Abstract
The human fungal pathogen Candida albicans switches from yeast to hyphal growth when exposed to serum or phagocytosed. However, the importance of this morphological switch for virulence remains highly controversial due to the lack of a mutant that affects hyphal morphogenesis only. Although many genes specifically expressed in hyphal cells have been identified and shown to encode virulence factors, none is required for hyphal morphogenesis. Here we report the first hypha-specific gene identified, HGC1, which is essential for hyphal morphogenesis. Deletion of HGC1 abolished hyphal growth in all laboratory conditions tested and in the kidneys of systemically infected mice with markedly reduced virulence. HGC1 expression is co-regulated with other virulence genes such as HWP1 by the cAMP/protein kinase A signaling pathway and transcriptional repressor Tup1/Nrg1. Hgc1 is a G1 cyclin-related protein and co-precipitated with the cyclin-dependent kinase (Cdk) CaCdc28. It has recently emerged that cyclin/Cdk complexes promote other forms of polarized cell growth such as tumor cell migration and neurite outgrowth. C. albicans seems to have adapted a conserved strategy to control specifically hyphal morphogenesis.
Keywords: C. albicans, Cdc28, hyphal morphogenesis, polarized growth, virulence
Introduction
Candida albicans (Ca) is one of the most important fungal pathogens of humans (Odds, 1988; Berman and Sudbery, 2002). It may cause serious infections when the host is immunocompromised by factors such as HIV infection and anticancer and immunosuppressive therapies. If the pathogen enters the blood stream, it may colonize internal organs leading to the death of patients. Understanding its biology, particularly the aspects that directly contribute to infection and virulence, is necessary for developing antifungal therapies.
Many microbial pathogens undergo morphological changes during infection, which may assist the pathogens to exploit and adapt to certain host environments and enhance their survival and infectivity (Lengeler et al, 2000; Gow et al, 2002). C. albicans yeast cells switch to hyphal growth when phagocytosed or exposed to host serum, which is thought to facilitate its invasion of host tissues and escape from phagocytotic destruction (Odds, 1988; Lo et al, 1997). Several signaling pathways mediate hyphal morphogenesis, among which the cAMP/protein kinase A (PKA) and mitogen-activated protein (MAP) kinase pathways are best defined (Liu et al, 1994; Lo et al, 1997; Liu, 2001). The cAMP/PKA and MAP kinase pathways target two transcription factors Efg1 and Cph1, respectively (Lo et al, 1997; Stoldt et al, 1997). The former seems to play a major role, because its inactivation blocks hyphal growth in most conditions. On the other hand, a transcriptional inhibitor Tup1 associates with DNA-binding protein Nrg1 or Rfg1 to repress the hyphal program in yeast cells (Braun and Johnson, 1997, 2000; Braun et al, 2001; Kadosh and Johnson, 2001; Murad et al, 2001b; Saville et al, 2003). TUP1 deletion causes constitutive filamentous growth (Braun and Johnson, 1997). Hypha-inducing conditions activate the expression of hypha-specific genes (HSGs), many of which are also derepressed in tup1Δ mutant (Braun and Johnson, 2000; Murad et al, 2001c). Although many HSGs identified so far encode virulence traits, surprisingly none seems to be required for hyphal morphogenesis (Berman and Sudbery, 2002; Gow et al, 2002).
A knowledge gap exists between the functions of the known HSGs and the machineries that control and execute morphogenesis. However, studies of Saccharomyces cerevisiae (Sc) morphogenesis may provide some hints. A currently predominant model is that cell cycle machinery controls morphogenesis in the budding yeast (Lew and Reed, 1993; Rua et al, 2001). The G1 cyclins, Cln1 and 2, in association with the Cdk Cdc28, promote apical bud elongation, whereas Clb2-Cdc28 kinase triggers a switch to isotropic bud expansion. A premature or delayed activation of this switch results in round or elongated morphology, respectively. In support of this, a delay in the G2/M transition was observed during pseudohyphal growth that produces chains of elongated cells (Kron et al, 1994; Rua et al, 2001).
Experiments have been carried out to assess the possibility that the cyclin/Cdks for cell cycle control may have a role in regulating C. albicans hyphal growth. Deletion of CaFKH2, a homolog of the transcription factors FKH1,2 that regulate CLB2 transcription in S. cerevisiae, caused pseudohyphal growth (Bensen et al, 2002). A similar phenotype was reported for mutants deleted of CaCDC5 (Bachewich et al, 2003) or CaMCM1 (Rottmann et al, 2003), genes having roles in cell cycle control. Cell cycle toxins, such as hydroxyurea and nocodazole, were found to cause cell elongation (Bai et al, 2002; Bachewich et al, 2003). While all these may suggest a possible role of cell cycle control in hyphal morphogenesis, there have also been contradicting observations. For example, deletion of the G1 cyclin gene, CaCLN1, has only limited effect on hyphal growth (Loeb et al, 1999), and no significant differences were detected in cell cycle progression between yeast and hyphal cells (Hazan et al, 2002). This confusion is further compounded by the recent reports that the initiation of hyphal growth could occur at any phase of a cell cycle and some polarity proteins, which normally localize to distinct sites in different cell cycle phases during yeast growth, exhibited cell cycle-independent tip localization during hyphal growth (Hazan and Liu, 2002; Hazan et al, 2002; Zheng et al, 2003). These results immediately challenge any model proposed on the basis of the cyclin/Cdk activities to explain how the periodic cyclin/Cdk activities promote the cell cycle-independent hyphal growth.
Here we report the identification of a novel gene HGC1 that has a specific and critical role in C. albicans hyphal morphogenesis. We describe the characterization of its properties and regulation, its role in controlling hyphal morphogenesis and its importance for virulence. The results provide valuable insights into the molecular mechanisms that control hyphal morphogenesis and help to settle the current debate over the importance of the yeast-to-hypha morphogenesis for virulence.
Results
Identification of a G1 cyclin-related protein in C. albicans
Based on the known roles of G1 cyclins in promoting bud elongation in S. cerevisiae and the need to explain the cell cycle-independent hyphal growth of C. albicans, we hypothesized that a Cln1- or 2-like protein that is activated by the hypha-inducing but not the cell cycle signals might be able to promote the persistent hyphal growth in C. albicans. A previous investigation did not detect significant differential expression of CaCLN1 and 2 between yeast and hyphal cells (Hazan et al, 2002). To explore the possibility that C. albicans may contain other G1 cyclin-related proteins, we used ScCln1 cyclin box sequence to search C. albicans genome database (http://www-sequence.standford. edu/group/candida). We found three significant matches. The highest identity of 47.3% was scored to a 785-amino-acid (aa) protein annotated Cln21 (contig6-2213, orf6.3156), followed by 39.1 and 34.5% to CaCln1 and 2, respectively. In the database, Cln21 is annotated as a G1 cyclin by homology with unknown function. Despite the high sequence identity over the cyclin box, Cln21 shares low overall identity to ScClns (∼20%). We next carried out more detailed sequence comparisons of all the cyclin family proteins found in C. albicans and S. cerevisiae. Like all other G1 cyclins, Cln21 contains one cyclin box near the amino terminus, distinct from the B-type cyclins each containing two cyclin boxes in the carboxyl-terminal half (Figure 1A). The cyclin box of Cln21 shares 28.7–47.3% identities to those of other G1 cyclins, which are significantly higher than the identities to those of the B-type cyclins ranging from 2.4 to 28.2%. Cln21 is also very different from the Pho85 cyclins, Pcls, in both the sizes of the proteins and the cyclin box sequence identities (4.5–12.4%). The amino-acid identities are also very low between the cyclin box of Cln21 and those of CaSrb11 (18%) and ScSbr11 (15.6%). A phylogenetic analysis of the cyclin box sequences revealed five well-separated clusters, with each embracing only the members of each type of cyclins (Figure 1B). Cln21 is enclosed in the cluster containing all other G1 cyclins. Taken together, the results of the sequence comparisons strongly suggest that Cln21 is a G1 cyclin-related protein.
