Abstract
Rapid advancements in the genetic manipulation of obligate intracellular bacterial pathogens have been made over the past two years. In this paper we attempt to summarize the work published since 2011 that documents these exciting accomplishments. While each genus comprising this diverse group of pathogens poses unique problems, requiring modifications of established techniques and the introduction of new tools, all appear amenable to genetic analysis. Significantly, the field is moving forward from a focus on the identification and development of genetic techniques to their application in addressing critical questions related to mechanisms of bacterial pathogenicity and the requirements of obligate intracellular growth.
Introduction
The obligate intracellular bacterial pathogens encompass a diverse group of genera (Anaplasma, Chlamydia, Coxiella, Ehrlichia, Orientia, Rickettsia) that are responsible for significant human disease both in terms of incidence (Chlamydia, Orientia) and severity (Rickettsia). These pathogens, with one recent exception, can replicate only within a eukaryotic host cell, but exploit different intracellular niches and use a variety of mechanisms for subverting the host cell's innate immune systems. Several genera (Anaplasma, Chlamydia, Coxiella, Ehrlichia) remodel the phagosome to prevent lysosomal fusion and promote bacterial growth within a sustaining vacuole, while others (Rickettsia, Orientia) find it more advantageous to escape the confines of a membranous enclosure and reside directly within the cytosol of the host cell. In contrast, Coxiella prefers the harsh acidic environment and flourishes within the phagolysosome. In 2009, ground breaking studies achieved host-cell-free growth of C. burnetii in a defined medium [1]. This “rescue from obligatism” [2] could mark its banishment from the group. However, due to its importance as a model system for the development and use of genetic systems and to the discussions surrounding what defines a facultative pathogen [3], we have retained it, for the purpose of this update, as a member of the host-cell dependent group. Finally, some of these bacteria (Anaplasma, Chlamydia, Coxiella, Ehrlichia) display an additional layer of complexity by manifesting biphasic developmental cycles that differ in infectivity and replicative ability, and perhaps, the capacity for genetic manipulation [2].
Obviously, the obligate intracellular existence of these fascinating pathogens places severe restrictions on genetic analysis. Not only must the bacteria, in most cases, be isolated from host-cell components, but protocols must be developed that permit genetic manipulations on purified bacteria without inhibiting their infectivity. For an obligate intracellular bacterium, an inability to enter a host cell is a dead-end proposition in the true sense of the word. Despite these challenges, great strides in the development and application of genetic systems have been made. These genetic advances are well described in the outstanding 2011 comprehensive review by Beare, et al. [2], which provides a temporal benchmark for the current update. Here, following the format of this earlier review, we take each genus in turn and focus on advances made since 2011 in the development and use of genetic systems for the obligate intracellular bacteria.
Anaplasma
Anaplasma species cause the tick-vectored disease anaplasmosis in both humans (A. phagocytophilium) and cattle (A. marginale) [4]. Exploiting the availability of a Himar1 transformation system, Pierle, et al. [5] recently reported the characterization of a specific transposon mutant of A. marginale that expresses a fluorescent protein (TurboGFP) and resistance to spectinomycin/streptomycin. Interestingly, the mutant was comparable to the wild type parent strain in its ability to infect cattle, to be transmitted by the tick vector, and to persist in immune competent animals. However, it differed from the wild-type in exhibiting a slow-growth phenotype that could be documented in both in vitro culture and in vivo infections. Analysis of this phenotype permitted the investigators to compare transcriptional pathways of the mutant and wild-type strains and identify changes specifically associated with the slow growth of the transformed A. marginale mutant. This work provides an excellent example of how the availability of genetic tools to generate specific mutants allows investigators to address the connection between a mutation and a significant intracellular growth phenotype of an obligate pathogen.
