Abstract
Several host systems are available for the production of recombinant proteins, ranging from Escherichia coli to mammalian cell-lines. This article highlights the benefits of using yeast, especially for more challenging targets such as membrane proteins. On account of the wide range of molecular, genetic, and microbiological tools available, use of the well-studied model organism, Saccharomyces cerevisiae, provides many opportunities to optimize the functional yields of a target protein. Despite this wealth of resources, it is surprisingly under-used. In contrast, Pichia pastoris, a relative new-comer as a host organism, is already becoming a popular choice, particularly because of the ease with which high biomass (and hence recombinant protein) yields can be achieved. In the last few years, advances have been made in understanding how a yeast cell responds to the stress of producing a recombinant protein and how this information can be used to identify improved host strains in order to increase functional yields. Given these advantages, and their industrial importance in the production of biopharmaceuticals, I argue that S. cerevisiae and P. pastoris should be considered at an early stage in any serious strategy to produce proteins.
Keywords: yeast, Saccharomyces cerevisiae, Pichia pastoris, recombinant protein, yield optimization, choice of expression host
RECOMBINANT PROTEIN PRODUCTION IN MICROBES: Escherichia COLI AS THE MOST POPULAR HOST
Proteins are essential components of living organisms and have a role in virtually every cellular process: they are enzymes; form cellular scaffolds and are central to signaling, transport, and regulatory functions. To study these diverse roles, it is necessary to be able to work with sufficient quantities (typically multi-milligram) of suitably stable and functional protein samples. While some proteins can be isolated from native sources for this purpose, many cannot because they are either intrinsically unstable or are present in impractically low quantities (Bill et al., 2011). Moreover, the study of mutant or truncated forms of a given protein is often central to understanding its structure and activity; such mutants must be synthesized recombinantly.
The biotechnological breakthrough required for recombinant gene expression was first demonstrated 40 years ago in the prokaryotic microbe, Escherichia coli (Cohen et al., 1973) and was soon followed by the recombinant production of human somatostatin (Itakura et al., 1977) and human insulin (Goeddel et al., 1979) in E. coli cultures. These innovations heralded the era of the recombinant biopharmaceutical: Humulin® synthesized in E. coli was launched by Eli Lilly and Company in 1982 (Altman, 1982); in 1987, Novo Nordisk started the industrial production of recombinant human insulin, Novolin®, using cultures of the eukaryotic microbe, Saccharomyces cerevisiae (Thim et al., 1986). Today, the recombinant production of biopharmaceuticals, particularly recombinant antibodies and vaccines, is a multi-billion dollar global business (Goodman, 2009), with more than 150 having been approved by the United States Food and Drug Administration to date (Ferrer-Miralles et al., 2009; Zhu, 2012). Approximately 20% of these biopharmaceutical proteins are produced in yeasts (the vast majority in S. cerevisiae), 30% in E. coli and 50% in mammalian cell-lines and hybridomas (Ferrer-Miralles et al., 2009; Mattanovich et al., 2012).
Research into the science of recombinant protein production is also thriving, both as an academic discipline in its own right and as a means to produce a myriad of proteins for further study (Lee et al., 2012). In 2010, it was reported that the proportion of recombinant genes expressed in E. coli, compared with those expressed in all hosts had remained constant, at roughly 60% per year during the 15 year period 1995–2009 (Sørensen, 2010). Table 1 includes the corresponding data for the other commonly used host cells; it shows that the proportion of recombinant genes expressed in E. coli has remained high to date and that approximately half of these genes are eukaryotic. For all other hosts, the absolute numbers are much smaller, but it is notable that the proportion of recombinant genes expressed in Pichia pastoris has steadily increased from 1995 to date, in contrast to all other host cells (Table 1). Coupled with the beginnings of a decline in usage for E. coli over the last 8 years, this could suggest that researchers are beginning to recognize the capacity of P. pastoris to produce more challenging recombinant targets.
Table 1.
