Abstract
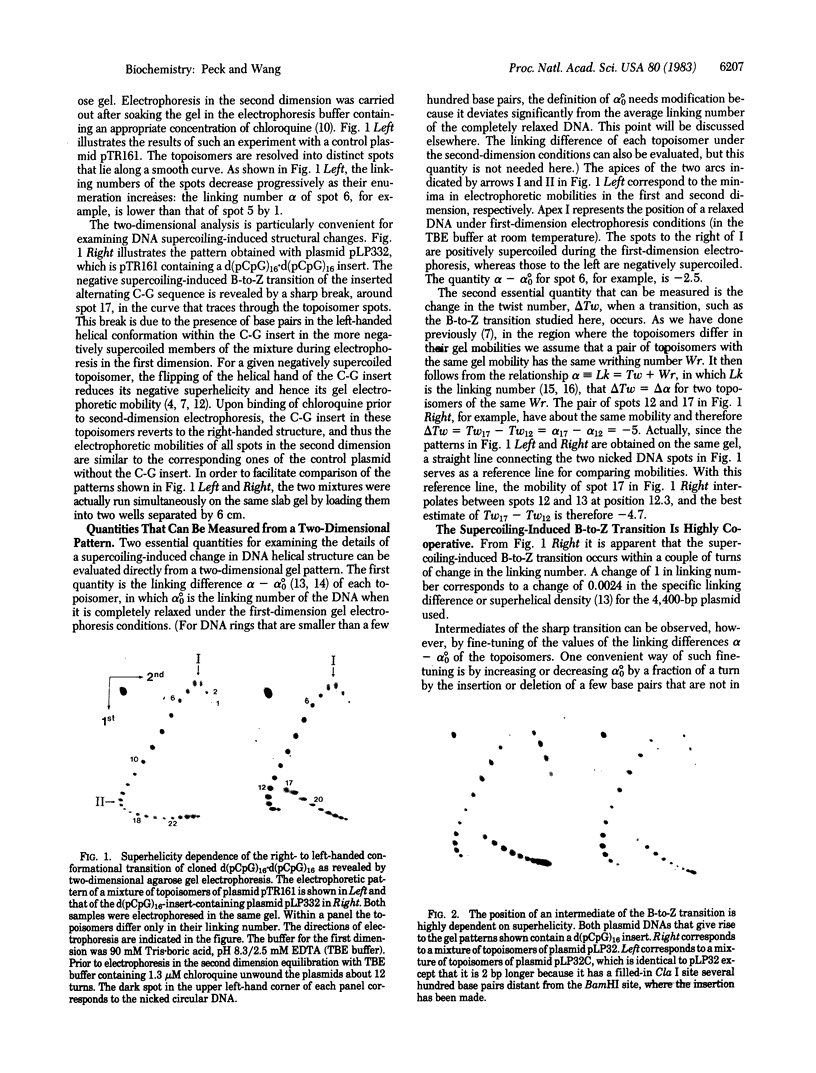
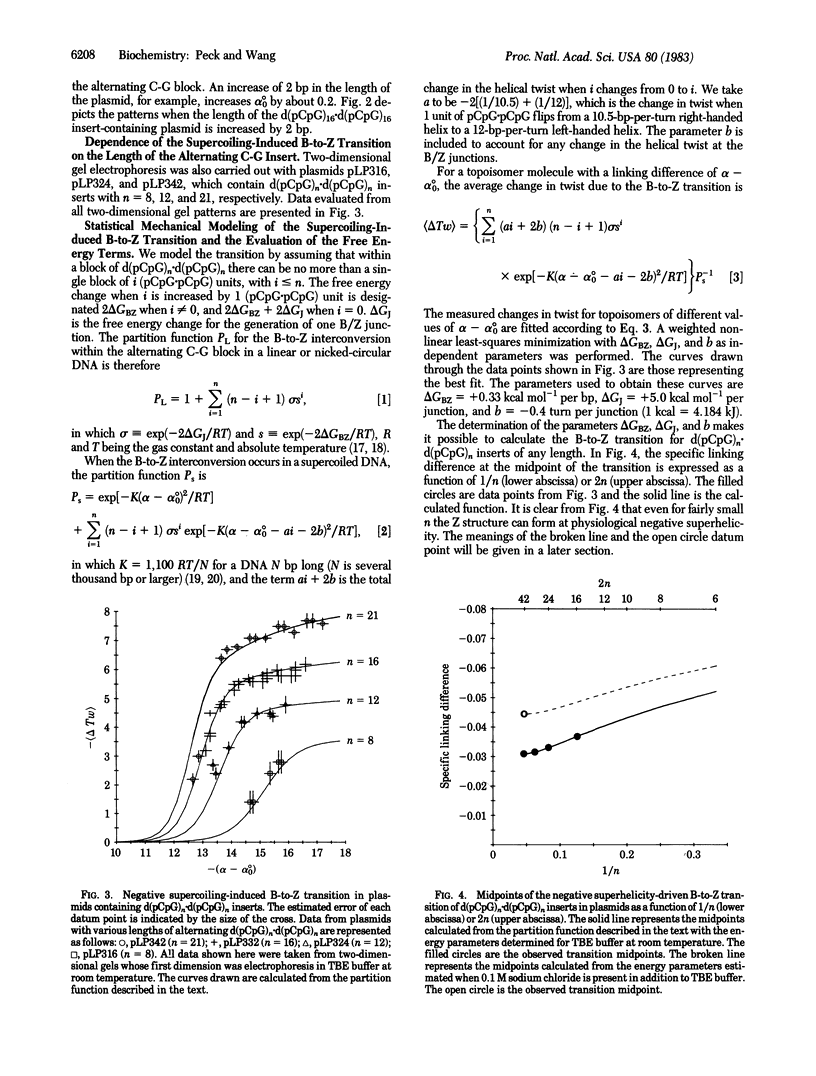
Analysis by two-dimensional gel electrophoresis of topoisomers of plasmids containing d(pCpG)n . d(pCpG)n inserts, in which n ranges between 8 and 21, shows that the B-to-Z transition within the alternating C-G is readily induced by negative supercoiling and is highly cooperative. The free energy parameters for the transition in dilute aqueous buffers have been evaluated from a statistical mechanical analysis of the data, and these parameters allow prediction of the superhelicities of plasmids at which the transition occurs in alternating C-G inserts over a wide range of lengths. In agreement with the crystal structures, the helical handedness of the B structure in solution and that of the Z structure are shown to be opposite to each other. Furthermore, it is found that the B form of the alternating C-G sequence in solution has a helical periodicity of 10.5 +/- 0.1 base pairs per turn, and the Z form has a helical periodicity of 11.6 +/- 0.3 base pairs per turn. There also appears to be a significant unwinding of the right-handed DNA duplex at each of the B/Z junctions.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azorin F., Nordheim A., Rich A. Formation of Z-DNA in negatively supercoiled plasmids is sensitive to small changes in salt concentration within the physiological range. EMBO J. 1983;2(5):649–655. doi: 10.1002/j.1460-2075.1983.tb01479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R., Conner B. N., Wing R. M., Fratini A. V., Kopka M. L. The anatomy of A-, B-, and Z-DNA. Science. 1982 Apr 30;216(4545):475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- Fuller F. B. The writhing number of a space curve. Proc Natl Acad Sci U S A. 1971 Apr;68(4):815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G., Bansal M., Sasisekharan V. Reversal of handedness in DNA: a stable link between RU and LZ helices. Biochem Biophys Res Commun. 1980 Dec 31;97(4):1258–1267. doi: 10.1016/s0006-291x(80)80002-7. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I., Minchenkova L. E., Minyat E. E., Schyolkina A. K. Cooperative transitions in DNA with no separation of strands. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):243–250. doi: 10.1101/sqb.1983.047.01.029. [DOI] [PubMed] [Google Scholar]