Chlamydia
Within the obligate intracellular group, chlamydial disease represents a major health burden, with Chlamydia trachomatis infections representing the most frequently reported sexually transmitted infections in the United States [6]. A concise synopsis of chlamydial research by Rockey and Valdivia provides an excellent starting point for those unfamiliar with the impact, history, and progression of chlamydial research [7]. In regard to genetic analysis, progress in the development and use of genetic systems to probe chlamydial biology has been especially rapid over the past two years. Both natural chlamydial gene exchange mechanisms and artificial transformation systems have been used to address basic mechanisms of chlamydial obligate intracellular growth and pathogenicity.
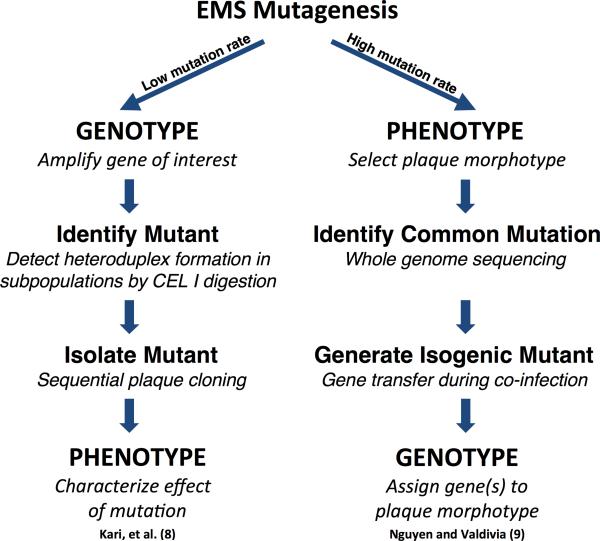
Two notable strategies exemplify the rapid progress made in isolating chlamydial chromosomal gene mutants. Both use chemical mutagenesis but differ in that one focuses on the phenotype associated with a targeted gene, while the other identifies genes associated with a targeted phenotype (Figure 1). Kari, et al. [8] made a major leap forward by employing, as noted by the authors, a labor-intensive approach, but one that can be used to target potentially any gene. The strategy uses low-level ethyl methanesulfonate (EMS) treatment to generate a preferred mutation frequency of one mutation per chlamydial genome. Subpopulations are generated and DNA isolated from each subpopulation is used as a template to amplify a target region (e.g trpAB operon). If a mutation has occurred within this region, denaturation and rehybridization of the DNA from multiple alleles yields mismatched heteroduplexes identifiable by their sensitivity to CEL I digestion. In a demonstration of this approach, the investigators generated a tryptophan synthase (trpB) null mutant isogenic in other genes to the wild-type. This mutant was sensitive to IFN-γ-induced tryptophan starvation, reinforcing the importance of tryptophan biosynthesis in chlamydial intracellular growth.
Figure 1.
A comparison of two chemical mutagenesis strategies for obtaining chlamydial chromosomal gene mutants.
In contrast, Nguyen and Valdivia [9] developed a strategy that begins with an identifiable phenotype and proceeds to identify the gene responsible. This strategy relies on a higher frequency of EMS-generated mutations per genome (3 to 20) and screening the population for mutants with common plaque phenotypes. Using whole genome sequencing the authors were able to identify specific gene mutations associated with targeted plaque phenotypes. To obtain a mutant with the desired genetic background, the authors took advantage of chlamydial interstrain gene exchange as described by DeMars and Weinfurter [10]. Jeffrey, et al. [11] characterized this genetic exchange and demonstrated that distinct phenotypes could be identified and segregated. Using this natural ability of chlamydia to exchange DNA, Nguyen and Valdivia co-infected cells with their selected mutants and wild-type chlamydiae. The resulting recombinants contained the desired mutation, were isogenic to the wild-type, and exhibited the targeted plaque phenotype. Importantly, several of these mutants exhibited altered Type II secretion and attenuated replication.