Recombinant gene expression in the most commonly used host cells.
| Year | All host cells | E. coli | S. cerevisiae | P. pastoris | Insect cells | Mammalian cell-lines |
|---|---|---|---|---|---|---|
| 1980 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1985 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1990 | 12 | 75% (9; 4E) | 8% (1) | 0 | 17% (2) | 0 |
| 1995 | 37 | 70% (26; 17E) | 5% (2) | 5% (2) | 5% (2) | 8% (3) |
| 2000 | 50 | 70% (35; 17E) | 0 | 4% (2) | 12% (6) | 12% (6) |
| 2005 | 121 | 85% (103; 53E) | 0 | 5% (6) | 6% (7) | 2% (2) |
| 2010 | 172 | 76% (131; 67E) | 0 | 9% (15) | 5% (6) | 5% (9) |
| 2013 | 128 | 73% (94; 54E) | 2% (2) | 11% (14) | 4% (5) | 4% (5) |
The proportion of recombinant genes expressed in E. coli, S. cerevisiae, P. pastoris, insect cells, and mammalian cell-lines was calculated according to Sørensen’s (2010) methodology; briefly, the PubMed Central database was searched for entries containing “expression purification” in the title field, which returned 1,847 articles. These articles were categorized by year of publication and expression host used and were then examined manually to confirm the categorization. The table shows the percentage of articles reporting recombinant gene expression in a given host cell and year with the actual number in parentheses; for proteins produced in E. coli the number of recombinant proteins of eukaryotic origin (E; ranging from unicellular protozoan to human proteins) is also noted. For all other hosts, the target proteins are exclusively eukaryotic. When percentages do not total 100% in a given year, less frequently used hosts (e.g., cell-free systems and other microbes) account for the remainder.
Escherichia coli stands out as the pre-eminent host cell for producing recombinant proteins in both commercial [50% of proteins; (Ferrer-Miralles et al., 2009; Mattanovich et al., 2012)] and research (>70% of proteins; Table 1) laboratories; it is quick and inexpensive to culture, making it ideal in many respects. However, it has been established that producing eukaryotic proteins in a prokaryotic host cell often results in inclusion body formation and/or low specific yields (Sørensen, 2010), which may be one reason for the slight decline in its more recent use (Table 1). An explanation for lower success rates with eukaryotic targets is that the rates of protein synthesis and folding are almost an order of magnitude faster in prokaryotes than they are in eukaryotes (Widmann and Christen, 2000). Furthermore, eukaryotic codons are often inefficiently expressed and authentic eukaryotic post-translational modifications cannot yet be achieved in E. coli (Sørensen, 2010). However, recent progress has been made in engineering defined glycosylation pathways in E. coli (Valderrama-Rincon et al., 2012), while the Keio collection of single-gene knockout mutants offers a route to understanding the molecular bottlenecks to high yields in this prokaryotic host (Baba et al., 2006).
In principle, the use of mammalian cell-lines should overcome the challenges of producing recombinant eukaryotic proteins in E. coli, especially with recent advances in stable recombinant gene expression (Bandaranayake and Almo, 2013; Kunert and Casanova, 2013). Furthermore, the authenticity of glycosylation performed by mammalian host cells is an important advantage over all other expression hosts. However, progress in the technologies that enable reproducible gene delivery and selection of stable clones continues to be slow (Bandaranayake and Almo, 2013). Moreover, specific yields from mammalian cell-lines are often low (Zhu, 2012) and Table 1 shows a declining trend in their use.
Eukaryotic microbes offer substantial advantages as host cells, despite their propensity to hyperglycosylate recombinant proteins. For example, an annotated genome sequence has been available for S. cerevisiae for almost two decades (Goffeau et al., 1996), an impressive range of deletion and over-expression strains are readily available for S. cerevisiae and the P. pastoris genome has been available since 2009 (De Schutter et al., 2009). Combining this wealth of molecular and genetic resources, with the fact that yeasts grow an order of magnitude more rapidly than mammalian cell-lines means that protein production and optimization can be done quickly and efficiently in yeast (Porro et al., 2011). Table 1 shows that for P. pastoris, at least, there is an increasing trend in its usage suggesting that these advantages have become more widely known. This is especially notable because P. pastoris is a relative new-comer, only having been first developed as a host system in 1985 (Cregg et al., 1985). Less elaborate hyperglycosylation, the availability of strains with humanized glycosylation pathways (Hamilton et al., 2003, 2006) and an increasing repertoire of molecular tools (Prielhofer et al., 2013) make this yeast an excellent alternative to S. cerevisiae. In particular, P. pastoris has been used with great success to produce challenging targets such as recombinant human G protein-coupled receptors and ion channels (Hedfalk, 2013); in total 19 high resolution structures have been resolved of recombinant eukaryotic membrane proteins produced in P. pastoris (Hedfalk, 2013). Table 1 shows that the number of recombinant proteins produced in S. cerevisiae is much smaller, despite the fact that this yeast species is an important industrial host for the production of biopharmaceuticals such as hormones (e.g., insulin and human growth hormone), vaccines (against e.g., hepatitis B and human papilloma viruses), and therapeutic adjuncts (human serum albumin) (Martinez et al., 2012); this may be a consequence of the search criteria used in generating Table 1 or possibly a perception that S. cerevisiae is not as amenable a host cell as P. pastoris.
