Abstract
Progress in our understanding of sociobiology has occurred with little knowledge of the genetic mechanisms that underlie social traits. However, several recent studies have described microbial genes that affect social traits, thereby bringing genetics to sociobiology. A key finding is that simple genetic changes can have marked social consequences, and mutations that affect cheating and recognition behaviors have been discovered. The study of these mutants confirms a central theoretical prediction of social evolution: that genetic relatedness promotes cooperation. Microbial genetics also provides an important new perspective: that the genome-to-phenome mapping of social organisms might be organized to constrain the evolution of social cheaters. This constraint can occur both through pleiotropic genes that link cheating to a personal cost and through the existence of phoenix genes, which rescue cooperative systems from selfish and destructive strategies. These new insights show the power of studying microorganisms to improve our understanding of the evolution of cooperation.
Introduction
The concepts of cooperation (see Glossary) and conflict are familiar in discussions of animal and human societies [1-4]. It is perhaps less appreciated that there are many examples of cooperation at other levels of biological organization [5,6]: these include (i) the coalitions of genes that constitute genomes [7]; (ii) the cells in multicellular organisms [8,9]; and (iii) the diverse cooperative actions carried out by groups of microorganisms, which are the focus of this article [10-15]. These lower-level forms of cooperation have clear differences from those among higher organisms, particularly humans, for whom intention and morality are so important [16]. Nevertheless, these simple systems have one of the fundamental properties of a cooperative system: a shared investment in a group resource [5,6]. For example, many microorganisms share molecules that are secreted into the environment, including enzymes that break down food, quorum-sensing signals, siderophores (which scavenge iron) and the protective polymeric slime that is released in bacterial biofilms [10-15,17] (Figure 1a,b). There is more sophisticated sociality too: this is epitomized by the dictyostelid amebae [18] and the Myxobacteria (also known as Myxococcales) [19], in both of which cells aggregate and many cells die so that others can be propagated as spores in terminally differentiated fruiting bodies (Figure 1c,d). Crucially, such traits have the potential for ‘cheaters’ to gain a selfish advantage by using the resources of others without themselves paying the full cost. Therefore, they are the subject of one of the central conundrums of sociobiology: how is cooperation stable in the face of selfishness and cheating [1,5,11,20-22]?
Figure 1.
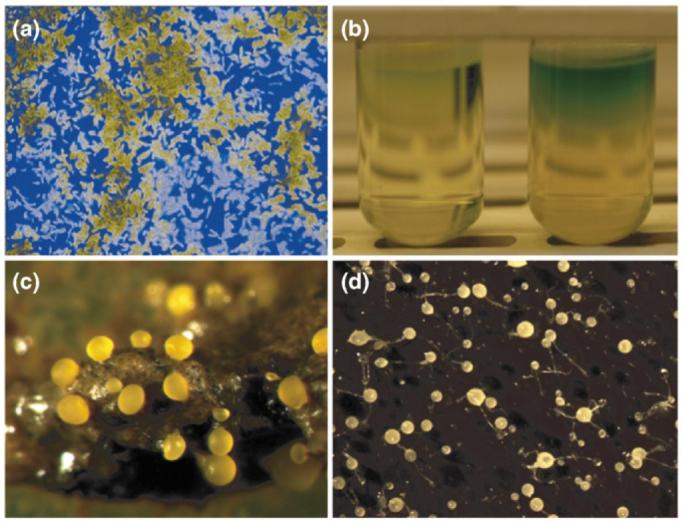
Microbial social behaviors. (a) Early-stage Pseudomonas aeruginosa biofilm containing two strains, one labeled with cyan fluorescent protein and the other with yellow fluorescent protein. Biofilms contain numerous shared group products, including the extracellular polymeric substances that produce a matrix surrounding the cells. (b) Two strains of P. aeruginosa in LB medium, showing the pigments of siderophores, which are extracellular molecules released by the bacteria to scavenge iron. (c) Fruiting bodies of the social bacterium Myxococcus xanthus. (d) Fruiting bodies of the social ameba (i.e. slime mold) Dictyostelium discoideum. Photo in part c kindly provided by Michiel Vos.
Microorganisms enable familiar sociobiological questions to be explored in a new system. Similar to any model system, they bring a unique set of advantages and disadvantages. The clearest disadvantage might be perceived to be the simplicity of their social behaviors. However, this can also be a blessing in a field that has been plagued by controversy over the nature-versus-nurture debate [23-25] and, in particular, the relative roles of culture and genetics in social behavior. The use of microorganisms elegantly side-steps this problem, because, by most definitions at least, culture does not drive the actions of microorganisms. Therefore, similar to the more simple animals [2,3], they provide the opportunity to evaluate the traits of nature in their purest form.