Although the Wyrick laboratory described the first chlamydial transformation system using electroporation in 1994, which was also the first reported transformation of any obligate intracellular bacterium, recombinant stability was problematic [12]. Fortunately, over the last two years, the development of transformation systems has been remarkable [13]. The breakthrough publication of Wang, et al. [14] describing the development of a shuttle vector that combined the chlamydial endogenous plasmid with an Escherichia coli plasmid origin of replication provided a solid foundation for further genetic analyses. The shuttle vector carries a β-lactamase gene for selection and was introduced into C. trachomatis Elementary Bodies (EBs) using an uncomplicated, room temperature CaCl2 protocol. With these innovations, the authors not only demonstrated the expression of a foreign protein in C. trachomatis (GFP), but also confirmed that the plasmid could restore the ability to store glycogen within inclusions (glycogen staining phenotype), confirming that this property is plasmid specific. This work established a model genetic system that is rapidly being characterized and exploited [15,16]. In a logical extension of plasmid characterization studies, Song, et al. [17], further examined the role of the endogenous chlamydial plasmid by generating derivatives that contained individual deletions of eight protein-encoding plasmid genes. Using the transformation system developed by Wang, et al. [14], each derivative was introduced into a plasmidless strain and transformants characterized for known phenotypes. For example, transformation with a plasmid containing a mutation in the pgp4 gene generated transformants that phenotypically mimicked a plasmidless strain in the appearance of abnormal inclusions and lack of glycogen accumulation. Following the isolation of this mutant, the authors examined transcriptional changes revealing that Pgp4 is a transcriptional regulator of a number of both plasmid and chromosomal genes, several of which are associated with virulence. Together these papers clearly demonstrate how the development of genetic tools for a previously intractable pathogen enables studies that can dissect the mechanisms of pathogenicity and intracellular growth. In addition, they set the stage for further vector characterization and development of protocols for targeted mutagenesis of chromosomal genes.
Most recently, Gerard, et al. developed a novel method for chlamydial transformation that does not require purification of the bacteria away from its host cell [18]. Instead, their method exploits the ability of dendrimers, synthetic molecules that can serve as a nucleic acid delivery vehicle, to introduce DNA into chlamydiae that are growing within the chlamydial host-cell inclusion. Previously, this group used dendrimers to deliver anti-sense oligonucleotides into chlamydiae to knockdown specific gene expression [19]. The authors were able to demonstrate attenuated expression of targeted genes, although the effect had a relatively limited duration. In their most recent publication, dendrimers were used to introduce a plasmid into C. pneumoniae. Using PCR they detected the vector through at least five rounds of harvesting EBs and reinfections. According to the authors, the dendrimer system can be used to generate specific gene knockouts of chromosomal genes, which would represent a significant advancement in chlamydial genetics.
Finally, following their success in developing an axenic medium capable of supporting Coxiella growth, the Hackstadt laboratory has developed a medium that supports chlamydial survival as well as metabolic and biosynthetic activities [20]. While replication and sustained growth has not yet been reported, development of an axenic growth medium would, as it did for Coxiella, provide a more versatile genetic tool box.
Coxiella
As mentioned in the Introduction and in the section above, one way to overcome the barriers associated with genetic manipulation of host-dependent bacteria is to release them from the restrictive bonds of intracellular replication and formulate a medium that will support axenic growth [2,21]. Genetic manipulation of Coxiella burnetii, the causative agent of Q fever, is discussed in detail in a recent book chapter written by Beare [22]. He describes the growing sophistication of Coxiella gene systems as they developed from host cell-associated techniques through the development of an axenic medium, to the use of classical plating and cloning techniques. The power of this latest development was recently demonstrated by the Heinzen laboratory [23] who described the development of two systems for generating gene deletions. Using suicide plasmids encoding SacB for positive selection, the authors developed two gene knockout strategies. One involved a multi-plasmid Cre-lox mediated recombination followed by sucrose counterselection while the other employed a single-plasmid, loop-in/loop-out strategy. The application of these techniques resulted in a significant milestone, the first targeted gene-specific deletions in Coxiella.