USING YEASTS TO INCREASE SUCCESS RATES IN RECOMBINANT PROTEIN PRODUCTION EXPERIMENTS
There is no universally applicable solution for the production of all recombinant proteins (Bill, 2001; Sørensen, 2010) and it is not yet possible to predict which host system is most likely to produce a given protein in high functional yields. To be effective, any protein production strategy should therefore encompass more than one host system.
Two main approaches are typically taken to design a new protein production experiment, preferably in combination with each other: (i) optimizing the corresponding gene sequence so it is more likely to be stably expressed and (ii) minimizing the metabolic burden on the chosen host cell(s) during recombinant protein production (Bonander and Bill, 2012). The first strategy may require that a mutant protein is produced; in support of this protein engineering approach there is an extensive literature on engineering stabilized proteins (Traxlmayr and Obinger, 2012; Scott et al., 2013). Codon optimization is also possible (Oberg et al., 2011) with more recent insights suggesting how this might aid functional expression (Halliday and Mallucci, 2014). In contrast, focusing on the host cell provides an opportunity to optimize the production of the native sequence; the principles of this second approach are broadly similar for all host cells, often requiring straightforward experimentation in the initial stages, such as optimizing culture conditions and induction protocols. Successful bioprocess engineering strategies such as these have been demonstrated to increase recombinant protein yields in cultures of both P. pastoris (Rebnegger et al., 2013; Spadiut et al., 2013) and E. coli (Jazini and Herwig, 2013). When a “Design of Experiments” (Bora et al., 2012) approach is used in this context, the effect of multiple parameters on the functional yield of recombinant protein can be examined simultaneously (Holmes et al., 2009); this is important since each input parameter is unlikely to exert an independent effect on functional protein yield (Bora et al., 2012). Successful implementation of such an approach in yeast has been shown to increase the productivity per cell by matching the methanol feed profile to the cellular metabolism (Holmes et al., 2009). In another approach, pulsing P. pastoris cells with methanol revealed the potential benefit of stress in increasing productivity (Dietzsch et al., 2011).
In the last few years, significant advances have been made in this second approach by understanding how a yeast cell responds to the stress of producing a recombinant protein at a molecular level, and how this information can be used to identify improved host strains (Bonander et al., 2009; Ashe and Bill, 2011; Bawa et al., 2011; Lee et al., 2012). Since S. cerevisiae is particularly amenable to studying the mechanistic basis of high-yielding recombinant protein production experiments using the tools of systems and synthetic biology, its more routine use is an obvious way to produce less tractable proteins recombinantly (Drew et al., 2008). Identifying or engineering yeast strains with improved yield characteristics may either be targeted toward one particular pathway or may take a more global approach (Ashe and Bill, 2011). Examples of the targeted approach are provided by the “humanization” of the yeast glycosylation (De Pourcq et al., 2010) and sterol (Kitson et al., 2011) pathways and modifying membrane phospholipid synthesis to proliferate intracellular membranes (Guerfal et al., 2013). Studies taking a more global approach in both S. cerevisiae (Bonander et al., 2005; Bonander and Bill, 2009) and P. pastoris (Baumann et al., 2011; Rebnegger et al., 2013) have identified the importance of the unfolded protein response (UPR; the cellular stress response activated in response to an accumulation of unfolded or misfolded protein) and reduced translational activity in high yielding cultures. In contrast to the mammalian UPR, the simpler UPR of yeast does not lead to down-regulation of translation to reduce protein synthetic load (Patil and Walter, 2001). We have previously noted that reducing protein synthetic capacity in yeast might be an effective way to improve recombinant protein yields since this capacity is unregulated in response to unfolded protein in cells (Ashe and Bill, 2011). Such insights, which are not yet possible in higher eukaryotic systems, have been used to select specific yeast strains that can substantially improve recombinant yields compared to wild-type cells (Bonander et al., 2009; Norden et al., 2011; Figure 1). The minimal use of S. cerevisiae as a host shown in Table 1 is therefore at odds with this unique potential for optimization; it is possible that the increasing popularity of P. pastoris has detracted from the use of S. cerevisiae. I suggest that this undervalued host system should therefore be revisited, especially in view of its success in the production of challenging targets (Drew et al., 2008).
