasing protein concentration, a second, strongly shifted spectral species appears that is in slow chemical exchange with the first species both in the presence (Figure 6A and B) and absence of calcium (Figure 6C and D). Assuming a bimolecular association, the KD of this reaction in the presence of calcium was fitted to a value of 0.72 mM from the dependence of the peak volumes of both spectral species (Figure 6E, solid points).
Differences in chemical shifts between the monomeric and associated forms are most pronounced for the amide 1H-15N groups of residues W2, I4, I7, Q23, K25, K73, D90, I94, V95, and I96 (∣Δ1HN∣>0.5 ppm or ∣Δ15N∣>1 ppm; Figure 6F and Supplementary Figure S1D). For the W2ɛ resonances, both a change in chemical shift and strong broadening at high concentrations are observed. Figure 6F depicts the affected residues highlighted in the crystallographic structure of ECAD12. Similar to the changes induced by the N-terminal processing, the residues are close to W2 or line the W2 binding pocket. However, the type of residues and the observed chemical shift changes are distinct between the two processes.
An analysis of 15N T1 and T2 relaxation times yields isotropic rotational diffusion times of ∼15 and ∼35 ns for the monomeric and the associated calcium-bound species at 25°C, respectively. These rotational correlation times are consistent with ECAD12 monomers and dimers (Häussinger et al, 2002), and we interpret the observed transition as the strand exchange homoassociation reaction in solution.
An off-rate koff of 0.7 s−1 for this process was derived from chemical exchange experiments under calcium-saturated conditions (data not shown). This off-rate is in very reasonable agreement with a value of 0.5 s−1 reported from biophysical flow chamber experiments on ECAD12 fragments immobilized onto glass beads (Perret et al, 2002). Clearly, the derived on-rate kon=koff/KD=0.9 × 103 M−1 s−1 is not consistent with diffusion-limited association with expected on-rates kondiff of about 108 M−1 s−1. Therefore, the dimerization kinetics are strongly hindered by an activation barrier on the order of RT ln(kondiff/kon)=7 kcal/mol.
In the absence of calcium, a similar concentration dependence and very similar spectral shifts are observed (Figure 6C and D). However, the affinity between monomers is strongly reduced (KD=10 mM; Figure 6E, open circles) and the monomolecular species shows several subconformations. Thus, it is clearly evident that calcium strongly shifts the equilibrium toward the aggregated form and also has a stabilizing effect on the structure of the ECAD12 monomers. Lateral homoassociation between several cadherin constructs in the absence of calcium has been reported based on chemical crosslinking and immunoprecipitation assays (Brieher et al, 1996; Chitaev and Troyanovsky, 1998; Troyanovsky et al, 1999), whereas no aggregation could be observed for the M-ECAD12 construct for concentrations of up to several millimolars (Häussinger et al, 2002).
Crystal structure of ECAD12 showing intermolecular strand exchange
The structure of ECAD12 was also characterized by X-ray diffraction. For this, ECAD12 was crystallized in the presence of 2 mM calcium and the structure determined (PDB code 1q1p) to a resolution of 3.2 Å by molecular replacement using the M-ECAD12 structure (PDB code 1ff5) as a search model (Table I and Supplementary data). Besides a small change (<2°) in the relative orientation of domains CAD1 and CAD2, the M-ECAD12 (Figure 7) and the ECAD12 (Figure 7) structures are virtually superimposable for residues I7–I214 with backbone heavy atom r.m.s.d.'s of 0.6 Å for residues within the single domains CAD1 and CAD2.
Table 1.