Figure 1.

Relationship of Cln21 with other cyclin family proteins in S. cerevisiae and C. albicans. (A) Localization and sequence relatedness of the cyclin box sequences. Cyclin boxes are identified using the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de) and are shown in black. The percentage values above each box were scored against Cln21. All the sequences were retrieved from the S. cerevisiae genome database (http://www.yeastgenome.org). (B) Phylogenetic relatedness of Cln21 to G1 cyclins. The tree was reconstructed using the Clustal W method in the DNASTAR sequence analysis software. (C) Alignment of the cyclin boxes of G1 cyclins. The invariable positions are shaded and other conserved positions are denoted by asterisks.
The cyclin box sequence alignment shows that Cln21 contains a stretch of 37 aa that is not present in CaCln1 and 2 (Figure 1C), which might suggest that Cln21 may have distinct functions. Interestingly, the cyclin boxes of ScCln1 and 2 contain 22 of the 37 aa, and 11 of the 22 are shared by all three sequences.
Hypha-specific expression of CLN21
To determine whether CLN21 expression is regulated in response to hyphal induction, RNA samples were prepared from C. albicans yeast and hyphal cells for Northern blot analysis (Figure 2A). Yeast cells were cultured in YPD at 30°C and hyphal growth was induced in YPD containing 10% serum at 37°C. We did not detect CLN21 expression in the yeast cells despite prolonged exposure of up to 3 days. However, an ∼3.5 kb mRNA, sufficiently large to encode the predicted protein, was clearly detected after 16-h exposure from the hyphal RNA. The level of this mRNA was largely constant from 0.5 to 5 h, suggesting a rapid activation of the gene upon hyphal induction. This mRNA was not detected in the cln21Δ mutant (see below for this mutant) under the same conditions.
Figure 2.
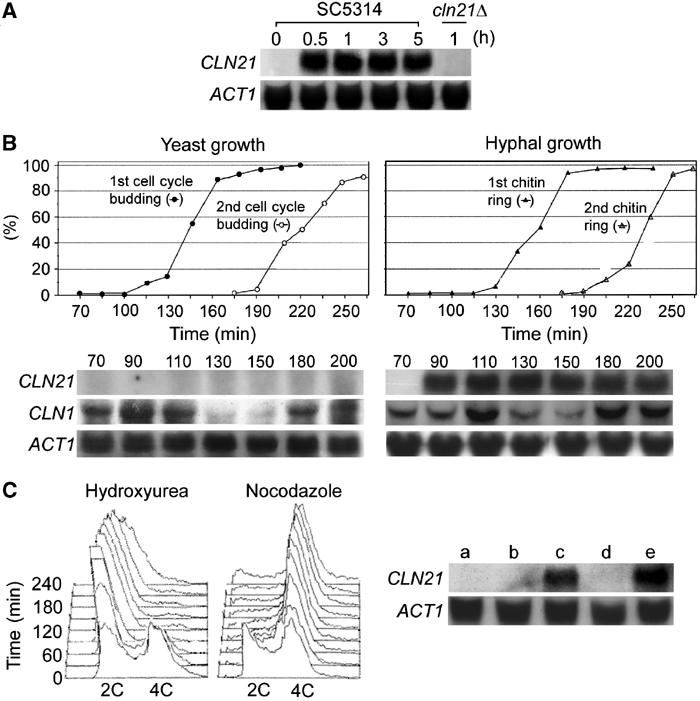
CLN21 expression. (A) Hypha-specific expression of CLN21. Overnight yeast cultures of SC5314 and cln21Δ were inoculated into fresh YPD at 2 × 106 cells/ml and grown at 30°C for 2 h before RNA preparation. For hyphal RNA analysis, overnight yeast cells were inoculated into YPD+10% serum at 2 × 105 cells/ml and grown at 37°C for the time indicated on top. (B) CLN21 expression in synchronous yeast (left) and hyphal (right) cells. SC5314 G1 yeast cells were prepared by centrifugal elutriation and released into YPD at 30°C at ∼2 × 105 cells/ml. Aliquots were collected every 15 min to score the percentage of budded cells (n=150). Guided by the budding index, aliquots of similar yeast cultures were harvested every 20 min from 70 to 200 min for RNA preparation. To obtain a synchronous hyphal culture, 10% serum was added to the synchronous yeast culture at the 70-min time point as described above and the culture was incubated at 37°C. Samples were taken every 15 min for Calcofluor staining to score the percentage of hyphal cells (n=150) containing the first and second chitin rings. For Northern analysis, samples were taken every 20 min between 70 and 200 min. (C) CLN21 expression in S and G2/M yeast cells. Hydroxyurea or nocodazole was added to a mid-log-phase yeast culture of SC5314 at 200 mM and 50 μM, respectively. Aliquots were harvested for FACS analysis every 30 min (left panel). For Northern blot analysis (right panel), RNA was prepared from the following cells: a, untreated yeast cells; b, yeast cells arrested with hydroxyurea for 2 h in YPD at 30°C; c, 10% serum was added to the arrested cells described in ‘b' for 30 more minutes of growth at 37°C; d, yeast cells treated with nocodazole for 2 h in YPD at 30°C; and e, 10% serum was added to the arrested cells described in ‘d' for 30 more minutes of growth at 37°C.
To assess the possibility that CLN21 might be expressed only transiently at a certain cell cycle stage, we prepared G1 yeast cells by centrifugal elutriation to grow a synchronous yeast culture. A budding index was first determined to approximate cell cycle progression (Figure 2B, left). The buds of the first and second cell cycles started to appear at around 100 and 185 min, respectively. We then collected cells for Northern analysis at 20-min intervals between 70 and 200 min. Again, at no time point was CLN21 mRNA detected. As a control, CaCLN1 mRNA was found to oscillate with cell cycle progression (Figure 2B), exhibiting high levels around the beginning of a cell cycle (90–110 and 180–200 min) and significantly lower levels at 130 and 150 min. To investigate whether CLN21 expression is cell cycle regulated during hyphal growth, the synchronous yeast cultures at 70 min described above were induced for hyphal growth. As described by Hazan et al (2002), the beginning of the first cell cycle was estimated to be around the time of the appearance of the first chitin ring along the germ tubes, and the appearance of the second chitin ring near the hyphal tips would indicate the beginning of the second cell cycle. By determining the percentage of cells with the first and second chitin rings at 15-min intervals (Figure 2B, right), we found that the first and second cell cycles started at ∼115 and 190 min, respectively. We then collected cells every 20 min from 70 to 200 min for RNA preparation. Northern analysis showed that during this period of time CaCLN1 exhibited cell cycle-dependent expression in a similar pattern as in the yeast cells, whereas CLN21 mRNA had reached near peak level at 90 min (20 min after the addition of serum) and remained rather constant throughout the experiment, indicating that CLN21 mRNA level is not regulated in a cell cycle-dependent manner during hyphal growth.