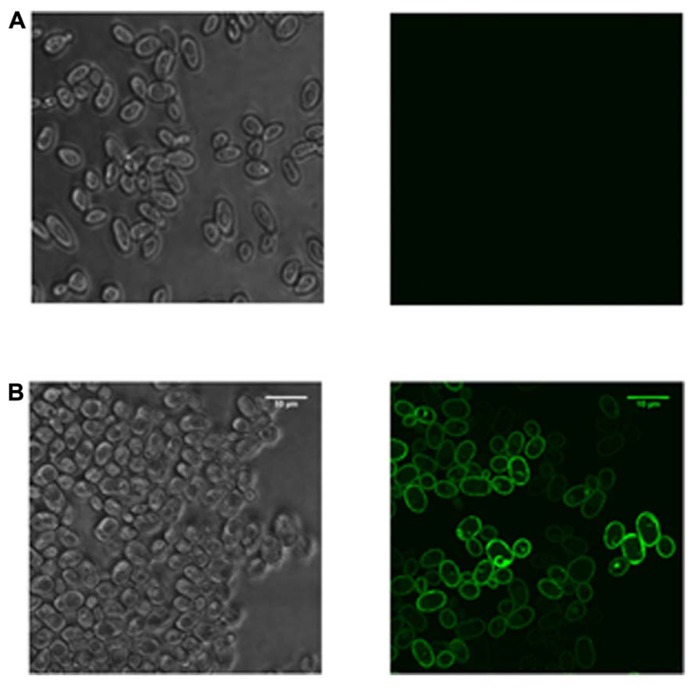
FIGURE 1.
Strain selection enables the production of a human membrane protein in S. cerevisiae. Yeast cells were transformed with a plasmid expressing a construct encoding a human membrane protein tagged with green fluorescent protein. Expression was driven from a constitutive promoter and cells were imaged using confocal microscopy with an upright Leica TCS SP5 system. The sample was excited with a visible argon laser at 488 nm and imaged using a 63× oil objective. The panels show confocal images with bright-field and fluorescence for (A) wild-type cells and (B) a mutant S. cerevisiae strain selected from a global screen for high yielding strains (Bonander et al., 2005). Only the mutant cells produced correctly localized protein.
YEASTS AS FIRST-CHOICE HOST CELLS IN RECOMBINANT PROTEIN PRODUCTION STRATEGIES
For the majority of researchers, E. coli is still the first host cell to be considered in any new protein production experiment; Table 1 shows it has been consistent in its usage for over 30 years, with the beginnings of a decline in the last 8 years. Large protein production initiatives such as NYSGRC1 and OPPF-UK2 use E. coli, insect, and mammalian cell-lines as routine hosts; yeast is still employed on an ad hoc basis and the reasons for that are unclear. Since individual research teams cannot typically afford the time and investment in the full range of available host systems, I propose that a laboratory with the ability to screen for the expression of recombinant genes in E. coli, S. cerevisiae, and P. pastoris would be well placed to produce most target proteins; Table 1 shows that since 2005, 85–90% of recombinant genes were expressed in these microbes. Data from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB3) show that, for soluble proteins in particular, the probability of successful expression in E. coli is sufficiently high to justify its premier position in Table 1 (Ferrer-Miralles et al., 2009). Complementing this, yeasts have the capacity to produce the most challenging proteins: Figure 1 strikingly demonstrates that the selection of a specific S. cerevisiae strain enables this type of bespoke optimization for a eukaryotic membrane protein tagged with green fluorescent protein that could not be produced in E. coli. The panels show confocal microscopy images with bright-field and fluorescence for wild-type cells and a mutant S. cerevisiae strain selected from a global screen for high yielding strains (Bonander et al., 2005). Only the mutant cells produced correctly localized protein. More broadly, it is notable that for eukaryotic membrane proteins, over half of all the structures deposited in the PDB obtained from recombinant material were from proteins synthesized in P. pastoris and S. cerevisiae (Bill et al., 2011). This lends further support to the use of these eukaryotic microbes alongside their prokaryotic counterpart for producing the majority of target proteins. Such a strategy also makes sense from a practical perspective, since working with bacteria and yeast require similar techniques, equipment, and approaches. Consequently, both hosts can be used within the same laboratory without the need for additional specialist investment. Yeasts should therefore be considered alongside E. coli at an early stage in any serious strategy to produce recombinant proteins.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Dr. Hans P. Sørensen, Taconic Europe A/S, Denmark, for his assistance with the analysis of recombinant host cell usage, Dr. Debasmita Sarkar and Charlotte Bland, Aston University, UK, for the images used in Figure 1 and Dr. Kristina Hedfalk, Gothenburg University, Sweden for critical comments on the manuscript. The confocal microscope used to generate Figure 1 is supported through the Aston Research Centre for Healthy Ageing (ARCHA).