X-ray data collection and refinement parameters
| Cell dimensions | |
| a, b, and c (Å) | 129.2, 48.6, 58.66 |
| β (deg) | 113.93 |
| Space group | C2 |
| Data collection statisticsa | |
| Resolution (Å) | 37.5–3.2 (3.37–3.2) |
| Observed reflections | 45.278 |
| Unique reflections | 5.632 |
| Completeness | 99.8 (97.2) |
| Rsym b | 13.1 (31.3) |
| 〈I/σI〉 | 8.3 (3.4) |
| Refinement statistics | |
| Rfactor c (%) | 25.87 (29.8) |
| Rfree (%) | 30.48 (37.6) |
| Mean B factor (Å2) | 22.2 |
| Bonds (Å)d | 0.012 |
| Angle (deg)d | 1.88 |
| Ramachandran plote |
75/22.3/2.2/0.5 |
| aFinal shell in parentheses. | |
| bRsym=∑∣I−〈I〉∣/∑I. | |
| cRfactor=∑∣∣Fobs∣−∣Fcalc∣∣/∑∣Fobs∣. | |
| dRoot-mean-square error. | |
| ePercentage of residues in most favored/additionally allowed/generously allowed/disallowed region. | |
Figure 7.
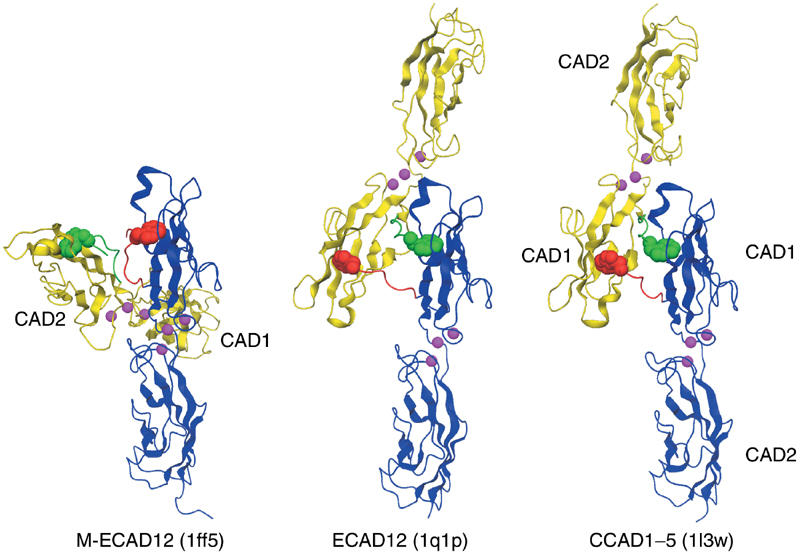
Comparison of the ECAD12, M-ECAD12, and CCAD1–5 crystal structures. The interacting monomers are shown in blue and yellow with their respective N-termini in red and green. The W2 residue is shown as space-fill. For better comparison, the blue CAD12 domains are shown in the same orientation for all three structures. Calcium atoms are indicated in magenta.
In contrast, remarkable differences between the two structures are found for N-terminal residues D1 to P6 as well as in the crystal contacts (Figure 7). M-ECAD12 contains two molecules in the asymmetric unit, which interact mainly at the calcium-binding region between the two domains to form an intertwisted X-shaped dimer. A very similar dimer arrangement was observed in the crystal structure of MR-ECAD12 (PDB code 1edh, Nagar et al, 1996); however, N-terminal residues MR-D1W2 were disordered. In the M-ECAD12 structure (1ff5), the side-chain of residue W2 is inserted into the W2 binding pocket on the same molecule. Conversely, the ECAD12 asymmetric unit contains a single molecule, which exchanges the first β-strand with a symmetry equivalent molecule. Thus, the side-chain of W2 is inserted into the pocket of the neighboring molecule such that an antiparallel L-shaped dimer is formed. Overall, this conformation of strand exchange, relative orientation of monomers, and crystal contacts is very similar to the previously reported structure of the five extracellular domains of C-cadherin (Figure 7) (Boggon et al, 2002).