Studying sociobiology by using microorganisms carries an additional key advantage: it is possible not only to confirm a genetic basis for social behaviors but also to identify the genes responsible [26]. Typically, the theories of social evolution rely on the assumption that social traits have a simple genetic basis that can be treated as a mechanistic ‘black box’ and largely ignored [27]. The main reason for this is the paucity of classical and molecular genetic tools available to biologists studying classical social organisms (Box 1). This problem is compounded by the probability that complex behaviors in higher organisms might not be regulated by, or ascribed to, single (master) genes. Therefore, although sociobiology has made considerable progress, it remains a challenge to understand the genetic basis of social traits, an enterprise the worthiness of which is apparent from the great dividends it has paid in others areas, such as cell and developmental biology [28,29].
Box 1. Sociobiology.
This brings us to the central theoretical problem of sociobiology: how can altruism, which by definition reduces personal fitness, possibly evolve by natural selection? (E.O. Wilson, Sociobiology, 1975)
E.O. Wilson's book Sociobiology [1] caused a storm when it was released in 1975 because of its discussion of the genetic and evolutionary basis of human social behavior. This idea was viewed as dangerous by some Marxist scholars, including Gould and Lewontin, because they thought it might be used to justify prejudice and racism [76]. (Not everyone agreed: Chomsky viewed it as a justification for accepting diversity.) Initially, the controversy overshadowed Wilson's achievement. In addition to making the fundamental point that there must be a genetic component to behavior, he reviewed the biology of a large range of social organisms and summarized many concepts needed to understand it. This was the real sociobiology.
Following the work of W.D. Hamilton [20], Wilson centered his discussion on the question of how sociality, and the related phenomena of cooperation and altruism, can evolve. This question has since received a great deal of attention, both empirically and theoretically [60]. One of the most studied animal groups is the social insects, for which much is now understood about cooperation and conflict [2]. Work has also centered on social mammals and birds [77] and, increasingly, on the evolution of cooperation between species [60,62]. Despite the initial taboo, sociobiology has now also returned in earnest to study human cooperation through both game-theory economics [4] and evolutionary psychology. Furthermore, there is now an explicit focus on genetics, with major efforts underway to identify the genes that determine social actions in many species (a field that is known as sociogenomics [65]). These studies are technically challenging in many species. Nevertheless, in humans, there has been a stream of published ‘genes for X’, which are typically based on a statistical association between a particular linkage group and a behavioral trait. However, the most progress in our understanding of the classical models of sociobiology has come from studying other animals [65]. For example, several genes that are important in the onset of foraging in the honeybee, Apis mellifera, have been identified. These findings are being followed by a concerted effort among social insect biologists to identify further genes and pathways that are important in social behavior, in particular the genes that regulate the balance between cooperation and conflict.
An important responsibility of biologists is to make it clear that this focus on genetics does not imply that all human behavior is genetically determined. As Wilson realized, the ultimate sociobiological synthesis will be made only when we learn how to combine genetic and cultural theories of behavior effectively.
Many microorganisms can be studied using the latest molecular genetic and post-genomic technologies, which, combined with the relatively simple actions of microorganisms, enables us to open the black box of the genetics underlying social traits. Nevertheless, it could be argued that, even for microorganisms, it is too early to evaluate the complex genetic interplay that ultimately drives social traits. In this article, we argue otherwise and draw attention to recent studies that combine the study of microbial genetics with social behavior. Importantly, there are many examples in which the mutation that causes a social trait has been identified and ascribed to a single gene, illustrating the considerable impact that single genes can have on the regulation of behavior (Figure 2). In addition, these studies offer an important new perspective on sociobiology and how stable cooperation can arise, which is the focus of this article.
Figure 2.
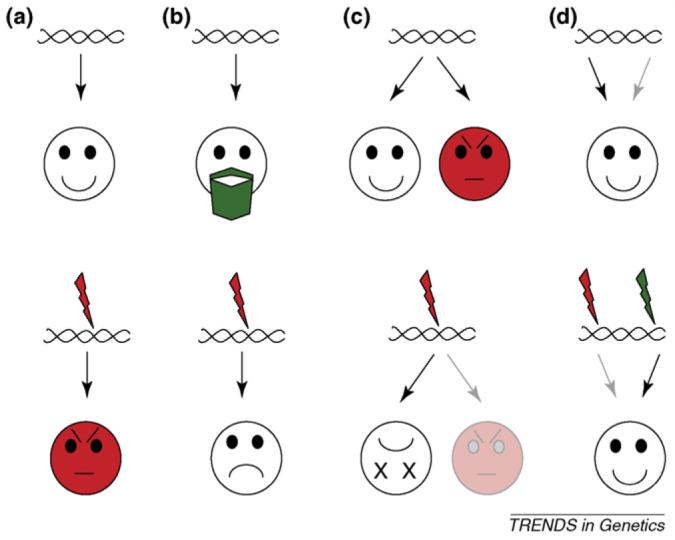
The four discussed gene types that determine social actions and the effects of their inactivation. The upper row shows the normal effect of the gene, and the lower row shows the effect of the gene after inactivation (red zigzag) or activation (green zigzag). (a) Cheater genes. Inactivation causes a selfish cheating strategy: for example, Saccharomyces cerevisiae SUC2. (b) Recognition genes. Inactivation causes exclusion from a social trait: for example, Dictyostelium discoideum csaA (a green-beard gene). (c) Pleiotropic genes that determine both cooperation and cheating. Inactivation causes a net loss of fitness: for example, D. discoideum dimA. (d) Phoenix genes. Inactivation of a cheater gene (left) is recovered by a change in expression of a phoenix gene (right): for example, Myxococcus xanthus strain PX. Note that some gene types were named after the resultant phenotype when inactive (a), and others were named after the resultant phenotype when active (b,d). Gray arrows indicate that the gene is not responsible for the social trait.