Footnotes
REFERENCES
- Altman L. K. (1982). A new insulin given approval for use in U.S. New York Times, October 30, 1982. [Google Scholar]
- Ashe M. P., Bill R. M. (2011). Mapping the yeast host cell response to recombinant membrane protein production: relieving the biological bottlenecks. Biotechnol. J. 6 707–714 10.1002/biot.201000333 [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2 1–11 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaranayake A. D., Almo S. C. (2013). Recent advances in mammalian protein production. FEBS Lett. 10.1016/j.febslet.2013.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K., Adelantado N., Lang C., Mattanovich D., Ferrer P. (2011). Protein trafficking, ergosterol biosynthesis and membrane physics impact recombinant protein secretion in Pichia pastoris. Microb. Cell Fact. 10 93 10.1186/1475-2859-10-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawa Z., Bland C. E., Bonander N., Bora N., Cartwright S. P., Clare M., et al. (2011). Understanding the yeast host cell response to recombinant membrane protein production. Biochem. Soc. Trans. 39 719–723 10.1042/BST0390719 [DOI] [PubMed] [Google Scholar]
- Bill R. M. (2001). Yeast–a panacea for the structure-function analysis of membrane proteins? Curr. Genet. 40 157–171 10.1007/s002940100252 [DOI] [PubMed] [Google Scholar]
- Bill R. M., Henderson P. J., Iwata S., Kunji E. R., Michel H., Neutze R., et al. (2011). Overcoming barriers to membrane protein structure determination. Nat. Biotechnol. 29 335–340 10.1038/nbt.1833 [DOI] [PubMed] [Google Scholar]
- Bonander N., Bill R. M. (2009). Relieving the first bottleneck in the drug discovery pipeline: using array technologies to rationalize membrane protein production. Expert Rev. Proteomics 6 501–505 10.1586/epr.09.65 [DOI] [PubMed] [Google Scholar]
- Bonander N., Bill R. M. (2012). Optimising yeast as a host for recombinant protein production (review). Methods Mol. Biol. 866 1–9 10.1007/978-1-61779-770-5_1 [DOI] [PubMed] [Google Scholar]
- Bonander N., Darby R. A., Grgic L., Bora N., Wen J., Brogna S., et al. (2009). Altering the ribosomal subunit ratio in yeast maximizes recombinant protein yield. Microb. Cell Fact. 8 10 10.1186/1475-2859-8-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonander N., Hedfalk K., Larsson C., Mostad P., Chang C., Gustafsson L., et al. (2005). Design of improved membrane protein production experiments: quantitation of the host response. Protein Sci. 14 1729–1740 10.1110/ps.051435705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bora N., Bawa Z., Bill R. M., Wilks M. D. (2012). The implementation of a design of experiments strategy to increase recombinant protein yields in yeast (review). Methods Mol. Biol. 866 115–127 10.1007/978-1-61779-770-5_11 [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. (1973). Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. U.S.A. 70 3240–3244 10.1073/pnas.70.11.3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J. M., Barringer K. J., Hessler A. Y., Madden K. R. (1985). Pichia pastoris as a host system for transformations. Mol. Cell Biol. 5 3376–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pourcq K., De Schutter K., Callewaert N. (2010). Engineering of glycosylation in yeast and other fungi: current state and perspectives. Appl. Microbiol. Biotechnol. 87 1617–1631 10.1007/s00253-010-2721-1 [DOI] [PubMed] [Google Scholar]
- De Schutter K., Lin Y. C., Tiels P., Van Hecke A., Glinka S., Weber-Lehmann J., et al. (2009). Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotechnol. 27 561–566 10.1038/nbt.1544 [DOI] [PubMed] [Google Scholar]