Discussion
NMR spectroscopic evidence for activation of ECAD12 by N-terminal processing
The present study on ECAD12 shows how the cleavage of an N-terminal HisXa-tag that mimics the native prosequence in ECAD triggers pronounced structural changes in the CAD1 domain and thereby activates homoassociation. Chemical shifts combined with NOE and relaxation data lead to the conclusion that HisXa-ECAD12 and M-ECAD12 adopt very similar structures in solution. In particular, the absence of NOEs between W2Hɛ and D90HN/M92HN (Figure 3A and B) must be interpreted in terms of a flexible N-terminus, where the W2 side-chain is not inserted firmly into the hydrophobic pocket of CAD1 in solution. This is in contrast to the crystal structure of M-ECAD12 (Pertz et al, 1999), where the W2 side-chain is inserted into the pocket of its own CAD1 domain. However, even in this crystal form, the insertion is not very tight, since co-crystallization with soluble tryptophan leads to the release of the W2 side-chain from the pocket and to the replacement by the added tryptophan derivative (Pertz et al, 1999). Thus, the difference between the soluble and the crystal conformations may be caused by crystal packing forces, which weakly favor the more compact inserted form of the W2 side-chain.
Removal of the HisXa-tag (Figure 2) results in the firm docking of the W2 side-chain as evidenced by the changes in the NOE pattern (Figure 3C and D) and the characteristic downfield shift of W2ɛ, indicating the formation of a strong H-bond to D90O. This process is accompanied by large chemical shift changes for the residues that line the hydrophobic pocket. We interpret these changes as a conformational rearrangement that optimizes W2 binding. Only for ECAD12, but not for HisXa-ECAD12, a protein concentration-dependent equilibrium is observed between a monomeric species, where W2 binds to the intramolecular pocket, and a spectrally very distinct associated form, which will be discussed further below.
Correlated with the intramolecular W2 side-chain insertion in solution, a strong intramolecular H-bond between V3HN and K25O is formed. No such H-bond is present in the solution forms of HisXa-ECAD12 and M-ECAD12 (Figure 3A and B) or in the crystal form of M-ECAD12 (Figure 4B). However, this H-bond is observed as an intramolecular H-bond in the ECAD1–internalin complex (Schubert et al, 2002) and as an intermolecular H-bond between V3HN and K25O′ in all crystal structures that show strand exchange, that is, NCAD1 (Shapiro et al, 1995), CCAD1–5 (Boggon et al, 2002), and ECAD12 (Figure 4A). This suggests an interdependence between the proper insertion of W2 and the formation of the V3HN–K25O H-bond in the intramolecular as well as in the intermolecular strand exchange form. Clearly, the absence of the intermolecular V3–K25 H-bond would strongly reduce the stability of the strand exchange dimer.
The crystal structure of M-ECAD12 (Figure 4B) shows that its kinked N-terminus pushes strands βA and βB apart such that the H-bond formation between V3 and K25 is sterically hindered. However, it is unclear as to why D1 and M-1 in M-ECAD12 do not adopt a similar conformation as in the extended N-terminus of ECAD1–internalin (Figure 4C). No sterical clashes are apparent for such a conformation (data not shown). Therefore, in addition to steric effects, also hydrophobic or electrostatic interactions may account for the observed differences in conformation.
Our data show that prevention of the V3HN/K25O H-bond and inhibition of the strand exchange dimerization take place simultaneously and in consequence lead to an inactive form of ECAD. Even a single additional methionine in M-ECAD12 can lead to the same effect as the comparatively large His10Xa-tag. This is in complete agreement with the observations of Ozawa and Kemler (1990), who reported no adhesive activity of ECAD after incorrect cleavage of the prosequence. The present study provides a structural rationale for these findings. We assume that through this mechanism, the prosequence in native ECAD prevents adhesive function in nonprocessed ECAD, thus allowing post-translational modification and incorporation into the membrane without aggregation.