- Kłysik J., Stirdivant S. M., Larson J. E., Hart P. A., Wells R. D. Left-handed DNA in restriction fragments and a recombinant plasmid. Nature. 1981 Apr 23;290(5808):672–677. doi: 10.1038/290672a0. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Mizusawa H., Kakefuda T. Unwinding of double-stranded DNA helix by dehydration. Proc Natl Acad Sci U S A. 1981 May;78(5):2838–2842. doi: 10.1073/pnas.78.5.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Panyutin I. G., Frank-Kamenetskii M. D. Evidence of cruciform structures in superhelical DNA provided by two-dimensional gel electrophoresis. FEBS Lett. 1983 Mar 21;153(2):298–302. doi: 10.1016/0014-5793(83)80628-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Lafer E. M., Peck L. J., Wang J. C., Stollar B. D., Rich A. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982 Dec;31(2 Pt 1):309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Pohl F. M. Salt-induced transition between two double-helical forms of oligo (dC-dG). Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):113–117. doi: 10.1101/sqb.1983.047.01.014. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. M., Kacich R., Ptashne M. A general method for maximizing the expression of a cloned gene. Proc Natl Acad Sci U S A. 1979 Feb;76(2):760–764. doi: 10.1073/pnas.76.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M., Pulleyblank D. E., Vinograd J. The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res. 1977;4(5):1183–1205. doi: 10.1093/nar/4.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang J. C. DNA: bihelical structure, supercoiling, and relaxation. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):29–33. doi: 10.1101/sqb.1979.043.01.006. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B. The three-dimensional structure of DNA. Annu Rev Biochem. 1982;51:395–427. doi: 10.1146/annurev.bi.51.070182.002143. [DOI] [PubMed] [Google Scholar]