- Dietzsch C., Spadiut O., Herwig C. (2011). A dynamic method based on the specific substrate uptake rate to set up a feeding strategy for Pichia pastoris. Microb. Cell Fact. 10 14 10.1186/1475-2859-10-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew D., Newstead S., Sonoda Y., Kim H., Von Heijne G., Iwata S. (2008). GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat. Protoc. 3 784–798 10.1038/nprot.2008.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Miralles N., Domingo-Espin J., Corchero J. L., Vazquez E., Villaverde A. (2009). Microbial factories for recombinant pharmaceuticals. Microb. Cell Fact. 8 17 10.1186/1475-2859-8-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeddel D. V., Kleid D. G., Bolivar F., Heyneker H. L., Yansura D. G., Crea R., et al. (1979). Expression in Escherichia coli of chemically synthesized genes for human insulin. Proc. Natl. Acad. Sci. U.S.A. 76 106–110 10.1073/pnas.76.1.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., et al. (1996). Life with 6000 genes. Science 274 563–547 10.1126/science.274.5287.546 [DOI] [PubMed] [Google Scholar]
- Goodman M. (2009). Market watch: sales of biologics to show robust growth through to 2013. Nat. Rev. Drug Discov. 8 837 10.1038/nrd3040 [DOI] [PubMed] [Google Scholar]
- Guerfal M., Claes K., Knittelfelder O., De Rycke R., Kohlwein S. D., Callewaert N. (2013). Enhanced membrane protein expression by engineering increased intracellular membrane production. Microb. Cell Fact. 12 122 10.1186/1475-2859-12-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday M., Mallucci G. R. (2014). Targeting the unfolded protein response in neurodegeneration: a new approach to therapy. Neuropharmacology 76(Pt A) 169–174 10.1016/j.neuropharm.2013.08.034 [DOI] [PubMed] [Google Scholar]
- Hamilton S. R., Bobrowicz P., Bobrowicz B., Davidson R. C., Li H., Mitchell T., et al. (2003). Production of complex human glycoproteins in yeast. Science 301 1244–1246 10.1126/science.1088166301/5637/1244 [DOI] [PubMed] [Google Scholar]
- Hamilton S. R., Davidson R. C., Sethuraman N., Nett J. H., Jiang Y., Rios S., et al. (2006). Humanization of yeast to produce complex terminally sialylated glycoproteins. Science 313 1441–1443 10.1126/science.1130256 [DOI] [PubMed] [Google Scholar]
- Hedfalk K. (2013). Further advances in the production of membrane proteins in Pichia pastoris. Bioengineered. 4 363–367 10.4161/bioe.23886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W. J., Darby R. A., Wilks M. D., Smith R., Bill R. M. (2009). Developing a scalable model of recombinant protein yield from Pichia pastoris: the influence of culture conditions, biomass and induction regime. Microb. Cell Fact. 8 35 10.1186/1475-2859-8-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura K., Hirose T., Crea R., Riggs A. D., Heyneker H. L., Bolivar F., et al. (1977). Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science 198 1056–1063 10.1126/science.412251 [DOI] [PubMed] [Google Scholar]
- Jazini M., Herwig C. (2013). Effects of temperature shifts and oscillations on recombinant protein production expressed in Escherichia coli. Bioprocess Biosyst. Eng. 36 1571–1577 10.1007/s00449-013-0927-1 [DOI] [PubMed] [Google Scholar]
- Kitson S. M., Mullen W., Cogdell R. J., Bill R. M., Fraser N. J. (2011). GPCR production in a novel yeast strain that makes cholesterol-like sterols. Methods 55 287–292 10.1016/j.ymeth.2011.09.023 [DOI] [PubMed] [Google Scholar]
- Kunert R., Casanova E. (2013). Recent advances in recombinant protein production: BAC-based expression vectors, the bigger the better. Bioengineered 4 258–261 10.4161/bioe.24060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Mattanovich D., Villaverde A. (2012). Systems metabolic engineering, industrial biotechnology and microbial cell factories. Microb. Cell Fact. 11 156 10.1186/1475-2859-11-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L., Liu L., Petranovic D., Nielsen J. (2012). Pharmaceutical protein production by yeast: towards production of human blood proteins by microbial fermentation. Curr. Opin. Biotechnol. 23 965–971 10.1016/j.copbio.2012.03.011 [DOI] [PubMed] [Google Scholar]