Equilibrium between monomeric and associated form of ECAD12 in solution
The V3HN/K25O H-bond is of intermolecular nature for the CAD1 domains in the crystal structures of NCAD1 (1ncg), CCAD1–5 (1l3w) as well as ECAD12 (1q1p) and therefore connects the two molecules forming the dimeric unit. For the low-concentration form of ECAD12 in solution, 15N-selected/15N-edited NOE experiments (Figure 5) prove that this H-bond is intramolecular. This fact, together with the relaxation data, allows the assignment of the low-concentration form of ECAD12 to a monomeric species. As evidenced by NOE connectivities, the H-bond pattern in the monomer must be very similar to the one observed in the monomeric ECAD1–internalin complex (Schubert et al, 2002) (Figure 4C, 1o6s). Molecular modelling of ECAD12 (data not shown) demonstrates that a virtually identical conformation can be adopted by ECAD12 without any steric clashes or violation of Ramachandran boundaries. The conformation of ECAD1 in the ECAD1–internalin complex can therefore be considered as a model for the correlation between V3HN/K25O H-bond formation and the proper insertion of the W2 side-chain. It should be pointed out, however, that further analogies to E-cadherin homoassociation are limited. First, in the tight complex, internalin covers the ECAD1 N-terminus and thus probably masks true terminal effects, such as the presence of the four non-native N-terminal residues GPLG and the replacement of the conserved N-terminal residue D1 by serine. Second, the tight complex formation with internalin prevents the formation of a strand exchange homodimer and also has a considerable influence on other parts of the ECAD1 structure (Schubert et al, 2002).
At high protein concentrations, an associated form of ECAD12 is the dominant species in solution. Based on relaxation data and chemical shift changes, which are strongest for W2 and for residues in the region of the W2 binding pocket, we suggest that this associated form adopts a strand exchange dimer conformation. However, higher aggregated forms cannot be ruled out completely. Due to the short relaxation times of the associated form of ECAD12, direct evidence for a strand exchange dimer or other aggregated forms could not be obtained from NOE data. Nevertheless, the strand exchange dimer arrangement found in the crystal structure for ECAD12 (Figure 7A) strongly supports the assumption of a similar dimeric form in solution.
Calcium dependence, thermodynamics, and kinetics of the equilibrium
The equilibrium between the monomeric and associated forms is strongly dependent on calcium concentration (Figure 6), and KD values of 0.7 and 10 mM were determined in the presence and absence of calcium, respectively. Obviously, calcium is necessary for the adhesiveness, which can be understood in terms of a rigidification of the calcium-binding hinge around E11, which connects the βA-strand to the β-barrel. This is corroborated by the observation of several subconformations for monomeric ECAD12 in the absence of calcium (Figure 6C and D), whereas in the presence of calcium only a single monomeric conformer is visible (Figure 6A and B).
The apparently weak affinities derived from the KD values are notable as they coincide with the observation of separate resonances for the monomeric and associated form of ECAD12, which indicates slow, that is, longer than millisecond, chemical exchange between the two species. This can only be understood if the formation of the strand exchange dimer is not a diffusion-controlled process, but instead is kinetically hindered by a considerable activation barrier. Following the dynamics of the process by exchange spectroscopy yields an off-rate koff of 0.7 s−1 in the presence of calcium, from which we estimate that the height of the activation barrier is approximately 7 kcal/mol. An activation energy of this size is very plausible considering that during the N-terminal β-strand exchange, a sizeable hydrophobic surface must be exposed to water and that a number of hydrogen bonds must be broken and re-established.
Homoassociation KD values in the presence of calcium in the range of 0.08–0.17 mM have been obtained by analytical ultracentrifugation for MR-ECAD12 and M-ECAD12 (Alattia et al, 1997; Koch et al, 1997), which both had non-native N-termini. Clearly, these values differ from our result of 0.7 mM obtained for the properly processed ECAD12. Also in marked contrast is the intermediate to fast chemical exchange on the NMR chemical shift time scale, that is, koff>103 s−1, observed earlier for homoassociation of M-ECAD12 (Häussinger et al, 2002) and the slow exchange, that is, koff=0.7 s−1, found for ECAD12. We ascribe these differences to the different association mechanisms for cadherins with or without foreign residues at the N-terminus. The fast exchange for M-ECAD12 is consistent with a purely diffusion-controlled process with no or only a negligible activation barrier (Häussinger et al, 2002). However, the slow exchange and the weak association constant for ECAD12 yields a kon rate of 0.9 × 103 M−1 s−1, which is much smaller than expected for purely diffusion-controlled processes and indicates the kinetic trapping by a substantial activation barrier. This is understandable, since strand exchange in ECAD12 requires substantial structural rearrangement, whereas the weak steric contacts in the M-ECAD12 or MR-ECAD12 dimer interface can be achieved by a simple diffusion-controlled association.