It has been documented that the response of C. albicans yeast cells to hyphal induction is cell cycle independent (Hazan et al, 2002). Although we have shown above that CLN21 expression can be induced in G1 cells, we wanted to examine whether it can also be induced in yeast cells in other cell cycle phases. We obtained S and G2/M cells by treating random yeast cultures with the cell cycle inhibitory drugs hydroxyurea (200 mM) and nocodazole (50 μM), respectively. Fluorescence-activated cell sorting (FACS) analysis showed that the two drugs at the concentrations used arrested the yeast cells in respective phases within 1 h and the arrest persisted for at least 3 more hours (Figure 2C, left), consistent with previous reports (Bai et al, 2002; Bachewich et al, 2003). We then added 10% serum to the yeast cells that had been arrested for 2 h for 30-min hyphal induction at 37°C in the presence of the drugs before RNA preparation. Northern blot analysis (Figure 2C, right) detected CLN21 expression in the arrested cells only after but not before the 30-min exposure to the hypha-inducing condition. Taken together, we conclude that CLN21 expression can be activated in the yeast cells of different cell cycle phases.
hgc1Δ was defective in hyphal growth
To better reflect its hypha-specific expression and relatedness to G1 cyclins, we propose to rename the gene HGC1. Next, we constructed HGC1 mutants to study its cellular functions. The two copies of HGC1 were sequentially replaced with ARG4 and HIS1 in strain BWP17. The heterozygous mutant (HGC1/hgc1Δ∷ARG4) exhibited normal growth rate, morphology and response to serum induction and thus was not studied further, whereas the homozygous mutant (hgc1Δ∷ARG4 hgc1Δ∷HIS1, referred to as hgc1Δ for simplicity throughout the paper) exhibited profound defects in hyphal morphogenesis. To compare HGC1 and hgc1Δ strains in otherwise identical genetic background, a Ura− hgc1Δ strain was transformed either with a CIp10 vector that contains a URA3 gene by integration at the RP10 locus yielding a Ura+ hgc1Δ strain (WYZ12.2) or with the CIp10 plasmid carrying a copy of HGC1 with its own promoter by integration at its original genomic locus generating a Ura+ hgc1Δ∷HGC1 strain (WYZ12.1). Southern blot verification of the mutants and WYZ12.1 is shown in Supplementary Figure 1. WYZ12.1 was found to be phenotypically indistinguishable from the wild-type strain SC5314 in all the experiments described below and thus is used to represent the wild type unless indicated otherwise. We examined the growth of hgc1Δ in a range of hypha-inducing and noninducing conditions. In all the noninducing liquid media used, such as YPD, GMM and Lee's medium (pH 4.0) at 30°C, HGC1 and hgc1Δ strains were similar in morphology (data not shown) and growth rate (Figure 3A). However, hgc1Δ did not form filaments in any of the liquid inducing media used, including serum, Lee's (pH 7.0) and RPMI at 37°C (Figure 3B). A closer examination revealed that hgc1Δ exhibited some limited response to hyphal induction. hgc1Δ G1 yeast cells formed a wide surface protrusion giving the cell a pear-like shape. The protrusion is presumably equivalent to the germ tubes of normal G1 cells, because they appeared at similar time and at random positions. Later, a bud emerged at the tip of the protrusion and grew apically to a length ∼1.5–2 times that of the mother cell, which is markedly shorter and wider than normal hyphal cells. The mother and daughter cells separated after cytokinesis and thus filaments were not formed. The results demonstrate that hgc1Δ is severely impaired in the control of germ tube/hyphal morphology and the maintenance of cell–cell attachment after cytokinesis. However, the results also indicate that hgc1Δ remains responsive to hyphal induction, capable of initiating hyphal growth with a largely normal spatiotemporal pattern. hgc1Δ was also defective in filamentous growth in solid hypha-inducing media, such as Lee's (pH 7.0), Spider's and serum, and when embedded in agar (Figure 3C). The morphology of the colonies and the cells removed from the colonies were examined daily for 1 week and the mutant did not show any hyphal growth at any time. Furthermore, the mutant grew almost exclusively as slightly elongated yeast cells in the kidneys of systemically infected mice in sharp contrast to the HGC1 strain that produced masses of long filaments (Figure 3D). Taken together, these results indicate that HGC1 function is critically required for hyphal growth in all the laboratory conditions tested and in systemically infected mice. Thus, HGC1 is the first HSG identified so far that is critically required for hyphal morphogenesis.
Figure 3.
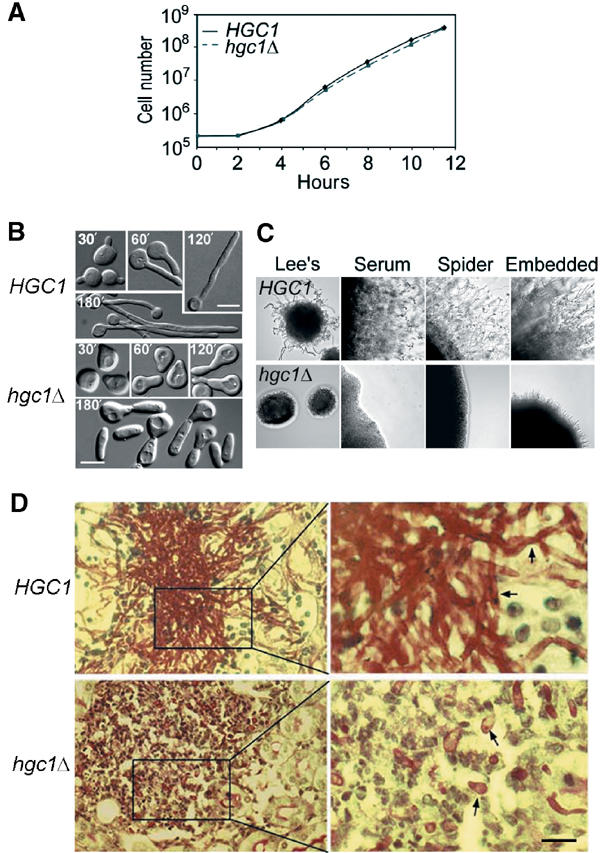
HGC1 is required for hyphal morphogenesis. (A) Growth curves of yeast cells. Strain WYZ12.2 (hgc1Δ) was compared with WYZ12.1 (HGC1). Overnight yeast culture was inoculated into YPD at 2 × 105 cells/ml and grown at 30°C. Cell numbers were counted every 2 h using a hemacytometer. (B) hgc1Δ was defective in hyphal growth in liquid media. Yeast cells of overnight cultures were inoculated into inducing media at 2 × 105 cells/ml and samples were examined at timed intervals. Photos were taken using differential interference phase contrast (DIC). The cells shown were induced in YPD+10% serum at 37°C and are representative of cells induced in GMM+10% serum, Lee's medium (pH 7.0) and RPMI. The scale bars indicate 5 μm throughout the paper. (C) hgc1Δ was defective in hyphal growth on solid media. The media used are shown on top and the cultures were 2 days old. (D) hgc1Δ was defective in hyphal growth in the kidneys of mice. Mice injected with 1 × 106 cells via the tail vein were killed after 2 days and kidneys were removed for histology. The photos show the cortex sections and the arrows denote C. albicans cells.
HGC1 is required for the filamentous phenotype of tup1Δ mutant
TUP1 encodes a transcriptional inhibitor for many HSGs, presumably including those required for hyphal growth (Braun and Johnson, 1997, 2000). Indeed, tup1Δ grows constitutively as branched filaments with highly elongated cells. However, it is not clear whether the mechanism that leads to this filamentous growth is the same as the one underlying normal hyphal growth, partly because information is incomplete on the identities of all the genes activated in the hyphal cells and those derepressed in tup1Δ. To determine whether HGC1 is required for the filamentous phenotype of tup1Δ, we deleted HGC1 together with TUP1. Strikingly, deletion of HGC1 abolished the constitutive cell elongation of tup1Δ and weakened cell–cell attachment after cytokinesis in both liquid and solid media, virtually converting the long branched filaments of tup1Δ to short chains of yeast cells (Figure 4). The growth rates of tup1Δ and tup1Δ hgc1Δ mutants were similar (data not shown) and the nuclear divisions in the double mutant appeared normal, suggesting that the loss of the constitutive filamentous growth in tup1Δ hgc1Δ is unlikely the result of the disruption of cell cycle progression. These results further established the critical role of HGC1 in hyphal morphogenesis and indicate that a common mechanism is responsible for both the normal hyphal growth and the filamentous growth caused by TUP1 deletion.
Figure 4.
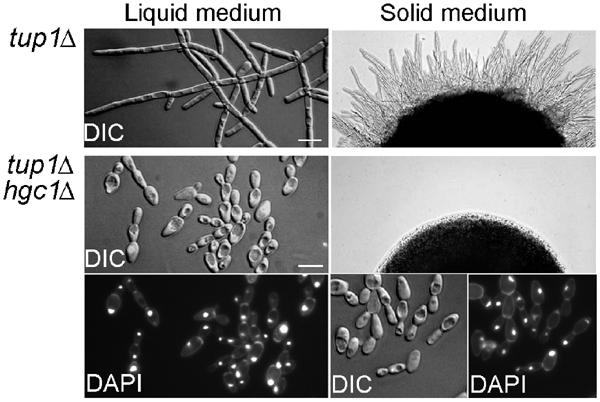
HGC1 is required for the filamentous phenotype of tup1Δ. Strains CaWY6 (tup1Δ) and WYZ15 (tup1Δ hgc1Δ) were grown in liquid GMM (left) at 30°C to mid-log phase or on GMM plates (right) at 30°C for 2 days. Nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI).
HGC1 is not required for the expression of HWP1, HYR1 and ECE1
In S. cerevisiae, a MAP kinase pathway activates the transcription factor Ste12, which in turn controls the expression of genes required for pseudohyphal growth (Liu et al, 1993, 1994). A cyclin/Cdk complex, Srb11/Srb10, is known to modulate Ste12 activity and thereby pseudohyphal growth (Nelson et al, 2003). In C. albicans, CaCLN1 is also needed for the expression of several HSGs in Lee's medium (Loeb et al, 1999). To determine whether HGC1 deletion blocks the signaling pathways that activate the expression of HSGs, we examined the expression of three well-known representative genes, HWP1 (Staab et al, 1999), HYR1 (Bailey et al, 1996) and ECE1 (Birse et al, 1993), in yeast and hyphal cells. We found that the expression patterns of these genes were unaffected in hgc1Δ (Figure 5A), indicating that the signaling pathways that activate their expression remain intact.
Figure 5.
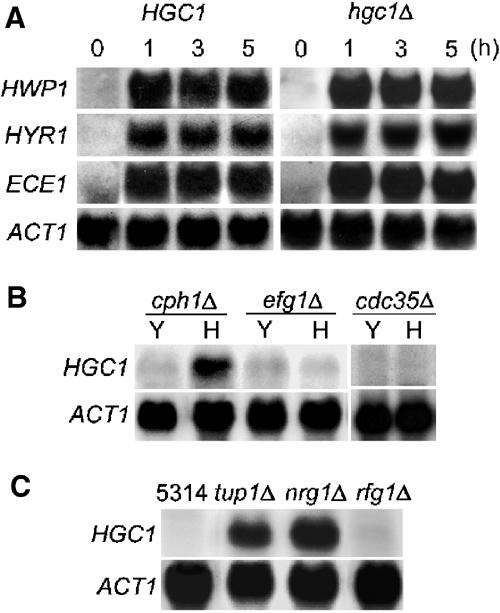
HGC1 expression is regulated by the cAMP/PKA pathway and transcriptional factors Efg1, Tup1 and Nrg1. (A) Deletion of HGC1 did not affect the expression of HWP1, HYR1 and ECE1. HGC1 and hgc1Δ yeast cells were grown in YPD at 30°C and hyphal growth was induced in YPD+10% serum at 37°C for 1, 3 and 5 h before RNA preparation. (B) HGC1 was not expressed in efg1Δ and Cacdc35Δ but induced normally in cph1Δ under inducing conditions. Cells of each strain (indicated on top) were grown in YPD at 30°C (Y) or YPD+10% serum at 37°C for 1 h (H). (C) HGC1 expression was derepressed in tup1Δ and nrg1Δ. All strains were grown in GMM at 30°C to mid-log phase.
HGC1 expression is regulated by the cAMP/PKA pathway and Tup1
The hypha-specific expression of HGC1 suggests that it may be co-regulated with other HSGs by the same signaling pathways. To identify the pathways that regulate HGC1 expression, we examined its expression in cph1Δ and efg1Δ mutants, in which the MAP kinase and cAMP/PKA pathways are blocked respectively (Liu et al, 1994; Lo et al, 1997; Murad et al, 2001c), and in tup1Δ, nrg1Δ and rfg1Δ mutants, in which many HSGs are constitutively expressed (Braun and Johnson, 1997, 2000; Braun et al, 2001; Murad et al, 2001c). Figure 5B shows that HGC1 mRNA was not detected in efg1Δ mutant under inducing conditions but induced normally in cph1Δ, suggesting that HGC1 expression may be activated via the cAMP/PKA pathway. This was confirmed by the absence of induced HGC1 expression in a strain deleted of CaCDC35, the gene encoding the adenylyl cyclase that catalyzes cAMP production (Rocha et al, 2001). Under noninducing conditions, HGC1 expression was readily detected in tup1Δ and nrg1Δ mutants but undetectable in rfg1Δ (Figure 5C), implying that HGC1 is repressed by Tup1 and Nrg1. These results suggest that HGC1 should be positioned at the same level as the typical HSGs, such as HWP1, HYR1 and ECE1, in the entire cascade of events activated by the hypha-inducing signals.
Constitutive overexpression of HGC1 alone is not sufficient to induce hyphal growth
Among the many known HSGs, HGC1 is the only one found to have a critical role in hyphal growth. To assess whether constitutive expression of HGC1 is sufficient to cause hyphal growth, we expressed HGC1 under the control of ACT1 promoter in hgc1Δ and CAI4 strains. HGC1 expression in the transformants was confirmed by Northern analysis (Supplementary Figure 2). Although the construct fully restored the induced hyphal growth in hgc1Δ, both strains grew as yeast under noninducing conditions, such as GMM and YPD at 30 or 37°C (data not shown), indicating that HGC1 expression alone is not sufficient for hyphal growth. We also wanted to assess whether other G1 cyclin genes, when made hypha-inducible or constitutively expressed, are able to suppress the defects of hgc1Δ. CaCLN1 or 2 was expressed in hgc1Δ under the control of either HGC1 or ACT1 promoter (Supplementary Figure 2). None of the constructs was able to suppress the hyphal growth defects of hgc1Δ (data not shown), implying that the function in promoting hyphal morphogenesis is specific to Hgc1.
Physical and functional interaction between Hgc1 and CaCdc28
Hgc1 shares high sequence identities to the G1 cyclins specific for Cdc28 but very low similarities to those for other Cdks, suggesting that it might be an interacting partner of CaCdc28. To determine whether Hgc1 and CaCdc28 physically interact with each other, we tagged CaCdc28 with myc and Hgc1 with hemagglutinin (HA) epitopes at the carboxyl and amino termini respectively, and performed co-immunoprecipitation. Both fusion proteins were expressed from their endogenous promoters. A strain expressing CaCdc28-myc alone was used as control. Similar levels of CaCdc28-myc, which is of the expected size of ∼44 kDa, were detected in both strains (Figure 6A, top). From the protein extract of the cells coexpressing CaCdc28-myc and HA-Hgc1, anti-HA antibody precipitated HA-Hgc1 (∼94 kDa) together with CaCdc28-myc (Figure 6A, middle) and the precipitant exhibited histone H1 kinase activity (Figure 6A, bottom). In comparison, neither CaCdc28-myc nor the kinase activity was detectable in the anti-HA precipitant from the cells expressing CaCdc28-myc alone. Together, these results indicate physical interaction between Hgc1 and CaCdc28. However, at this stage, we do not know whether this interaction is required for hyphal morphogenesis. We observed that the HA-Hgc1 band appeared diffuse (Figure 6A, middle), suggesting possible post-translational modification of this protein. We are currently investigating the nature and significance of this modification.