Despite its novelty, it is reassuring that results from this emerging blend of sociobiology and microbial genetics fit broadly into the existing theories. Accordingly, we organize this article around two main types of explanation for cooperation: first, genetic relatedness among individuals [1,20,30]; and, second, coercion or constraints that prevent rebellion [31]. This follows a recent review of conflict resolution in insect societies [2] and is intended to illustrate that nontrivial analogies exist between microbial cooperation and the cooperation that occurs in the more traditional model systems for sociobiology (Box 1).
Genetic relatedness
Cheater genes
The importance of genetic relatedness in the evolution of cooperation has been underlined by several studies that have generated ‘cheater’ mutants in the laboratory. These mutants are so named because they behave like social ‘cheaters’: that is, individuals that reap the benefit of social traits while contributing less than average to the cost (Figure 1a). In the simplest cases, these are mutants that do not contribute a secreted chemical that is shared by all cells (an example of a ‘public good’). For example, strains of the yeast Saccharomyces cerevisiae with a loss-of-function mutation in the gene SUC2 fail to produce invertase, a secreted enzyme that breaks down extracellular sucrose [32]. These mutants outcompete wild-type cells when grown in mixtures. Similarly, strains of the pathogenic bacterium Pseudomonas aeruginosa that do not produce the siderophores that are required to scavenge insoluble iron from the environment [33,34] also fare better than wild-type cells in mixtures. In addition, in societies of the bacterium Myxococcus xanthus [19,35-37] and the slime mold Dictyostelium discoideum [38], mutants have been identified that overproduce spores at the expense of other strains in the fruiting body. The ability to generate these strains is of great importance, because it shows that crude genetic changes, including whole-gene knockouts, can result in exploitative behaviors that threaten social groups.
What, therefore, prevents the rampant spread of these mutants into natural systems and the resultant collapse of the social traits? One answer might be that there are hidden costs to the cheater strains, such as a cost that is pleiotropically linked to the expression of the cheating trait (see the section Pleiotropic genes). However, another major explanation is genetic relatedness, which is the probability above chance that individuals (of the same species) have identical alleles at one or more loci (reviewed in Refs [3] and [11]). Ever since the seminal work of W.D. Hamilton [20], it has been realized that cooperation and even altruism can be naturally selected when a social partner is related. This process occurs because when two individuals have the same alleles, one individual can increase the frequency of its alleles that are carried by the next generation by helping the other individual to reproduce. Note that the important measure of relatedness that determines the evolutionary trajectory of an allele for a social action is relatedness at its locus [3,11]. Consequently, it is possible that two bacterial strains are genetically identical at most loci but are unrelated with respect to the gene that determines the social action, if they have different alleles at this locus.
The effect can be easily understood in the context of cheater mutants such as SUC2− S. cerevisiae, which do not produce invertase [32], or strains of P. aeruginosa that fail to produce siderophores [33,34]. Only when individuals are not highly related (e.g. when the cheaters mix with cells that produce invertase or siderophores, respectively) do cheaters gain a selfish benefit and would therefore be favored by natural selection. However, when cheaters only meet genetically identical cells, there are no cells to exploit, and they do badly (i.e. have reduced fitness) [10-15]. It seems probable that the failure of certain cheater alleles of D. discoideum [38] and M. xanthus [19,35-37] to spread might be similarly explained. Therefore, the ability to create cheater mutants has provided unique and powerful ways to test a central prediction of sociobiology: that relatedness promotes cooperation [3,11]. These and other studies have provided experimental confirmation of the idea that cheaters only succeed under conditions in which individuals are not highly related [19,33-35,38].
Recognition genes
In setting out the basis for what became known as kin-selection theory, Hamilton [20] envisaged that the relatedness that drives cooperation would arise through groupings of relatives (or kin), such as a family group in a colony of social insects or a growing bacterium that is surrounded by clones [11]. However, he also outlined a thought experiment for another way that relatedness might arise: if a gene that determines cooperation could identify other copies of itself in others. These hypothetical genes became known as ‘green beard’ genes after Dawkins developed the analogy of a gene that encodes both green beards and an altruistic act directed only towards other green-bearded individuals [39]. The existence of green-beard genes was hypothetical until the discovery in the red fire ant (Solenopsis invicta) of the gene Gp-9, which encodes a pheromone-binding protein [40,41]. Workers that carry the b allele of this gene kill BB queens (which lack the b allele) in colonies with multiple queens. However, it was not until a genetically characterized and tractable microorganism was studied that the first genuine single-gene green beard was found. (The red fire ant phenotype is probably driven by a supergene complex.) The gene csaA (also known as csA) of D. discoideum encodes a homophilic cell-adhesion molecule, which recognizes and adheres to the same molecule displayed by other individuals [42]. Importantly, it has been shown that in D. discoideum aggregations on soil, cells that do not express the gene fail to aggregate and are excluded from the group. Therefore, carrying the gene confers both the ability to recognize other individuals that carry it and the ability to interact cooperatively with them in the aggregation.