As the interaction of W2 with its binding pocket and the β-sheet connectivities of the first β-strand are very similar in the monomeric and dimeric forms of ECAD12, the energetic difference between these two forms is likely to be very small. Indeed, it was pointed out in a recent review (Liu and Eisenberg, 2002) that such free energy differences between monomers and domain-swapped dimers are small in general, because they result from only minor interactions in the hinge regions, release of strain, and entropic effects. In contrast, activation barriers for the swapping are usually high. This completely coincides with our finding for ECAD12, which has a weak affinity for dimerization, that is, low ΔG, but a high activation barrier, that is, ΔG‡.
Cis–trans interactions in cadherin homoassociation
Although proteolytic activation of E-cadherin is the main focus of the present work, the widely accepted concept of cis and trans interactions of cadherins is briefly discussed. The current model of cadherin-mediated cell adhesion proposes initial dimerization of cadherin ectodomains on the cell surface (cis) as a prerequisite of subsequent adhesive interaction involving cadherin dimers on opposing cells (trans). A large number of publications describe the existence and importance of cis and trans interactions for cadherin function (Brieher et al, 1996; Tomschy et al, 1996; Pertz et al, 1999; Takeda et al, 1999; Shan et al, 2000; He et al, 2003). The salient features of these investigations are the electron microscopic detection of four cadherin strands meeting with their N-terminal ends, the observation that dimer formation precedes tetramer formation, and the fact that dimerization at the cell surface is a prerequisite for the adhesive contact. Despite these findings, a definitive description of cis and trans interaction surfaces, which may also explain the specificity of the cadherin homoassociation, is still lacking.
In the NMR experiments, the transition from the low to the high concentration form of ECAD12 causes strong chemical shift changes that are close to W2 or line the W2 binding pocket of CAD1. We interpret these data as the strand exchange homoassociation reaction in solution. It should be emphasized that inherent to the solution NMR experiments, there is no distinction between molecules originating from the same or from different cell surfaces. Therefore, it is not possible to characterize the observed interaction surface as either a cis or a trans contact based on the NMR experiments.
The new crystal structure of ECAD12 very much resembles the crystal structure of CCAD1–5 (Boggon et al, 2002). In the latter work, the intermolecular strand exchange was primarily interpreted as the adhesive trans interface; however, due to a certain plasticity of the strand dimer, its role as a cis interaction was also discussed. The CCAD1–5 structure contains a second, cis-oriented crystal contact between the face of CAD1 that is distal to W2 and CAD2. Although its location suggests an involvement in the cis interaction, no physiological significance for this CAD1/CAD2 contact has been proven so far.
An identical cis-oriented contact is also observed in the present crystal structure of ECAD12 (1q1p), in M-ECAD12, and MR-ECAD12 (Boggon et al, 2002). Albeit concentration-dependent chemical shift changes were found in the region of this CAD1/CAD2 contact for M-ECAD12 (Häussinger et al, 2002), these changes were much smaller than the ones observed for the strand exchange reaction in the present study of ECAD12. In contrast, concentration-dependent chemical shift changes in the region of the cis-oriented crystal CAD1/CAD2 contact were considerably weaker for ECAD12 than for M-ECAD12 for protein concentrations of up to 2 mM (data not shown). Thus, the NMR data on ECAD12 do not provide evidence for a strong association via this CAD1/CAD2 contact.
Clearly, the extended strand exchange dimer interfaces in the crystal structures of ECAD12 (1q1p), CCAD1–5 (1l3w), and NCAD1 (1ncg) differ substantially from the M-ECAD12 (1ff5) and MR-ECAD12 (1edh) structures, in which cross-like structures form by weak contacts in the calcium-binding region. The latter arrangement had been discussed previously in the context of a potential cis interaction (Pertz et al, 1999). However, in the new ECAD12 crystal structure without foreign N-terminus, no such interaction is observed. Thus the weak contact in the calcium-binding region of M-ECAD12 and MR-ECAD12 may be a crystal artifact.