- Mattanovich D., Branduardi P., Dato L., Gasser B., Sauer M., Porro D. (2012). Recombinant protein production in yeasts. Methods Mol. Biol. 824 329–358 10.1007/978-1-61779-433-9_17 [DOI] [PubMed] [Google Scholar]
- Norden K., Agemark M., Danielson J. A., Alexandersson E., Kjellbom P., Johanson U. (2011). Increasing gene dosage greatly enhances recombinant expression of aquaporins in Pichia pastoris. BMC Biotechnol. 11:47 10.1186/1472-6750-11-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg F., Sjohamn J., Conner M. T., Bill R. M., Hedfalk K. (2011). Improving recombinant eukaryotic membrane protein yields in Pichia pastoris: the importance of codon optimization and clone selection. Mol. Membr. Biol. 28 398–411 10.3109/09687688.2011.602219 [DOI] [PubMed] [Google Scholar]
- Patil C., Walter P. (2001). Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13 349–355 10.1016/S0955-0674(00)00219-2 [DOI] [PubMed] [Google Scholar]
- Porro D., Gasser B., Fossati T., Maurer M., Branduardi P., Sauer M., et al. (2011). Production of recombinant proteins and metabolites in yeasts: when are these systems better than bacterial production systems? Appl. Microbiol. Biotechnol. 89 939–948 10.1007/s00253-010-3019-z [DOI] [PubMed] [Google Scholar]
- Prielhofer R., Maurer M., Klein J., Wenger J., Kiziak C., Gasser B., et al. (2013). Induction without methanol: novel regulated promoters enable high-level expression in Pichia pastoris. Microb. Cell Fact. 12 5 10.1186/1475-2859-12-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebnegger C., Graf A. B., Valli M., Steiger M. G., Gasser B., Maurer M., et al. (2013). In Pichia pastoris, growth rate regulates protein synthesis and secretion, mating and stress response. Biotechnol. J. 10.1002/biot.201300334 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. J., Kummer L., Tremmel D., Pluckthun A. (2013). Stabilizing membrane proteins through protein engineering. Curr. Opin. Chem. Biol. 17 427–435 10.1016/j.cbpa.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Sørensen H. P. (2010). Towards universal systems for recombinant gene expression. Microb. Cell Fact. 9 27 10.1186/1475-2859-9-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadiut O., Zalai D., Dietzsch C., Herwig C. (2013). Quantitative comparison of dynamic physiological feeding profiles for recombinant protein production with Pichia pastoris. Bioprocess Biosyst. Eng. 10.1007/s00449-013-1087-z [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thim L., Hansen M. T., Norris K., Hoegh I., Boel E., Forstrom J., et al. (1986). Secretion and processing of insulin precursors in yeast. Proc. Natl. Acad. Sci. U.S.A. 83 6766–6770 10.1073/pnas.83.18.6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxlmayr M. W., Obinger C. (2012). Directed evolution of proteins for increased stability and expression using yeast display. Arch. Biochem. Biophys. 526 174–180 10.1016/j.abb.2012.04.022 [DOI] [PubMed] [Google Scholar]
- Valderrama-Rincon J. D., Fisher A. C., Merritt J. H., Fan Y. Y., Reading C. A., Chhiba K., et al. (2012). An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat. Chem. Biol. 8 434–436 10.1038/nchembio.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann M., Christen P. (2000). Comparison of folding rates of homologous prokaryotic and eukaryotic proteins. J. Biol. Chem. 275 18619–18622 10.1074/jbc.C000156200 [DOI] [PubMed] [Google Scholar]
- Zhu J. (2012). Mammalian cell protein expression for biopharmaceutical production. Biotechnol. Adv. 30 1158–1170 10.1016/j.biotechadv.2011.08.022 [DOI] [PubMed] [Google Scholar]