If csaA is an example of a green-beard gene that drives altruism, then colicin toxins are an excellent example of spite [43]. Colicins are produced by bacteria that carry a plasmid consisting of tightly linked genes that encode both colicins and proteins that provide resistance to the toxic effects of colicins [44]. The release of a colicin by one cell benefits related cells by poisoning those that do not carry the colicin-encoding genes. This is an example of spite [43,45] because it is an action that harms another individual but does not benefit the cell that releases the toxin, which lyses in the process. Both examples show how a simple genetic system can result in a mechanism that directly identifies the genetic identity of others, and they are also a further testament to the ability of a small number of genes to cause marked social effects.
Coercion and constraint
Pleiotropic genes
Another type of explanation for cooperation is traits that coerce individuals into cooperating or, equally, traits that constrain them from rebelling [31]. In human societies, coercion is evident in policing systems, a phenomenon that has striking analogies with both insect societies [2] and vertebrate societies [46]. Single-gene studies of microorganisms are now revealing another important piece of the puzzle: the particulate nature of the genome [47] provides an underlying structure that can help to constrain cheating.
A single gene is called pleiotropic when it influences multiple, apparently independent, phenotypic traits. Recent genome-wide studies of the architecture of genetic networks reveal that pleiotropy is more widespread than was previously thought [48,49]. Consequently, new mutations often exert effects on multiple traits simultaneously, meaning that pleiotropy can constrain adaptation because adaptive change in one trait might be compromised by others [50-52]. However, this constraint can be an advantage for cooperation when it prevents the origination of cheaters. This raises the intriguing possibility that the mapping between the genomes and the phenomes in social species might be organized such that it limits rebellions and thereby promotes the maintenance of cooperation [53]. Although, at first, this might seem farfetched, several recent studies provide support for this idea.
Before D. discoideum cells form a fruiting body, they aggregate and organize into a migrating slug that moves to the soil surface. This structure contains prespore cells, which are destined to become spores, and prestalk cells, most of which vacuolate and die in the fruiting-body stalk. Interestingly, this differentiation seems to involve coercion, because prespore cells release a chemical known as differentiation-inducing factor 1 (DIF-1), which induces prestalk cells to differentiate. So what prevents the pre-stalk cells from cheating by ignoring the DIF-1 signal? The answer, at least in part, is pleiotropy. The gene dimA is central to the response of prestalk cells to DIF-1, and a mutant with this gene knocked out creates cells that behave like cheaters and preferentially become prespore cells in slugs when mixed with wild-type cells [53-55]. However, another effect of inactivating dimA ultimately prevents cheating: the ability of cells to enter the spore head of the fruiting body and produce spores is inhibited [53]. This pleiotropic link between stalk and spore formation ensures that dimA is expressed and helps to prevent stalk cells from evading the coercive signal of the prespore cells. It remains to be determined whether this link arose through natural selection or simply through chance (see the section How do pleiotropic constraints arise?), but several other examples indicate that such effects might be common.
Pleiotropy is also seen in the D. discoideum green-beard mutant, csaA−. When csaA− cells develop on an agar sub-stratum, they aggregate effectively when in mixtures with wild-type cells. Furthermore, altered cell adhesion enables the mutant cells to cheat and become greatly over-represented in the spores. It is only when developed on the more challenging substratum of soil that the pleiotropic deficiency is observed: without the adhesion molecule CsaA, aggregation of the mutant cells is greatly impaired. Another example is Escherichia coli with the GASP (growth at stationary phase) phenotype, which divides rapidly and selfishly in nutrient-limited cultures and interrupts the normal cooperative slowing of growth in stressed E. coli cells [56]. This success, however, is short lived. The main mutation that endows cells with the GASP phenotype is a partial loss-of-function allele of rpoS, which encodes the stationary-phase σ factor (σS). The protein encoded by this gene is a global regulator of gene expression that is required to coordinate the large range of responses that cells undergo to adapt to stationary-phase conditions. Consequently, without a fully functioning σS, mutant cells cannot tolerate the acidic conditions that are usually encountered in a key niche, the mammalian stomach.
Finally, pleiotropy has a central role in promoting the cooperative bioluminescence caused by Vibrio fischeri, a bacterial symbiont that lives in the light-emitting organ of the bobtail squid, Euprymna scolopes. The symbiotic relationship is based on the bacterium providing bioluminescence that might aid the squid in darkness or camouflage the squid from below against the bright ocean surface. In return, the squid provides nutrients for bacterial growth. However, light production is probably costly for the bacterium, because it requires the synthesis of numerous proteins; bioluminescence is controlled by at least eight lux genes [57]. Consequently, strains that fail to bioluminesce are expected to cheat wild-type cells in mixed colonies in light organs, because such mutants would not pay the cost of light production. Three such strains, with targeted single-gene mutations that abolish bioluminescence, have been generated, and their behavior has been tested. All mutants showed little bioluminescence; however, no clone could cheat. This is because the expression of lux genes is also required for efficient colonization [58,59]. Analogous to the case of dimA, this seems to be a system in which pleiotropic constraints go hand-in-hand with coercion. In this case, the squid appears to create an environment that enforces light production, which the bacteria are pleiotropically constrained from avoiding. This system is also an example of ‘partner choice’ [60-62], in which one species in a mutualistic relationship selects for more-cooperative individuals in its partner species.
Phoenix genes
Tracing the evolutionary steps that resulted in a modern trait is a classic and persistent problem in evolutionary biology. The use of microorganisms offers a possible solution, because microorganisms can be made to evolve in the laboratory and adaptation followed as it occurs, with the potential for identifying the causal genetic changes. The elegance of this approach was recently shown by an experimental evolution study of the bacterium M. xanthus [19,37], which, similar to D. discoideum, forms fruiting bodies on starvation (Figure 1c,d). Cheater mutants were pitted against wild-type cells in mixed groups over multiple cycles of fruiting-body formation. In many cases, the populations became extinct, because the cheater mutants could dominate but could not sporulate when cultured alone. However, in one example, a new strain – phoenix – arose from the social collapse. This strain could form spores, resist the cheater mutant, and even produced more spores than the ancestral (wild-type) strain. Whole-genome sequencing revealed that it took just a single base change to go from the cheater strain to the new super strain [63]. The mutation increases expression of a gene that is predicted to encode a member of the Gcn5-related N-acetyltransferase superfamily, some members of which regulate both global and specific gene expression [64]. Although the mechanism by which the phoenix strain suppressed the cheater strain has not been completely determined, it is clear that a single mutation could rescue the social system from the brink of collapse. Similar to the studies on pleiotropy, this finding is consistent with underlying genetics that enables cheaters to be easily suppressed. It is an open question, given its success in the laboratory, why phoenix does not seem to have spread in nature.
Concluding remarks and future studies
The emergence of the terms sociogenomics [65] and sociomicrobiology [11] is a testament to the excitement surrounding the interface of sociobiology, microbiology and genetics. A great deal has already been learned from these emerging disciplines. This includes new verification of old sociobiological theories, which, importantly, were ideas developed for organisms in a different kingdom. In addition, there are novel hypotheses, such as a role for pleiotropic and phoenix genes in social evolution. The scale of the importance of such genes remains to be established, both in microorganisms and in other organisms. Nevertheless, it seems clear that microbial genetics will continue to bear fruit for sociobiology, and there are several frontiers that are particularly promising.
Social-gene evolution in nature
The identification of genes that determine social actions enables us, for the first time, to track directly genetic changes over space and time. This goal is helped by the small size of many microbial genomes and by the emergence of economical sequencing methods [63]. One hypothesis is that genes that are involved in competitive interactions undergo rapid evolution owing to evolutionary arms races [66,67]. However, another hypothesis is that genes that are required for cooperation are stabilized by pleiotropy and related effects, and evolve slowly. Clearly, more data are needed, but it is tantalizing that, among strains of S. cerevisiae, SUC genes are highly polymorphic, varying both in copy number and chromosomal location [32]. Is this the signature of rapid selective change?
How do pleiotropic constraints arise?
From the genetic studies of microorganisms that are discussed here, it is clear that pleiotropic constraints can help to stabilize cooperation. However, what are the evolutionary steps that result in such pleiotropic relationships [37]? In the simplest case, it might be an intrinsic property of all biological systems that it is difficult for cheaters to arise. However, it might be that such pleiotropic relationships have arisen through evolution: for example, specific post hoc modifiers that appear in the original social strain after a cheater has arisen. It could also be envisaged that various pleiotropic relationships between social actions and other traits have arisen throughout history, but only those that could stabilize cooperation have persisted. Such high-level trait or species selection [68,69] is thought to be important in the evolution of sexual reproduction [70,71] and might also make pleiotropic constraints on cheating common. It remains a daunting challenge for the field to distinguish between these explanations. An associated question is how much of microbial cooperation is explained by such constraints, and how much by relatedness [72]?
Subtle mutants
Studying microorganisms has provided an important insight for sociobiologists: that marked changes in social phenotype can result from a change in a single gene. However, it is possible that these early studies might not be representative, because genetic screens are more likely to identify the mutants with the largest effect. Therefore, a challenge for the future is to identify the more subtle mutants, because these mutants might tell a different story. This idea is supported by a study of a knockout mutant of V. fischeri that has a subtle defect in bioluminescence [73]. This litR− mutant shows only a slight delay in the onset of detectable bioluminescence when colonizing the light organ of the host squid. Unlike lux knockouts, which show a more marked effect, litR− mutants can colonize the light organ and even outcompete the wild type. There might be other fitness costs at other stages of the life cycle, but, at face value, this seems to be an intriguing exception to the pleiotropy hypothesis. It remains to be seen whether more-sophisticated genetic screens will unearth similar mutants in other social microorganisms.
Multigene effects and robustness
An obvious but fascinating open question relates to multigene effects and genetic interactions. We have discussed how pleiotropy can limit cheating by linking it to a personal cost. But, unless there are many more traits than genes, multiple genes will affect many traits. This can lead to redundancy such that changes in one gene have little phenotypic effect, whereas changes in multiple genes have a marked effect (a process known as aggravating epistasis [74]). This property might help to stabilize sociality, because it makes genetic networks robust to single gene changes. For a cooperative trait, it would mean that many mutational changes need to occur before the trait is fully disrupted, ensuring that sociality is not quickly lost and providing time for compensatory mutations to arise [19,37].
From genome to phenome
Many genes with social effects have now been identified, but, in some cases, we have little idea of the path from genotype to social phenotype. One of the main goals for the future is a global understanding of gene functions, genetic interactions and the downstream effects that ultimately determine the phenotype. We can expect that there is a common ontology among many genes associated with social actions, because it is already clear that there is enrichment for certain functional groups of genes, including those responsible for cell–cell recognition, protein and small-molecule secretion, and development. However, in each group, there undoubtedly will be a bewildering diversity of interactions and effects. As is so often the case [75], we expect the devil to be in the details.
Acknowledgements
We thank Dave Queller and Joan Strassmann for the metaphor of a ‘black box’ in the genetics of social behavior, for the prediction that genes with social effects might evolve rapidly and for comments on the manuscript. We also thank the following: Andrew Murray and Jeff Smith for insights into the pleiotropy hypothesis; Greg Velicer, Matthew Cobb and Jason Wolf for stimulating discussion; and Stuart West and the referees for comments. K.R.F. is supported by the National Institutes of Health Center Grant 5P50 GM068763-01. C.R.L.T. and K.P. are supported by the Medical Research Council and a Lister Institute of Preventive Medicine Research Prize.
Glossary
- Altruism
carrying out an action that benefits a recipient (i.e. increases the evolutionary fitness of the recipient) at a cost to an individual's own lifetime fitness (from the effect on personal reproduction, i.e. direct fitness).
- Cheater
an individual that benefits from a social trait but pays a below-average share of the cost. When alone, cheaters produce suboptimal group adaptations.
- Cooperation
carrying out an action that benefits a recipient.
- Evolutionary arms race
a series of adaptations and counteradaptations that occurs in two classes of individual that are in evolutionary conflict, such as between predator and prey, cheater and cooperators, or parasite and host.
- Mutualistic
an interaction that benefits both parties. This term is typically used for interactions between multiple species.
- Partner choice
the preferential interaction with more-cooperative individuals of the same or a different species.
- Quorum sensing
the ability to respond to local population density. For microorganisms, this occurs by the secretion of quorum-sensing molecules. The concentration of these molecules is an indicator of local population density.
- Relatedness
a genetic correlation between individual loci or organisms.
- Social behavior
an action that affects other individuals.
- Sociobiology
the study of the biology and evolution of social behavior.
- Spite
carrying out an action that harms a recipient but does not increase an individual's own direct fitness.
References
- 1.Wilson EO. Sociobiology. Belknap; 1975. [Google Scholar]
- 2.Ratnieks FLW, et al. Conflict resolution in insect societies. Annu. Rev. Entomol. 2006;51:581–608. doi: 10.1146/annurev.ento.51.110104.151003. [DOI] [PubMed] [Google Scholar]
- 3.Bourke AFG, Franks NR. Social Evolution in Ants. Princeton University Press; 1995. [Google Scholar]
- 4.Henrich J, et al. Foundations of Human Sociality: Economic Experiments and Ethnographic Evidence from Fifteen Small-scale Societies. Oxford University Press; 2004. [Google Scholar]
- 5.Keller L, editor. Levels of Selection in Evolution. Princeton University Press; 1999. [Google Scholar]
- 6.Maynard Smith J, Szathmáry E. The Major Transitions in Evolution. W.H. Freeman; 1995. [Google Scholar]
- 7.Burt A, Trivers RL. Genes in Conflict: the Biology of Selfish Genetic Elements. Harvard University Press; 2006. [Google Scholar]
- 8.Michod RE. Darwinian Dynamics: Evolutionary Transitions in Fitness and Individuality. Princeton University Press; 1999. [Google Scholar]
- 9.Buss LW. The Evolution of Individuality. Princeton University Press; 1987. [Google Scholar]
- 10.Kreft JU. Biofilms promote altruism. Microbiology. 2004;150:2751–2760. doi: 10.1099/mic.0.26829-0. [DOI] [PubMed] [Google Scholar]
- 11.West SA, et al. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 12.Foster KR. Hamiltonian medicine: why the social lives of pathogens matter. Science. 2005;308:1269–1270. doi: 10.1126/science.1108158. [DOI] [PubMed] [Google Scholar]
- 13.Crespi BJ. The evolution of social behavior in microorganisms. Trends Ecol. Evol. 2001;16:178–183. doi: 10.1016/s0169-5347(01)02115-2. [DOI] [PubMed] [Google Scholar]
- 14.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 15.Velicer GJ. Social strife in the microbial world. Trends Microbiol. 2003;11:330–337. doi: 10.1016/s0966-842x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- 16.Khalil EL. What is altruism? J. Econ. Psychol. 2004;25:97–123. [Google Scholar]
- 17.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strassmann JE, et al. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature. 2000;408:965–967. doi: 10.1038/35050087. [DOI] [PubMed] [Google Scholar]
- 19.Fiegna F, et al. Evolution of an obligate social cheater to a superior cooperator. Nature. 2006;441:310–314. doi: 10.1038/nature04677. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton WD. The genetical evolution of social behaviour. I and II. J. Theor. Biol. 1964;7:1–52. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 21.Foster KR. Diminishing returns in social evolution: the not-so-tragic commons. J. Evol. Biol. 2004;17:1058–1072. doi: 10.1111/j.1420-9101.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 22.Travisano M, Velicer GJ. Strategies of microbial cheater control. Trends Microbiol. 2004;12:72–78. doi: 10.1016/j.tim.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Barlow GW. Nature–nurture and the debates surrounding ethology and sociobiology. Am. Zool. 1991;31:286–296. [Google Scholar]
- 24.Meaney MJ. Nature, nurture, and the disunity of knowledge. Ann. New York Acad. Sci. 2001;935:50–61. doi: 10.1111/j.1749-6632.2001.tb03470.x. [DOI] [PubMed] [Google Scholar]
- 25.Sykora P. Is sociology afraid of biology? Sociologia. 1999;31:375–396. [Google Scholar]
- 26.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Grafen A. How not to measure inclusive fitness. Nature. 1982;298:425–426. doi: 10.1038/298425a0. [DOI] [PubMed] [Google Scholar]
- 28.Hartwell LH, et al. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. U. S. A. 1970;66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 30.Foster KR, et al. Kin selection is the key to altruism. Trends Ecol. Evol. 2006;21:57–60. doi: 10.1016/j.tree.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 31.Frank SA. Repression of competition and the evolution of cooperation. Evolution Int. J. Org. Evolution. 2003;57:693–705. doi: 10.1111/j.0014-3820.2003.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 32.Greig D, Travisano M. The Prisoner's Dilemma and polymorphism in yeast SUC genes. Proc. R. Soc. Lond. B Biol. Sci. 2004;271(Suppl. 3):S25–S26. doi: 10.1098/rsbl.2003.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffin AS, et al. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 34.Harrison F, et al. Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 2006;4:21. doi: 10.1186/1741-7007-4-21. DOI: 10.1186/1741-7007-4-2 ( www.biomedcentral.com/bmcbiol) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiegna F, Velicer GJ. Competitive fates of bacterial social parasites: persistence and self-induced extinction of Myxococcus xanthus cheaters. Proc. R. Soc. Lond. B Biol. Sci. 2003;270:1527–1534. doi: 10.1098/rspb.2003.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velicer GJ, et al. Developmental cheating in the social bacterium Myxococcus xanthus. Nature. 2000;404:598–601. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- 37.Foster KR. Sociobiology: the Phoenix effect. Nature. 2006;441:291–292. doi: 10.1038/441291a. [DOI] [PubMed] [Google Scholar]
- 38.Ennis HL, et al. Dictyostelium amoebae lacking an F-box protein form spores rather than stalk in chimeras with wild type. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3292–3297. doi: 10.1073/pnas.050005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawkins R. The Selfish Gene. Oxford University Press; 1976. [Google Scholar]
- 40.Keller L, Ross KG. Selfish genes: a green beard in the red fire ant. Nature. 1998;394:573–575. [Google Scholar]
- 41.Krieger MJ, Ross KG. Identification of a major gene regulating complex social behavior. Science. 2002;295:328–332. doi: 10.1126/science.1065247. [DOI] [PubMed] [Google Scholar]
- 42.Queller DC, et al. Single-gene greenbeard effects in the social amoeba Dictyostelium discoideum. Science. 2003;299:105–106. doi: 10.1126/science.1077742. [DOI] [PubMed] [Google Scholar]
- 43.Gardner A, et al. Bacteriocins, spite and virulence. Proc. R. Soc. Lond. B Biol. Sci. 2004;271:1529–1535. doi: 10.1098/rspb.2004.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 2002;56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 45.Foster KR, et al. Spite: Hamilton's unproven theory. Ann. Zool. Fenn. 2001;38:229–238. [Google Scholar]
- 46.Flack JC, et al. Policing stabilizes construction of social niches in primates. Nature. 2006;439:426–429. doi: 10.1038/nature04326. [DOI] [PubMed] [Google Scholar]
- 47.Fisher RA. The Genetical Theory of Natural Selection. Clarendon Press; 1930. [Google Scholar]
- 48.Anholt RR, Mackay TF. Quantitative genetic analyses of complex behaviours in Drosophila. Nat. Rev. Genet. 2004;5:838–849. doi: 10.1038/nrg1472. [DOI] [PubMed] [Google Scholar]
- 49.Featherstone DE, Broadie K. Wrestling with pleiotropy: genomic and topological analysis of the yeast gene expression network. BioEssays. 2002;24:267–274. doi: 10.1002/bies.10054. [DOI] [PubMed] [Google Scholar]
- 50.Otto SP. Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc. R. Soc. Lond. B Biol. Sci. 2004;271:705–714. doi: 10.1098/rspb.2003.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen TF. Is modularity necessary for evolvability? Remarks on the relationship between pleiotropy and evolvability. BioSystems. 2003;69:83–94. doi: 10.1016/s0303-2647(02)00132-6. [DOI] [PubMed] [Google Scholar]
- 52.Barton NH. Pleiotropic models of quantitative variation. Genetics. 1990;124:773–782. doi: 10.1093/genetics/124.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foster KR, et al. Pleiotropy as a mechanism to stabilize cooperation. Nature. 2004;431:693–696. doi: 10.1038/nature02894. [DOI] [PubMed] [Google Scholar]
- 54.Huang E, et al. bZIP transcription factor interactions regulate DIF responses in Dictyostelium. Development. 2006;133:449–458. doi: 10.1242/dev.02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson CR, et al. A bZIP/bRLZ transcription factor required for DIF signaling in Dictyostelium. Development. 2004;131:513–523. doi: 10.1242/dev.00939. [DOI] [PubMed] [Google Scholar]
- 56.Vulic M, Kolter R. Evolutionary cheating in Escherichia coli stationary phase cultures. Genetics. 2001;158:519–526. doi: 10.1093/genetics/158.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meighen EA, Dunlap PV. Physiological, biochemical and genetic control of bacterial bioluminescence. Adv. Microb. Physiol. 1993;34:1–67. doi: 10.1016/s0065-2911(08)60027-2. [DOI] [PubMed] [Google Scholar]
- 58.Visick KL, et al. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lupp C, et al. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 2003;50:319–331. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- 60.Sachs JL, et al. The evolution of cooperation. Q. Rev. Biol. 2004;79:135–160. doi: 10.1086/383541. [DOI] [PubMed] [Google Scholar]
- 61.Foster KR, Kokko H. Cheating can stabilize cooperation in mutualisms. Proc. R. Soc. Lond. B Biol. Sci. 2006;273:2233–2239. doi: 10.1098/rspb.2006.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Foster KR, Wenseleers T. A general model for the evolution of mutualisms. J. Evol. Biol. 2006;19:1283–1293. doi: 10.1111/j.1420-9101.2005.01073.x. [DOI] [PubMed] [Google Scholar]
- 63.Velicer GJ, et al. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8107–8112. doi: 10.1073/pnas.0510740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vetting MW, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 65.Robinson GE, et al. Sociogenomics: social life in molecular terms. Nat. Rev. Genet. 2005;6:257–270. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- 66.Hurst LD, et al. Genetic conflicts. Q. Rev. Biol. 1996;71:317–364. doi: 10.1086/419442. [DOI] [PubMed] [Google Scholar]
- 67.Dawkins R, Krebs JR. Arms races between and within species. Proc. R. Soc. Lond. B Biol. Sci. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- 68.Foster KR. Balancing synthesis with pluralism in sociobiology. J. Evol. Biol. 2006;19:1394–1396. doi: 10.1111/j.1420-9101.2006.01188.x. [DOI] [PubMed] [Google Scholar]
- 69.Nunney L. Lineage selection and the evolution of multistage carcinogenesis. Proc. R. Soc. Lond. B Biol. Sci. 1999;266:493–498. doi: 10.1098/rspb.1999.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maynard Smith J. The Evolution of Sex. Cambridge University Press; 1978. [Google Scholar]
- 71.Bell G. The Masterpiece of Nature: the Evolution and Genetics of Sexuality. Croom Helm; 1982. [Google Scholar]
- 72.Wenseleers T, Ratnieks FLW. Animal behaviour: enforced altruism in insect societies. Nature. 2006;444:50. doi: 10.1038/444050a. [DOI] [PubMed] [Google Scholar]
- 73.Fidopiastis PM, et al. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 2002;45:131–143. doi: 10.1046/j.1365-2958.2002.02996.x. [DOI] [PubMed] [Google Scholar]
- 74.Segre D, et al. Modular epistasis in yeast metabolism. Nat. Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 75.Beekman M, Ratnieks FLW. Power over reproduction in social Hymenoptera. Philos. Trans. R. Soc. London Ser. B. 2003;358:1741–1753. doi: 10.1098/rstb.2002.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Segerstrale U. Defenders of the Truth: the Battle for Science in the Sociobiology Debate and Beyond. Oxford University Press; 2000. [Google Scholar]
- 77.Griffin AS, West SA. Kin discrimination and the benefit of helping in cooperatively breeding vertebrates. Science. 2003;302:634–636. doi: 10.1126/science.1089402. [DOI] [PubMed] [Google Scholar]