However, the question on the nature of the cis contact region remains open. Cis, that is, parallel, dimer interactions were clearly observed in the presence of calcium in electron microscopic studies of chimeric ECAD1–5/COMP pentamers that contained the native ECAD1 N-termini (Tomschy et al, 1996; Pertz et al, 1999). The formation of such contacts preceded the formation of antiparallel (trans) tetramers. An interesting new explanation for such cis and trans interactions has been proposed in a very recent electron tomography study on desmosomal cadherins (He et al, 2003) where the observed cis and trans contacts between the CAD1 domains were both interpreted as the result of strand exchange. Nonetheless, an experimental proof for this suggestion is missing.
In summary, this study presents evidence for major structural changes that are induced by N-terminal processing of E-cadherin. A properly processed N-terminus is a crucial prerequisite for the adhesive activity of E-cadherin and likely also of other prosequence-containing cadherins. The presence of additional amino acids or even of one single methionine residue can prevent proper insertion of W2 into intra- and intermolecular binding pockets. A similar obstruction is expected from the N-terminal prodomain, which prevents cadherin aggregation during post-translational modification and transport in the Golgi apparatus. The release of the native N-terminus by cleavage of the preceding polypeptide chain leads to a major reorganization of the CAD1 domain with optimized docking and fixation of the W2 side-chain. Only under these conditions, β-strand exchange dimerization is possible in solution and in the crystalline state. The close similarity of strand exchange and relative monomer orientations observed in the crystalline arrangements of epithelial, neuronal, and C-cadherins suggests that this dimer forms the essential functional part of the cadherin adhesion complex. Correct proteolytic maturation is a necessary condition for the formation of this strand exchange dimer. This explains the stringent connection between N-terminal processing and adhesive activity at the structural level.
Materials and methods
Protein expression and purification
The DNA fragment containing the coding sequence of the 219 N-terminal amino-acid residues of ECAD preceded by the His-tag/factor Xa cleavage site (MGH10SSGHID4KHMIEGR) was obtained by PCR using a cDNA of murine E-cadherin (Häussinger et al, 2002) and the oligonucleotides 5′ ATA CTT TCA TAT GAT CGA AGG TCG TGA CTG GGT CAT CCC 3′ and 5′ GTG GTC TCG AGT TAT TAA GGA GCG TTG TCA TTA ATA TCC 3′. The fragment was cloned into NdeI/XhoI restriction sites of the pET-19b vector (Novagen). Uniformly (>98%) 15N- or 15N/13C-labelled, deuterated (>85% 2H, for nonexchanging hydrogen nuclei) protein was expressed in Escherichia coli strain BL21 (DE3) as described (Häussinger et al, 2002). Purification was carried out by Ni-affinity chromatography under denaturing conditions (8 M urea). The protein was then refolded on the column using 5 mM Tris–HCl, pH 7.9. Factor Xa (Quiagen) digestion (40 h at room temperature, 20 mM Tris–HCl, pH 6.5, 10 mM NaCl, 1 mM EDTA, 15 mg/ml protein, and 100 U/ml factor Xa) resulted in the formation of the mature N-terminus (DWVIP) of ECAD12 as confirmed by Edman degradation. A further Ni-affinity chromatography step to remove the cleaved-off His-tag was followed by dialysis (20 mM Tris–HCl, 10 mM NaCl, and 1 mM EDTA, pH 7.8) in order to remove traces of nickel. The final product was kept under reducing conditions with TCEP (1 mM, pH 7.8).
NMR samples of the various ECAD12 constructs were prepared in the presence of 10 mM DTT, 10 mM NaCl, 0.02% NaN3, and 5 mM Tris–HCl (pH 7.8) as 280 μl volumes in Shigemi microtubes. Calcium-free or calcium-saturated conditions were achieved by dialysis with such buffer solutions containing in addition either 1 mM EDTA or 1 mM CaCl2, respectively.