Ehrlichia
In 2013, the Ganta and Munderloh laboratories reported on a genetic system for generating mutants of Ehrlichia chaffeensis, the causative agent of human monocytic ehrlichiosis (HME) [24]. This paper exemplifies the steps needed to develop a useful system, which includes identification of an effective antibiotic selection, construction of plasmid vectors, and generation of electroporation protocols. Interestingly, while targeted mutagenesis techniques that relied on homologous recombination generated the desired recombinants, as confirmed by PCR, the recombinants were only stable in culture for 8 days. This underscores the difficulties in identifying and isolating viable mutants. However, random mutagenesis, using the Himar1 transposon, was more successful. For the first time, stable, insertional mutants of E. chaffeensis were identified and, most importantly, associated with a phenotype; the inability to infect their natural deer reservoir and be acquired by the tick vector.
Orientia
Genetic systems for the analysis of Orientia tsutsugamushi, the causative agent of scrub typhus [25], have proved elusive. Orientia is noted for its genome plasticity and the presence of large numbers of repeats, transposons, and conjugative elements [26-28]. In addition, sequence comparisons between different isolates have revealed a high level of recombination and genetic diversity [28]. This strongly suggests horizontal gene transfer in vivo and the existence of homologous recombination systems. Thus, many of the advances described for the other obligates should be applicable to Orientia and investigators will find a fertile field for employing genetic systems to explore the biology of an interesting and distinctive pathogen.
Rickettsia
The past two years have witnessed several major accomplishments in the use of genetic systems to dissect members of this genus, a genus that includes the causative agents of such serious human diseases as epidemic typhus and Rocky Mountain Spotted Fever. In a first for the rickettsiae, the Hackstadt laboratory, using a transposon delivery system, demonstrated complementation of a mutation in a relA/spoT-like gene that manifested as a distinct plaque phenotype [29]. This laboratory was also able to determine a transformation frequency for the transposon-based transformation of Rickettsia rickettsii (1 X 10−7) [30].
Exploiting available systems, the Welch laboratory used a modified Himar1 transposon to express a FLAG-tagged variant of the actin-polymerizing protein, RickA of R. parkeri [31]. This permitted the detection and localization of RickA using commercial antibodies. In addition, this paper described modifications of the Himar1 delivery vehicle, pMW1650 [32] that generated a plasmid (pRIE) with additional sites for the insertion of a fluorescent protein gene of choice and a gene under native promoter control [31]. These transposon modifications further expand the utility of the Himar1 transposon mutagenesis system.
Additional tools have been added to the rickettsiologist's toolbox through the pioneering studies on rickettsial plasmids by the Munderloh laboratory. Many rickettsial species harbor native plasmids and the Munderloh laboratory identified and characterized several of these plasmids in order to develop shuttle vectors for the transformation of diverse species [33]. These vectors include derivatives of a plasmid isolated from R. amblyommii that was used in the establishment of the first replicative plasmid in R. prowazekii [34]. Combining the power of site-directed mutagenesis and the ability to complement knockouts with plasmids or transposons will allow investigators to address basic questions of rickettsial gene function and essentiality.
Future Directions
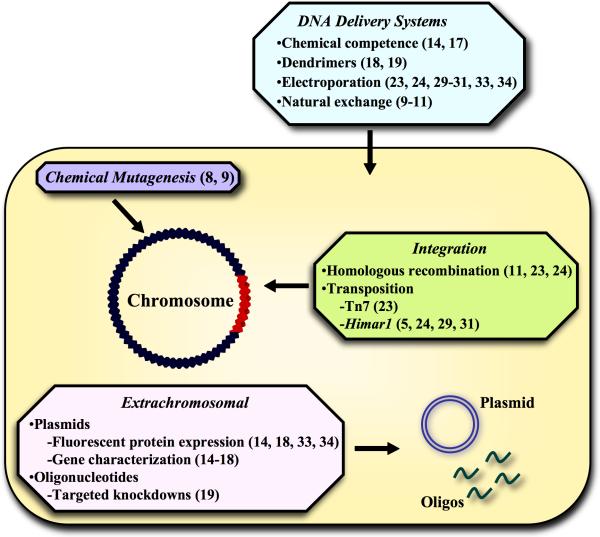
Based on the accomplishments over the past two years, it is obvious that obstacles to the genetic analysis of the obligate intracellular bacterial pathogens are becoming less formidable. We have witnessed rapid advancements in developing methods for introducing DNA, generating mutants, constructing plasmid vectors, and establishing cloning protocols (Figure 2). Each genus poses unique problems, but all appear amenable to genetic manipulation in one form or another. Certainly, for some of the obligates, problems associated with the lack of selectable markers, barriers to selection of rare transformants, and isolation of pure clones remain to be solved. In fact, the major obstacle in the genetic analysis of these bacteria is only a trivial concern when dealing with facultative bacteria; namely, the isolation of pure clones. While labor intensive methods such as limiting dilution [35] can be successful for organisms that fail to form plaques, the expression of fluorescent proteins by transformants and the detection and sorting of cells containing fluorescent bacteria using a fluorescence activated cell sorter may provide an efficient alternative.
Figure 2.
Genetic analysis of obligate intracellular pathogens; an overview of recent advances. Selected references are shown in parentheses.
Significantly, the field is moving forward from focusing on the identification and development of genetic techniques to their application in addressing critical questions of pathogenicity and intracellular growth. With tools in hand, it is now possible to develop newer methods for manipulating gene expression of these organisms. These include the tagging of proteins with domains that would allow their selective degradation (e.g. dd-degradation domain [36]) or the expression of a system that could selectively regulate specific gene expression such as CRISPR/Cas [37,38]. While the necessity of a eukaryotic host cell for bacterial replication and growth will always make the study of obligate intracellular bacteria technically more difficult, this lifestyle no longer excludes any of these pathogens from the illuminating scrutiny of the bacterial geneticist.
Highlights.
We review recent studies on genetic systems of obligate intracellular pathogens.
We identify key advances in the use of genetic systems over the past two years.
We speculate on future directions in the genetic analysis of the obligate pathogens.
Acknowledgments
Work in D.O. Wood's laboratory is supported by National Institutes of Health (NIH) grants AI020384 and AI103272. We thank Paul Brett, Mary Burtnick, and John Foster for critical reading of the manuscript and for insightful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
*of special interest
**outstanding interest
- 1.Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci USA. 2009;106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2**.Beare PA, Sandoz KM, Omsland A, Rockey DD, Heinzen RA. Advances in genetic manipulation of obligate intracellular bacterial pathogens. Front Microbiol. 2011;2:97. doi: 10.3389/fmicb.2011.00097. [The authors provide an outstanding, comprehensive review of the challenges faced when attempting to transform obligate intracellular bacteria and detail the advances made for each genus.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silva MT. Classical labeling of bacterial pathogens according to their lifestyle in the host: inconsistencies and alternatives. Front Microbiol. 2012;3:71. doi: 10.3389/fmicb.2012.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rikihisa Y. Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin Microbiol Rev. 2011;24:469–489. doi: 10.1128/CMR.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5*.Pierle SA, Hammac GK, Palmer GH, Brayton KA. Transcriptional pathways associated with the slow growth phenotype of transformed Anaplasma marginale. BMC Genomics. 2013;14:272. doi: 10.1186/1471-2164-14-272. [An A. marginale transformant exhibiting a slow growth phenotype was found to have significant alteration of translation, translation elongation, and purine biosynthetic pathways.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention . Chlamydia - fact sheet. Centers for Disease Control and Prevention. Atlanta, GA: http://www.cdc.gov/std/chlamydia/stdfact-chlamydia.htm. [Google Scholar]
- 7.Rockey D, Valdivia R. Microbe Magazine. American Society for Microbiology; Sep, 2012. A New Dawn for Chlamydia Research. [Google Scholar]
- 8*.Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, et al. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci USA. 2011;108:7189–7193. doi: 10.1073/pnas.1102229108. [Chemical mutagenesis was used to generate mutants with distinguishable phenotypes including a tryptophan synthase null mutant sensitive to IFN-γ-induced tryptophan starvation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9*.Nguyen BD, Valdivia RH. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci USA. 2012;109:1263–1268. doi: 10.1073/pnas.1117884109. [Chemical mutagenesis was used to generate mutants with distinguishable phenotypes including an attenuated strain with a Type II secretion defect.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMars R, Weinfurter J. Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance. J Bacteriol. 2008;190:1605–1614. doi: 10.1128/JB.01592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeffrey BM, Suchland RJ, Eriksen SG, Sandoz KM, Rockey DD. Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiol. 2013;13:142. doi: 10.1186/1471-2180-13-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam JE, Davis CH, Wyrick PB. Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can J Microbiol. 1994;40:583–591. doi: 10.1139/m94-093. [DOI] [PubMed] [Google Scholar]
- 13.Hudson AP. A major advance in elucidating the biology/pathobiology of Chlamydia trachomatis. Infect Immun. 2013;81:622–624. doi: 10.1128/IAI.00012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14**.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 2011;7:e1002258. doi: 10.1371/journal.ppat.1002258. [The authors describe a simple method for transformation of chlamydial elementary bodies and performed proof-of-principle experiments with shuttle vectors.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Cutcliffe LT, Skilton RJ, Persson K, Bjartling C, Clarke IN. Transformation of a plasmid-free, genital tract isolate of Chlamydia trachomatis with a plasmid vector carrying a deletion in CDS6 revealed that this gene regulates inclusion phenotype. Pathog Dis. 2013;67:100–103. doi: 10.1111/2049-632X.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Persson K, Bjartling C, Clarke IN. Genetic transformation of a clinical (genital tract), plasmid-free isolate of Chlamydia trachomatis: engineering the plasmid as a cloning vector. PLoS One. 2013;8:e59195. doi: 10.1371/journal.pone.0059195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, et al. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun. 2013;81:636–644. doi: 10.1128/IAI.01305-12. [Employing an elementary body transformation system, the authors dissect the function of plasmid-encoded genes of the endogenous chlamydial plasmid and associate a specific gene with a virulence phenotype.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Gerard HC, Mishra MK, Mao G, Wang S, Hali M, Whittum-Hudson JA, Kannan RM, Hudson AP. Dendrimer-enabled DNA delivery and transformation of Chlamydia pneumoniae. Nanomedicine: NBM. 2013;xx1-13 doi: 10.1016/j.nano.2013.04.004. [Using a dendrimer DNA delivery system, the authors transform C. pneumoniae growing within chlamydial inclusions.] [DOI] [PubMed] [Google Scholar]
- 19.Mishra MK, Gerard HC, Whittum-Hudson JA, Hudson AP, Kannan RM. Dendrimer-enabled modulation of gene expression in Chlamydia trachomatis. Mol Pharm. 2012;9:413–421. doi: 10.1021/mp200512f. [DOI] [PubMed] [Google Scholar]
- 20.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci USA. 2012;109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omsland A, Heinzen RA. Life on the outside: the rescue of Coxiella burnetii from its host cell. Annu Rev Microbiol. 2011;65:111–128. doi: 10.1146/annurev-micro-090110-102927. [DOI] [PubMed] [Google Scholar]
- 22.Beare PA. Genetic manipulation of Coxiella burnetii. Adv Exp Med Biol. 2012;984:249–271. doi: 10.1007/978-94-007-4315-1_13. [DOI] [PubMed] [Google Scholar]
- 23**.Beare PA, Larson CL, Gilk SD, Heinzen RA. Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol. 2012;78:4580–4589. doi: 10.1128/AEM.00881-12. [Building on advances in axenic growth, the authors describe the first targeted gene deletions in C. burnetii.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Cheng C, Nair AD, Indukuri VV, Gong S, Felsheim RF, Jaworski D, Munderloh UG, Ganta RR. Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog. 2013;9:e1003171. doi: 10.1371/journal.ppat.1003171. [This is the first report of the transformation of E. chaffeensis and the isolation of insertional mutants that exhibit a defined infection phenotype.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapsang AG, Bhattacharyya P. Scrub typhus. Indian J Anaesth. 2013;57:127–134. doi: 10.4103/0019-5049.111835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama K, Yamashita A, Kurokawa K, Morimoto T, Ogawa M, Fukuhara M, Urakami H, Ohnishi M, Uchiyama I, Ogura Y, et al. The Whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008;15:185–199. doi: 10.1093/dnares/dsn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho NH, Kim HR, Lee JH, Kim SY, Kim J, Cha S, Kim SY, Darby AC, Fuxelius HH, Yin J, et al. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc Natl Acad Sci USA. 2007;104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duong V, Blassdell K, May TT, Sreyrath L, Gavotte L, Morand S, Frutos R, Buchy P. Diversity ofOrientia tsutsugamushi clinical isolates in Cambodia reveals active selection and recombination process. Infect Genet Evol. 2013;15:25–34. doi: 10.1016/j.meegid.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 29**.Clark TR, Ellison DW, Kleba B, Hackstadt T. Complementation of Rickettsia rickettsiiRelA/SpoT restores a nonlytic plaque phenotype. Infect Immun. 2011;79:1631–1637. doi: 10.1128/IAI.00048-11. [This is the first description of genetic complementation in a rickettsial species using transposon insertion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark TR, Lackey AM, Kleba B, Driskell LO, Lutter EI, Martens C, Wood DO, Hackstadt T. Transformation frequency of a mariner-based transposon in Rickettsia rickettsii. J Bacteriol. 2011;193:4993–4995. doi: 10.1128/JB.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch MD, Reed SC, Lamason RL, Serio AW. Expression of an epitope-tagged virulence protein inRickettsia parkeri using transposon insertion. PLoS One. 2012;7:e37310. doi: 10.1371/journal.pone.0037310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z-M, Tucker AM, Driskell LO, Wood DO. mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl Environ Microbiol. 2007;73:6644–6649. doi: 10.1128/AEM.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burkhardt NY, Baldridge GD, Williamson PC, Billingsley PM, Heu CC, Felsheim RF, Kurtti TJ, Munderloh UG. Development of shuttle vectors for transformation of diverse Rickettsia species. PLoS One. 2011;6:e29511. doi: 10.1371/journal.pone.0029511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood DO, Hines A, Tucker AM, Woodard A, Driskell LO, Burkhardt NY, Kurtti TJ, Baldridge GD, Munderloh UG. Establishment of a replicating plasmid in Rickettsia prowazekii. PLoS One. 2012;7:e34715. doi: 10.1371/journal.pone.0034715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rachek LI, Hines A, Tucker AM, Winkler HH, Wood DO. Transformation of Rickettsia prowazekii to erythromycin resistance encoded by the Escherichia coli ereB gene. J Bacteriol. 2000;182:3289–3291. doi: 10.1128/jb.182.11.3289-3291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madeira da Silva L, Owens KL, Murta SM, Beverley SM. Regulated expression of the Leishmania major surface virulence factor lipophosphoglycan using conditionally destabilized fusion proteins. Proc Natl Acad Sci USA. 2009;106:7583–7588. doi: 10.1073/pnas.0901698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter H, Randau L, Plagens A. Exploiting CRISPR/Cas: Interference Mechanisms and Applications. Int J Mol Sci. 2013;14:14518–14531. doi: 10.3390/ijms140714518. [DOI] [PMC free article] [PubMed] [Google Scholar]