Figure 6.
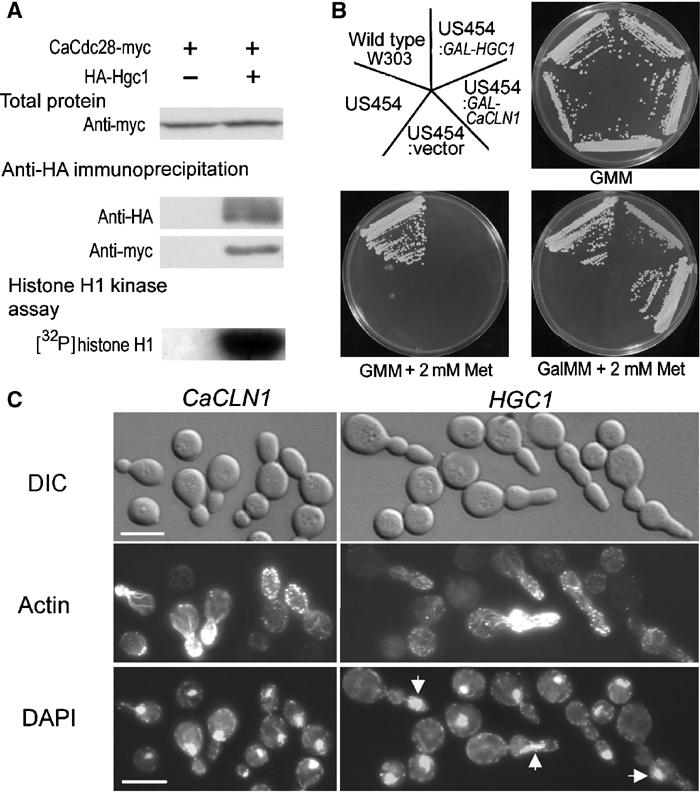
Interactions of Hgc1 with Cdc28. (A) Hgc1 co-immunoprecipitates with CaCdc28. The strain expressing CaCdc28-myc alone (WYZ19) and that coexpressing CaCdc28-myc and HA-Hgc1 (WYZ20) were induced in serum for 1 h. Protein extracts (top) and the immunoprecipitant pulled down by anti-HA antibody (middle) were Western-blotted and probed by anti-myc or anti-HA antibody. The immunoprecipitant was assayed for histone H1 kinase activity (bottom). (B) HGC1 supports the growth of an S. cerevisiae mutant without endogenous G1 cyclins (strain US454). Strains were grown on minimal medium plates containing 2% glucose (GMM) or galactose (GalMM) with or without methionine (Met) at 30°C for 2 days. (C) US454 cells expressing HGC1 are highly elongated. The US454 cells transformed with HGC1 or CaCLN1 under the control of Gal1-10 promoter were first cultured in GMM before being transferred to GalMM containing 2 mM methionine and grown for 6–8 h. Rhodamine–phalloidin was used to stain actin structures and DAPI for nuclei (arrows).
In S. cerevisiae, any one of the three G1 cyclins alone is sufficient to sustain cell growth (Andrews and Measday, 1998). We thought if HGC1 may do likewise, it would provide further evidence supporting functional as well as physical interaction of Hgc1 with CaCdc28. Our repeated efforts in constructing a C. albicans strain with HGC1 as the only G1 cyclin were not successful, probably due to technical difficulties in deleting multiple copies of the CaCLNs from this diploid organism. Alternatively, we tested this in an S. cerevisiae strain US454 (Li and Cai, 1999) deleted of all three G1 cyclin genes, which was kept alive by a copy of ScCLN3 under the control of MET3 promoter. HGC1 or CaCLN1 was introduced into this strain under the control of Gal1-10 promoter on a centromeric plasmid. All the strains grew on GMM plate lacking methionine (Figure 6B), a condition that allows ScCLN3 expression, while no strain, except the wild type, grew on GMM containing 2 mM methionine where the expressions of ScCLN1, HGC1 and CaCLN1 were suppressed. When glucose in the GMM+Met medium was replaced by galactose, only the strains expressing either CaCLN3 or HGC1 grew, although the strain expressing HGC1 grew considerably more slowly. The same test in liquid media produced similar results (data not shown). Interestingly, ∼50% of the cells expressing HGC1 as the only G1 cyclin exhibited significantly elongated morphology, with multiple constrictions and with actin patches and cables polarized toward the bud tip (Figure 6C). We also noticed that ∼70% of the polarized cells contained a single nuclear mass, suggesting possible defects in cell cycle progression. However, these features were not observed in the cells expressing CaCLN1 as the only G1 cyclin. Although these results show that Hgc1 may be only partially functional in driving cell cycle progression, they nevertheless support the interaction between Hgc1 and Cdc28 and suggest functional differences between Hgc1 and CaCln1. The polarized morphology of the S. cerevisiae cells expressing HGC1 alone might be a reflection of its intrinsic ability in promoting morphogenesis, but we cannot exclude the possibility that it may be caused by problems in cell cycle progression (Lew and Reed, 1993; Rua et al, 2001).
HGC1 is required to maintain hyphal tip localization of actin and CaSpa2
Some polarity proteins localize persistently to the hyphal tips (Hazan and Liu, 2002; Hazan et al, 2002; Zheng et al, 2003). Actin and CaSpa2 are two examples. To determine how HGC1 deletion affects the cellular localization of these proteins, we used rhodamine–phalloidin to stain the actin structures and tagged CaSpa2 with the green fluorescent protein (GFP) at its carboxyl terminus. The CaSpa2GFP fusion protein is functional (Zheng et al, 2003). Synchronous G1 yeast cells were treated with serum at 37°C and examined at regular intervals. In response to hyphal induction, both actin and CaSpa2GFP in hgc1Δ cells rapidly polarized to a site at the cell surface and later were observed at the tip of the broad surface protrusion and bud throughout most of the cell cycle (Figure 7). However, they disappeared from the bud tip and appeared at the bud neck at the time of cytokinesis. In contrast, both proteins localized to the tips throughout hyphal growth in HGC1 cells. The results show that under inducing conditions hgc1Δ is able to localize CaSpa2 and actin to the site of polarized growth through a large part of the cell cycle but loses this ability around the time of cytokinesis. It suggests that Hgc1 may have a role in maintaining actin and CaSpa2 at the hyphal tips when the cells undergo cytokinesis, which is probably required to ensure persistent hyphal tip growth and prevent cell separation.
Figure 7.
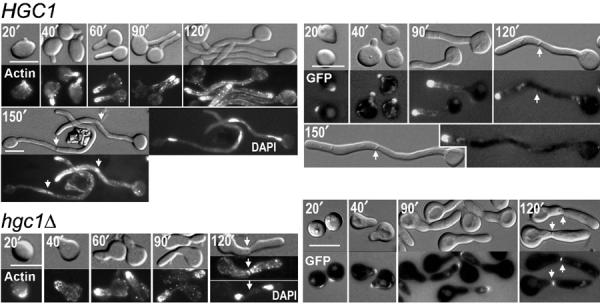
Role of Hgc1 in maintaining tip localization of actin and CaSpa2. G1 yeast cells of strains WYZ5 (HGC1) and WYZ14 (hgc1Δ) expressing CaSpa2GFP were induced for hyphal growth in YPD+10% serum and samples were harvested at regular intervals. The DIC photos are shown on the top and the fluorescence ones for actin staining (left) and CaSpa2GFP (right) are shown at the bottom. The cells that have completed cell division (the 150′ sample for HGC1 and 120′ sample for hgc1Δ, left panel) were also stained with DAPI. The arrows indicate the position of septum.
HGC1 is required for virulence
The importance of hyphal morphogenesis for C. albicans virulence remains controversial due to its co-regulation with many genes encoding other virulence traits (Berman and Sudbery, 2002; Gow et al, 2002). Our results above show that HGC1 deletion abolished hyphal morphogenesis without detectable effect on the yeast growth and the expression of HWP1, HYR1 and ECE1. Thus, hgc1Δ should serve as the best test so far available to evaluate the role of hyphal morphogenesis in virulence. Using the mouse systemic infection model, we found that 70% of the mice infected with 1 × 106 yeast cells of hgc1Δ remained alive 30 days post infection and the rest died between days 20 and 30, whereas the same inoculum killed all the mice infected by an isogenic strain carrying a copy of HGC1 within 7 days (Figure 8). For comparison, efg1 cph1 mutant did not kill any of the mice at the same inoculum. These results show that HGC1 is required for C. albicans virulence, strongly supporting the view that the yeast-to-hypha morphogenesis is important for pathogenesis. However, the results also indicate that other factors, which are blocked in efg1 cph1, are also required for the full virulence.
Figure 8.
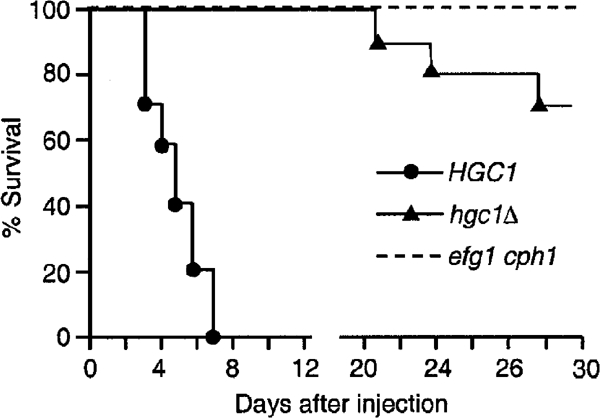
hgc1Δ exhibits markedly reduced virulence. Yeast cells of strains WYZ12.1 (HGC1), WYZ12.2 (hgc1Δ) and WYNR1 (efg1 cph1) were grown in YPD to mid-log phase. Each mouse was injected via the tail vein with 1 × 106 cells and monitored for death for 30 days. The test was performed twice yielding nearly identical trends.
Discussion
This study discovered and characterized the first hypha-specific gene identified so far that is critically required for C. albicans hyphal morphogenesis. We showed that the deletion of HGC1 blocked hyphal growth under a range of experimental conditions that are thought to activate distinct signal sensing/transducing mechanisms that converge to promote hyphal morphogenesis, such as serum, neutral pH, and embedment in agar matrix and the kidneys of mice. We also demonstrated that hgc1Δ was significantly weakened in virulence. HGC1 expression was found to be specifically induced during hyphal growth, and its regulation, like many other known HSGs, was mediated by the cAMP/PKA pathway and transcriptional inhibitors Tup1 and Nrg1. As one of many HSGs, the effect of deleting HGC1 was observed to be limited primarily, if not entirely, to hyphal morphogenesis. HGC1 encodes a G1 cyclin-related protein and exhibited physical and functional interaction with the cell cycle regulator CaCdc28, allowing us to propose a highly plausible mechanism for the control of hyphal morphogenesis.
Role of Hgc1 in virulence
C. albicans yeast-to-hypha transition has been shown in many studies to facilitate its invasion of host tissues and escape from phagocytosis, two processes important for microbial virulence (Odds, 1988; Lo et al, 1997; Berman and Sudbery, 2002). The observed specific function of HGC1 in promoting C. albicans hyphal morphogenesis and its requirement for virulence suggests that the role of HGC1 in virulence is most likely attributable to its activity in regulating hyphal growth, although we cannot exclude the possibility that some other virulence attributes may be compromised in the HGC1 deletion mutants. The transcriptional co-regulation of HGC1 with the genes encoding other virulence traits further supports its intimate involvement in virulence. The signaling pathways for hyphal growth activate the expression of a group of proteins responsible for diverse, infection-related functions such as adhesins, secreted aspartyl proteinases, and iron acquisition (Staab et al, 1999; Braun and Johnson, 2000; Kapteyn et al, 2000; Ramanan and Wang, 2000; Naglik et al, 2003). Hgc1, a new member of this group, seems to be responsible for the morphogenetic aspect of infection. It is evident that the evolution of C. albicans has coordinated a range of biochemically distinct cellular functions in a ‘pathogenesis program' under the same transcriptional control because of each one's specific contribution to different stages of infection. Hence, these genes are collectively responsible for the full virulence of the pathogen. This is consistent with the various degrees of partial loss of virulence when each individual function is blocked and the nearly complete loss of virulence in efg1 cph1 in which most of these functions are compromised.
Role of Hgc1 in hyphal morphogenesis
The core mechanisms underlying universal cellular processes in eucaryotes are often highly conserved. Cell cycle progression and polarized cell growth are two well-known examples (Drubin, 2000; Nurse, 2000). In S. cerevisiae, the Cln1,2/Cdc28 kinases have been repeatedly demonstrated to promote bud elongation, and mutants that enhance or prolong the activities of Cln1,2/Cdc28 sometimes cause significant bud elongation that morphologically resembles the germ tubes of C. albicans (Lew and Reed, 1993; Rua et al, 2001). Recently, the cell cycle regulator Cdc2–cyclin B2 was shown to promote actin assembly and cell migration in mammalian cells (Manes et al, 2003), and Cdk5 kinase was found to promote neurite outgrowth (Nikolic et al, 1996). The involvement of cell cycle regulators in promoting distinct forms of polarized cell growth in yeast and mammals suggests that a highly conserved mechanism may be specifically modified in different cell types or organisms to control different forms of polarized cell growth.
Our discovery of Hgc1 and the characterization of its sequence features, expression pattern and cellular functions suggest that C. albicans may have adapted the mechanism that normally promotes the apical bud elongation during the G1 phase in the yeast cells to control specifically hyphal growth. Here we propose that Hgc1 might have diverged from other G1 cyclins having lost its function in cell cycle control but retaining the function of promoting apical bud extension like the corresponding activity of ScCln1 and 2 (Lew and Reed, 1993; Rua et al, 2001). Concurrently, its transcriptional regulation has been placed under the control of the hypha-inducing signals instead of the cell cycle signals that regulate the periodic expression of other cyclin genes. The loss of cell cycle function and the gain of the hypha-specific expression would ensure that Hgc1 is produced at any time of a cell cycle when the cells are exposed to hypha-inducing signals and remains continuously present as long as the inducing conditions are maintained with minimal or no effect on the cell cycle progression. Once produced, Hgc1 may recruit a fraction of Cdc28 to promote hyphal morphogenesis, leaving the rest of the G1 and mitotic cyclins to control cell cycle progression. The Cdks for cell cycle control are well documented to associate with different cyclins to regulate simultaneously multiple cellular processes (Nurse, 2000). If Hgc1 activity should require interaction with Cdc28, the constant presence of Cdc28 in the cell would permit its availability. In addition, Hgc1 might also play an important role in preventing cell separation by sequestering some polarity proteins from the site of cytokinesis. This model accommodates well the main properties of Hgc1 characterized in this study and agrees in principle with the currently prevailing model on the mechanism underlying cell morphogenesis in S. cerevisiae. It also provides a satisfactory explanation for the cell cycle-independent occurrence of hyphal growth and the lack of effect of CaCLN1 deletion on hyphal growth (Loeb et al, 1999). The pseudohyphal growth reported in mutants deleted of CaFKH2 (Bensen et al, 2002), CaCDC5 (Bachewich et al, 2003) or CaMCM1 (Rottmann et al, 2003) or in cells treated with cell cycle toxins (Bai et al, 2002; Bachewich et al, 2003) is likely the consequence of altering the normal cell cycle control, which may not involve Hgc1.
Other factors are also required for hyphal growth
Although Hgc1 is of critical importance, we found that its constitutive expression is not sufficient to cause hyphal growth, indicating that other unknown proteins or cellular processes coactivated with Hgc1 by the hypha-inducing signals are also needed. The germ tube emergence in hgc1Δ cells, although morphologically aberrant, exhibited nearly normal spatiotemporal control, suggesting that other factors are responsible for the initiation and limited maintenance of the polarized growth. Potential candidates may include the polarity proteins Rho3 and Bem2, whose expression was recently shown to be rapidly elevated upon hyphal induction (Enjalbert et al, 2003). Stochastic spontaneous polarization might be another possible mechanism that initiates the randomized germ tube emergence (Wedlich-Soldner et al, 2003). The molecular links between Hgc1 activity and the proteins that execute polarized growth, such as the Rho family GTPase Cdc42, its regulators and the actin assembly machinery (Drubin, 2000; Etienne-Manneville and Hall, 2002) remain to be established. It will be interesting to identify the substrates or other interacting partners of Hgc1/CaCdc28.
The evolution of C. albicans seems to have incorporated a conserved mechanism into a complex program that coordinates multiple events required for infection and virulence. Elucidation of the underlying mechanisms may provide valuable insights into a fundamental biological process—polarized cell growth—as well as new means to develop novel anti-Candida therapies.
Materials and methods
Strains and culture conditions
All the strains used are listed in Table I. C. albicans yeast was grown at 30°C in YPD (2% yeast extract, 1% bactopeptone and 2% glucose) or in GMM (2% glucose and 1 × yeast nitrogen base) or GMM supplemented with nutrients. For hyphal growth, the yeast cells were inoculated into YPD+10% serum, Lee's (pH 7.0) medium (Lee et al, 1975), RPMI-1640 or Spider's medium and grown at 37°C. Solid media contain 1.5% agar. G1 yeast cells were prepared by centrifugal elutriation as described (Zheng et al, 2003).
Table 1.
C. albicans and S. cerevisiae strains used in this study
| Strains | Relevant genotype | Source |
|---|---|---|
| C. albicans | ||
| SC5314 | Clinical isolate from London Mycological Reference Laboratory | |
| CAI4 | ura3∷imm434/ura3∷imm434 | Fonzi and Irwin (1993) |
| BWP17 | ura3∷imm434/ura3∷imm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG | Enloe et al (2000) |
| CR153 | cacdc35D/cacdc35D | Rocha et al (2001) |
| CaWY5 | rfg1Δ∷HIS1/rfg1Δ∷URA3 | Hu et al (2002) |
| CaWY6 | tup1Δ∷HIS1/tup1Δ∷URA3 | Hu et al (2002) |
| CaWY7 | nrg1Δ∷HIS1/nrg1Δ∷URA3 | Hu et al (2002) |
| JKC19 | cph1Δ∷hisG/cph1Δ∷hisG-URA3-hisG | Liu et al (1994) |
| HLC52 | efg1Δ∷hisG/efg1Δ∷hisG-URA3-hisG | Lo et al (1997) |
| WYNR1 | efg1Δ∷hisG/efg1Δ∷hisG cph1Δ∷hisG/cph1Δ∷hisG-URA3-hisG | This study |
| WYZ5 | CaSPA2/Caspa2∷CaSPA2GFP URA3 | Zheng et al (2003) |
| WYZ12 | ura3∷imm434/ura3∷imm434 hgc1Δ∷ARG4/hgc1Δ∷HIS1 | This study |
| WYZ12.1 | hgc1Δ∷ARG4/hgc1Δ∷HIS1, HGC1 URA3 | This study |
| WYZ12.2 | hgc1Δ∷ARG4/hgc1Δ∷HIS1, URA3 | This study |
| WYZ13 | his1∷hisG/his1∷hisG hgc1Δ∷ARG4/hgc1Δ∷hisG-URA3-hisG | This study |
| WYZ13.1 | ura3∷imm434/ura3∷imm434 his1∷hisG/his1∷hisG hgc1Δ∷ARG4/hgc1Δ∷hisG | This study |
| WYZ14 | hgc1Δ∷ARG4/hgc1Δ∷HIS1, CaSPA2/Caspa2∷CaSPA2GFP URA3 | This study |
| WYZ15 | hgc1Δ∷ARG4/hgc1Δ∷HisG tup1Δ∷HIS1/tup1Δ∷URA3 | This study |
| WYZ16 | hgc1Δ∷ARG4/hgc1Δ∷HIS1, ACT1-promoter-HGC1 URA3 | This study |
| WYZ17 | hgc1Δ∷ARG4/hgc1Δ∷HIS1, HGC1-promoter-CaCLN1 URA3 | This study |
| WYZ18 | hgc1Δ∷ARG4/hgc1Δ∷HIS1, ACT1 promoter-CaCLN1 URA3 | This study |
| WYZ19 | hgc1Δ∷ARG4/hgc1Δ∷HisG, CaCDC28-6MYC HIS1 | This study |
| WYZ20 | hgc1Δ∷ARG4/hgc1Δ∷HisG, CaCDC28-6MYC HIS1 6HA-HGC1 URA3 | This study |
| S. cerevisiae | ||
| W303 | MATa ade2 ura3 leu2 trp1 his3 | Li and Cai (1999) |
| US454 | MATa cln1Δ cln2Δ cln3Δ ade2 ura3 (pMET3-CLN3) | Li and Cai (1999) |
| WYZ21 | MATa cln1 cln2 cln3 ade2 ura3 (pMET3-CLN3) pGAL-HGC1 | This study |
| WYZ22 | MATa cln1 cln2 cln3 ade2 ura3 (pMET3-CLN3) pGAL-CaCLN1 | This study |
C. albicans gene deletion
Deletion of HGC1 copies with ARG4 and HIS1 cassettes from BWP17 produced strain WYZ12 by following the strategy described by Zheng et al (2003). HGC1 coding region with ∼1000-bp 5′- and 1700-bp 3′-UTR was PCR-amplified and cloned between the KpnI and SpeI sites of plasmid CIp10 (Murad et al, 2001a). This plasmid (pCIp10HGC1) was linearized at the XbaI site in the promoter and integrated into HGC1 promoter in WYZ12, yielding strain WYZ12.1. CIp10 alone was also introduced into WYZ12 at the RP10 locus to yield strain WYZ12.2. A second hgc1Δ strain was constructed from BWP17 using ARG4 and HisG-URA3-HisG cassettes yielding WYZ13 and WYZ13.1 after looping out URA3 on 5-FOA plate. WYZ13.1 was used to construct strains WYZ15, 19 and 20.
Chitin, actin and nuclear staining and fluorescence microscopy
Chitin, actin and nuclear stainings were carried out as described (Zheng et al, 2003). A Leica DMR fluorescence microscope and a Hamamatsu digital camera interfaced with METAMORPH software (Universal Imaging) were used for imaging.
Mouse systemic infection
Systemic infection of mice and histology were performed as described (Bai et al, 2002). Each animal was injected with 1 × 106 yeast cells and two groups of 12 mice were used for each strain. Two animals were killed 2 days after injection for histology and the rest were kept for 30 days to monitor survival.
Co-immunoprecipitation and Western blot
The nucleotide sequences for × 6HA (YPYDVPDYA) and × 6myc (LDEESILKQE) were synthesized with restriction sites added to the ends. The × 6myc sequence was inserted in front of the stop codon of CaCDC28 in a DNA fragment corresponding to the 3′ half of the gene containing a unique XhoI site. The DNA fragment was cloned in a modified pCIp10 in which the CaURA3 gene had been replaced by HIS1. After XhoI cleavage, the plasmid was used to replace the 3′ end of a copy of CaCDC28 in WYZ13.1 yielding WYZ19. The × 6HA tag was inserted after the start codon of HGC1 in pCIp10HGC1, linearized with XbaI and integrated at the promoter in WYZ13.1 yielding WYZ20. Both CaCdc28-myc and HA-Hgc1 are functional, because they could support normal cell growth as the sole allele. Protein extract was prepared as described (Surana et al, 1993). For immunoprecipitation of HA-Hgc1, ∼1 mg of protein was incubated with rabbit anti-HA antibody Y-11 (Santa Cruz Biotechnology Inc.) for 1 h at 4°C and then with protein A–sepharose beads (Pharmacia) for 1 h. Anti-HA and -myc antibodies were used to detect HA-Hgc1 and CaCdc28-myc (Clontech) in Western blot. Histone H1 kinase was assayed as described (Surana et al, 1993). HGC1 and CaCLN1 were cloned in a CEN plasmid under the control of Gal1-10 promoter.
Supplementary Material
Supplemental Figure 1
Supplemental Figure 2
Acknowledgments
We thank A Brown, M Whiteway and U Sarana for providing C. albicans and yeast strains, and WJ Hong and W Chia for comments. This work was supported by the Agency for Science, Technology and Research of Singapore. Y Wang is an adjunct associate professor in the Department of Microbiology, National University of Singapore.
References
- Andrews B, Measday V (1998) The cyclin family of budding yeast: abundant use of a good idea. Trends Genet 14: 66–72 [DOI] [PubMed] [Google Scholar]
- Bachewich C, Thomas DY, Whiteway M (2003) Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol Biol Cell 14: 2163–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Ramanan N, Wang YM, Wang Y (2002) Spindle assembly checkpoint component CaMad2p is indispensable for Candida albicans survival and virulence in mice. Mol Microbiol 45: 31–44 [DOI] [PubMed] [Google Scholar]
- Bailey DA, Feldmann PJ, Bovey M, Gow NA, Brown AJ (1996) The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol 178: 5353–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Filler SG, Berman J (2002) A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot Cell 1: 787–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J, Sudbery PE (2002) Candida albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet 3: 918–930 [DOI] [PubMed] [Google Scholar]
- Birse CE, Irwin MY, Fonzi WA, Sypherd PS (1993) Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun 61: 3648–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD (1997) Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277: 105–109 [DOI] [PubMed] [Google Scholar]
- Braun BR, Johnson AD (2000) TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155: 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Kadosh D, Johnson AD (2001) NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J 20: 4753–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D (2000) Frontiers in Molecular Biology: Cell Polarity. Oxford: Oxford Press [Google Scholar]
- Enjalbert B, Nantel A, Whiteway M (2003) Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol Biol Cell 14: 1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enloe B, Diamond A, Mitchell AP (2000) A single-transformation gene function test in diploid Candida albicans. J Bacteriol 182: 5730–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420: 629–635 [DOI] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY (1993) Isogenic strain construction and gene mapping in Candida albicans. Genetics 134: 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NA, Brown AJ, Odds FC (2002) Fungal morphogenesis and host invasion. Curr Opin Microbiol 5: 366–371 [DOI] [PubMed] [Google Scholar]
- Hazan I, Liu H (2002) Hyphal tip-associated localization of Cdc42 is F-actin dependent in Candida albicans. Eukaryot Cell 1: 856–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan I, Sepulveda-Becerra M, Liu H (2002) Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol Biol Cell 13: 134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Bai C, Wang YM, Wang Y (2002) Characterization and functional analysis of the siderophore-iron transporter CaArn1p in Candida albicans. J Biol Chem 277: 30598–30603 [DOI] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD (2001) Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol Cell Biol 21: 2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn JC, Hoyer LL, Hecht JE, Muller WH, Andel A, Verkleij AJ, Makarow M, Van Den Ende H, Klis FM (2000) The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol Microbiol 35: 601–611 [DOI] [PubMed] [Google Scholar]
- Kron SJ, Styles CA, Fink GR (1994) Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell 5: 1003–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KL, Buckley HR, Campbell CC (1975) An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13: 148–153 [DOI] [PubMed] [Google Scholar]
- Lengeler KB, Davidson RC, D'souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev 64: 746–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed S (1993) Morphogenesis in the yeast cell cycle, regulation by Cdc28 and cyclins. J Cell Biol 120: 1305–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai M (1999) Recovery of the yeast cell cycle from heat shock-induced G(1) arrest involves a positive regulation of G(1) cyclin expression by the S phase cyclin Clb5. J Biol Chem 274: 24220–24227 [DOI] [PubMed] [Google Scholar]
- Liu H (2001) Transcriptional control of dimorphism in Candida albicans. Curr Opin Microbiol 4: 728–735 [DOI] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR (1994) Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266: 1723–1726 [DOI] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR (1993) Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262: 1741–1744 [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR (1997) Nonfilamentous C. albicans mutants are avirulent. Cell 90: 939–949 [DOI] [PubMed] [Google Scholar]
- Loeb JD, Sepulveda-Becerra M, Hazan I, Liu HA (1999) G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol Cell Biol 19: 4019–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manes T, Zheng DQ, Tognin S, Woodard AS, Marchisio PC, Languino LR (2003) Alpha(v)beta3 integrin expression up-regulates Cdc2, which modulates cell migration. J Cell Biol 161: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad AM, d'Enfert C, Gaillardin C, Tournu H, Tekaia F, Talibi D, Marechal D, Marchais V, Cottin J, Brown AJ (2001c) Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol Microbiol 42: 981–993 [DOI] [PubMed] [Google Scholar]
- Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ (2001a) CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16: 325–327 [DOI] [PubMed] [Google Scholar]
- Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, Schnell N, Talibi D, Marechal D, Tekaia F, d'Enfert C, Gaillardin C, Odds FC, Brown AJ (2001b) NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J 20: 4742–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B (2003) Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67: 400–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C, Goto S, Lund K, Hung W, Sadowski I (2003) Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421: 187–190 [DOI] [PubMed] [Google Scholar]
- Nikolic M, Dudek H, Kwon YT, Ramos YF, Tsai LH (1996) The Cdk5/p35 kinase is essential for neurite outgrowth during neuronal differentiation. Genes Dev 10: 816–825 [DOI] [PubMed] [Google Scholar]
- Nurse P (2000) A long twentieth century of the cell cycle and beyond. Cell 100: 71–78 [DOI] [PubMed] [Google Scholar]
- Odds FC (1988) Candida and Candidosis, 2nd edn London: Baillière Tindall [Google Scholar]
- Ramanan N, Wang Y (2000) A high-affinity iron permease essential for Candida albicans virulence. Science 288: 1062–1064 [DOI] [PubMed] [Google Scholar]
- Rocha CR, Schroppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E (2001) Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell 12: 3631–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann M, Dieter S, Brunner H, Rupp S (2003) A screen in Saccharomyces cerevisiae identified CaMCM1, an essential gene in Candida albicans crucial for morphogenesis. Mol Microbiol 47: 943–959 [DOI] [PubMed] [Google Scholar]
- Rua D, Tobe BT, Kron SJ (2001) Cell cycle control of yeast filamentous growth. Curr Opin Microbiol 4: 720–727 [DOI] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL (2003) Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2: 1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab JF, Bradway SD, Fidel PL, Sundstrom P (1999) Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283: 1535–1538 [DOI] [PubMed] [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker CE, Ernst JF (1997) Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J 16: 1982–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K (1993) Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J 12: 1969–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner R, Altschuler S, Wu L, Li R (2003) Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299: 1231–1235 [DOI] [PubMed] [Google Scholar]
- Zheng XD, Wang YM, Wang Y (2003) CaSPA2 is important for polarity establishment and maintenance in Candida albicans. Mol Microbiol 49: 1391–1405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1
Supplemental Figure 2