NMR experiments
All NMR experiments were performed at 25°C on Bruker DRX-600 or DRX-800 spectrometers equipped with triple-axis pulsed field gradient 1H/15N/13C probeheads optimized for proton detection as described previously for M-ECAD12 (Häussinger et al, 2002). NMR data were processed and analyzed using the NMRPipe package (Delaglio et al, 1995) and the program PIPP (Garrett et al, 1991), respectively.
Crystallization and X-ray structure determination
Crystallization was carried out at room temperature by the vapor diffusion method. Hanging droplets were made by mixing 2 μl of ECAD12 solution (12 mg/ml) with 0.2 M ammonium chloride, 0.2 M Tris–HCl (pH 7.6), and 17% PEG 8000. The crystals belong to space group C2 and contain one molecule in the asymmetric unit (VM=3.4 Å3/Da). The crystals were cryoprotected by 20% (v/v) glycerol and flash frozen in liquid nitrogen.
An X-ray diffraction data set was collected at 100 K with monochromatized CuKα radiation (λ=1.5418 Å) from an Elliot GX-20 rotating anode on a MAR imaging plate detector. Diffraction images were processed using the program suite MOSFLM (Leslie, 1994), and the structure factors were scaled and reduced using SCALA from the CCP4 package (CCP4, 1994).
Molecular replacement was performed using the AMORE program of the CCP4 package with a poly-serine model of M-ECAD12 (PDB code 1ff5) as a search template. The structure was refined with REFMAC using a TLS refinement protocol (CCP4, 1994). A total of 10% of the reflections were excluded for use in a crossvalidation set. Refinement with REFMAC was alternated with manual electron density refitting of side-chains and terminal regions using MAIN (Turk, 1992). The final model contains amino-acid residues W2-D213, three calcium atoms coordinated at the interface between both subunits, and 234 water molecules. The final structure was analyzed with PROCHECK (Laskowsky et al, 1993), and statistics of the data set and the refinement are summarized in Table I.
Supplementary Material
SUPPLEMENTARY MATERIAL
Acknowledgments
We thank Therese Schulthess and Marco Rogowski for their expertise and help in growing and purifying ECAD12, and Hans-Jürgen Sass for help with structure calculations and valuable discussions. We are also grateful to Alexander W Koch, McGill University, Montreal, for sharing data prior to publication. This work was supported by SNF grant 31-61′757.00 to SG and by SNF grant 31-67′206.01 to JE.
References
- Alattia JR, Ames JB, Porumb T, Tong KI, Heng YM, Ottensmeyer P, Kay CM, Ikura M (1997) Lateral self-assembly of E-cadherin directed by cooperative calcium binding. FEBS Lett 417: 405–408 [DOI] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L (2002) C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science 296: 1308–1313 [DOI] [PubMed] [Google Scholar]
- Brieher WM, Yap AS, Gumbiner BM (1996) Lateral dimerization is required for the homophilic binding activity of C-cadherin. J Cell Biol 135: 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) Collaborative Computing Project No. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chitaev NA, Troyanovsky SM (1998) Adhesive but not lateral E-cadherin complexes require calcium and catenins for their formation. J Cell Biol 142: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) nmrPipe—a multidimensional spectral processing system based on Unix pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Garrett DS, Powers S, Gronenborn AM, Clore GM (1991) A common-sense approach to peak picking in 2-dimensional, 3-dimensional, and 4-dimensional spectra using automatic computer-analysis of contour diagrams. J Magn Reson 95: 214–220 [DOI] [PubMed] [Google Scholar]
- Gumbiner BM (1996) Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84: 345–357 [DOI] [PubMed] [Google Scholar]
- Gumbiner BM (2000) Regulation of cadherin adhesive activity. J Cell Biol 148: 399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häussinger D, Ahrens T, Sass HJ, Pertz O, Engel J, Grzesiek S (2002) Calcium-dependent homoassociation of E-cadherin by NMR spectroscopy: changes in mobility, conformation and mapping of contact regions. J Mol Biol 324: 823–839 [DOI] [PubMed] [Google Scholar]
- He W, Cowin P, Stokes DL (2003) Untangling desmosomal knots with electron tomography. Science 302: 109–113 [DOI] [PubMed] [Google Scholar]
- Koch AW, Bozic D, Pertz O, Engel J (1999) Homophilic adhesion by cadherins. Curr Opin Struct Biol 9: 275–281 [DOI] [PubMed] [Google Scholar]
- Koch AW, Pokutta S, Lustig A, Engel J (1997) Calcium binding and homoassociation of E-cadherin domains. Biochemistry 36: 7697–7705 [DOI] [PubMed] [Google Scholar]
- Laskowsky RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Leslie AGW (1994) MOSFLM Users Guide, MRC-LMB, Cambridge [Google Scholar]
- Liu Y, Eisenberg D (2002) 3D domain swapping: as domains continue to swap. Protein Sci 11: 1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar B, Overduin M, Ikura M, Rini JM (1996) Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature 380: 360–364 [DOI] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F (2000) Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol 299: 551–572 [DOI] [PubMed] [Google Scholar]
- Overduin M, Harvey TS, Bagby S, Tong KI, Yau P, Takeichi M, Ikura M (1995) Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science 267: 386–389 [DOI] [PubMed] [Google Scholar]
- Ozawa M (2002) Lateral dimerization of the E-cadherin extracellular domain is necessary but not sufficient for adhesive activity. J Biol Chem 277: 19600–19608 [DOI] [PubMed] [Google Scholar]
- Ozawa M, Kemler R (1990) Correct proteolytic cleavage is required for the cell adhesive function of uvomorulin. J Cell Biol 111: 1645–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret E, Benoliel AM, Nassoy P, Pierres A, Delmas V, Thiery JP, Bongrand P, Feracci H (2002) Fast dissociation kinetics between individual E-cadherin fragments revealed by flow chamber analysis. EMBO J 21: 2537–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O, Bozic D, Koch AW, Fauser C, Brancaccio A, Engel J (1999) A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J 18: 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert WD, Urbanke C, Ziehm T, Beier V, Machner MP, Domann E, Wehland J, Chakraborty T, Heinz DW (2002) Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 111: 825–836 [DOI] [PubMed] [Google Scholar]
- Shan WS, Tanaka H, Phillips GR, Arndt K, Yoshida M, Colman DR, Shapiro L (2000) Functional cis-heterodimers of N- and R-cadherins. J Cell Biol 148: 579–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, Colman DR (1998) Structural biology of cadherins in the nervous system. Curr Opin Neurobiol 8: 593–599 [DOI] [PubMed] [Google Scholar]
- Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grubel G, Legrand JF, Als-Nielsen J, Colman DR, Hendrickson WA (1995) Structural basis of cell–cell adhesion by cadherins. Nature 374: 327–337 [DOI] [PubMed] [Google Scholar]
- Takeda H, Shimoyama Y, Nagafuchi A, Hirohashi S (1999) E-cadherin functions as a cis-dimer at the cell–cell adhesive interface in vivo. Nat Struct Biol 6: 310–312 [DOI] [PubMed] [Google Scholar]
- Takeichi M (1995) Morphogenetic roles of classic cadherins. Curr Opin Cell Biol 7: 619–627 [DOI] [PubMed] [Google Scholar]
- Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L (1998) Structure–function analysis of cell adhesion by neural (N-) cadherin. Neuron 20: 1153–1163 [DOI] [PubMed] [Google Scholar]
- Tomschy A, Fauser C, Landwehr R, Engel J (1996) Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO J 15: 3507–3514 [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky RB, Klingelhofer J, Troyanovsky S (1999) Removal of calcium ions triggers a novel type of intercadherin interaction. J Cell Sci 112: 4379–4387 [DOI] [PubMed] [Google Scholar]
- Turk DC (1992), PhD thesis, Technische Universität München Munich, Germany
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIAL


