Abstract
CRISPR-Cas systems protect prokaryotes from viruses and plasmids and function primarily as an adaptive immune system in these organisms. Recent discoveries, however, revealed unexpected roles for CRISPR loci as barriers to horizontal gene transfer and as modulators of gene expression. We review how both of these functions of CRISPR-Cas systems can affect the emergence and virulence of human bacterial pathogens.
Introduction
Outnumbered by their viral predators by a factor of 10 [1], prokaryotes have evolved a diverse assortment of defense mechanisms to survive bacterial virus (phage) attack. Historically, the best studied of such mechanisms include restriction modification, abortive infection, and toxin-antitoxin systems [2]. More recently, a prokaryotic immune system has been identified that uses small guide RNAs to combat phage and prevent the stable incorporation of mobile genetic elements [3–5]. This immune system is composed of clustered regularly interspaced short palindromic repeat (CRISPR) loci and flanking CRISPR-associated (cas) genes (Fig. 1).
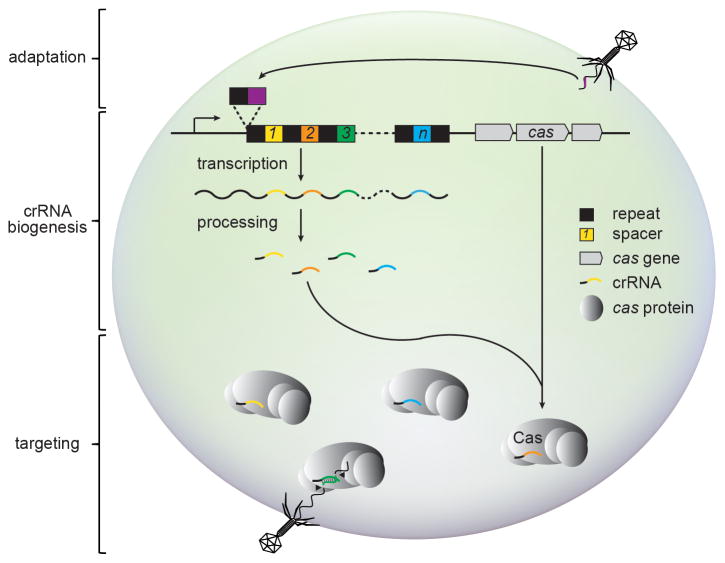
Figure 1. The three stages of CRISPR-Cas immunity.
During adaptation, foreign nucleic acids stimulate prokaryotes to insert invader-derived sequences called spacers (colored, numbered squares) in between DNA repeats (black squares) found in the CRISPR locus. During crRNA biogenesis, these repeats and spacers are transcribed into one contiguous precursor crRNA, which is processed to liberate small interfering crRNAs containing a single spacer sequence. During targeting, these mature crRNAs assemble with cas gene products and direct the destruction of matching invasive nucleic acids. CRISPR immunity against both phage and plasmid DNA, as well as RNA have been observed.
While CRISPR-Cas immune systems in different organisms exhibit marked phylogenetic and mechanistic diversity, they all block the stable entry of foreign nucleic acids in three common steps: adaptation, CRISPR RNA (crRNA) biogenesis, and targeting (Fig. 1). During adaptation, viral or plasmid challenge stimulates the incorporation of short (24–48 nucleotide) invader-derived sequences in between equally-short DNA repeats found in the CRISPR locus [6–9]. These unique sequences, called spacers, provide a historical account of past invaders and specify future targets of CRISPR immunity. Although the majority of spacers match viruses and other mobile genetic elements, many also match chromosomal sequences [10,11]. Therefore CRISPR-Cas systems seem incapable of discriminating between DNA molecules of different origins and have been proposed to act as a barrier to horizontal gene transfer [12]. During crRNA biogenesis, transcription of the CRISPR locus generates a long RNA precursor containing repeats and spacers in one contiguous array. This precursor is cleaved within repeats to separate individual spacers and liberate mature crRNAs that each define a single nucleic acid target [13–17]. During targeting, crRNAs assemble with Cas proteins to form a surveillance complex, which destroys invading genetic elements antisense to the crRNA it carries [18–20].
Considerable variations of the CRISPR immunity pathway and the Cas proteins that execute it have been observed in different organisms, thus spurring efforts to unite these systems under one simple classification scheme. Recent comparative analyses of Cas protein sequences, cas gene content, and genomic organization of CRISPR-Cas loci have lead to their consolidation into three distinct types (I-III) and eleven further subtypes (IA-F, IIA-C and IIIA-B) [21–23]. The only genes universally present in all CRISPR-Cas systems are cas1 and cas2, which together mediate adaptation [9]. CrRNA biogenesis and targeting require various other cas genes, and specific signature genes have been identified among these to distinguish each type and subtype [24].
Figure 2 depicts the families of Cas proteins that mediate crRNA biogenesis and targeting in each of the three main Types of CRISPR-Cas systems. Types I and III systems exhibit the most similarities (Fig. 2 A and C). In both systems, crRNA biogenesis requires Cas6 family members to cleave the repeat sequences of the crRNA precursor, and targeting is facilitated by a large, multi-subunit ribonucleoprotein complex. The targeting complex in Type I systems is called CASCADE (CRISPR-associated complex for antiviral defense) [13], which contains a signature subunit from the Cas8 family, along with members of Cas5, Cas6, and Cas7 families. A distinct small subunit (SS, Fig. 2A), may also be present in Type I complexes. Nucleolytic cleavage of the target DNA is carried out by a Cas3 family nuclease, which is not a member of the complex. In Type III CRISPR-Cas systems, following the cleavage of the crRNA precursor by Cas6, crRNAs are further trimmed at the 3′ end by an unknown nuclease [16,25]. The main difference between Types III-A and III-B is the chemistry of the target nucleic acid: while genetic data indicates that Type III-A CRISPR-Cas systems cleave DNA molecules in vivo [26], type III-B CRISPR-Cas systems cleave RNA targets in vitro [19]. In both cases, targeting requires a large ribonucleoprotein complex, the Cas10-Csm complex in Type III-A [27] and the CMR complex in Type III-B [19]. Both targeting complexes possess the signature subunit Cas10, along with Cas5, multiple distinct Cas7 family members, and a distinct small subunit (Fig. 2C). The nucleases that attack DNA and RNA targets are yet unknown. In contrast to Types I and III systems, Type II systems require minimal Cas machinery for immunity (Fig. 2B). For crRNA biogenesis, these systems utilize a trans-encoded CRISPR RNA (tracrRNA), a small RNA that shares partial complementarity with CRISPR repeats [15]. Pairing between the tracrRNA and the precursor crRNA generates a double-stranded substrate that is cleaved by the host-encoded RNase III to liberate the small crRNAs. Cleavage of the DNA target in Type II systems is carried out by a single large multidomain protein, Cas9. This is an RNA-guided double-stranded DNase with two independent nuclease domains, HNH and RuvC, each of which cleaves one strand of the target DNA [28].
Figure 2. The Cas protein families required for crRNA biogenesis and targeting in the three CRISPR-Cas types.
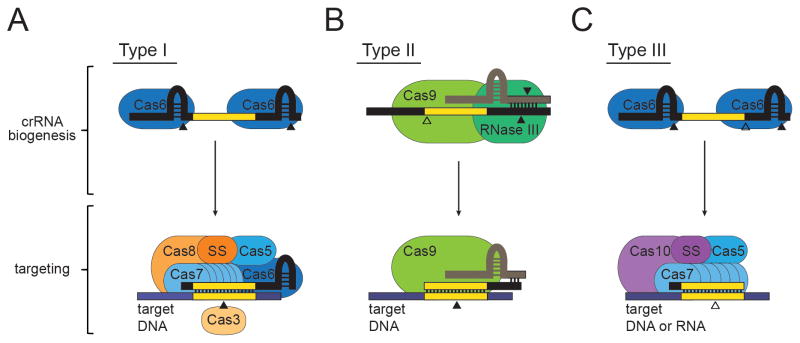
(A) In Type I CRISPR-Cas systems, a Cas6 endoribonuclease cleaves within repeat sequences (colored black) to generate mature crRNAs. CrRNAs then assemble with a targeting complex that includes Cas6, the Type I signature subunit Cas8, and members from the Cas5 and Cas7 families. An independent small subunit (SS) may be present in some subtypes, or found fused to Cas8. The DNA target (colored blue) is cleaved by Cas3, a protein that is not associated with the complex. (B) In Type II CRISPR-Cas systems, a trans-encoded CRISPR-RNA (tracrRNA, colored beige) binds crRNA repeats through base-pair complementarity, and facilitates RNase III-mediated cleavage of both RNAs. CrRNA biogenesis requires additional cleavage by an unknown nuclease to generate mature crRNAs. A single large multi-domain protein, Cas9, is required for both crRNA biogenesis and DNA target cleavage. (C) In Type III CRISPR-Cas systems, crRNA biogenesis is accomplished by Cas6-mediated cleavage within repeats and additional trimming by an unknown nuclease. The Type III targeting complex contains Cas10, the large signature subunit, Cas5, and distinct members of the Cas7 family. An additional subtype-specific small subunit (SS) is also a present in Type III complexes. Targeting against DNA and RNA in Type III systems is catalyzed by an unknown nuclease. Solid arrows represent nucleolytic cleavage events carried out by the protein on top of which they appear, and open arrows indicate cleavage events carried out by unknown nucleases.
CRISPRs reside in bacteria and bacterial pathogens alike
Pervasive in the prokaryotic world, CRISPR-Cas systems have been identified in nearly all archaea and over 40% of bacteria, including numerous bacterial pathogens. This overall prevalence of CRISPRs in all sequenced bacteria is reflected in a subset of common bacterial pathogens: of 438 selected pathogenic strains reported in the CRISPRs database [29], a web-based tool that scores the CRISPR content in sequenced bacteria, ~45% (198) harbor at least one CRISPR system (Table 1). Type I CRISPR-Cas systems are the most common in these pathogens (37%, Table 1), congruent with the general abundance of Type I systems among all sequenced bacteria (~38%) [23]. Similarly, Type III systems are present in ~10% of the listed pathogens (compared to the 15% prevalence of Type III systems in all sequenced bacteria). In contrast, Type II systems are the least abundant in bacteria (found in ~10% of sequenced strains) [23], but seem to be twice as prevalent in our selection of human pathogens, residing in ~20% of them (Table 1). In the following sections, we review the studies that have addressed the role of CRISPR-Cas systems in human pathogens.
Table 1.
CRISPR loci and their subtypes in common bacterial pathogens
| Species | Disease | CRISPRa | Subtypeb |
|---|---|---|---|
| Bacillus anthracis | Cutaneous, pulmonary or gastrointestinal anthrax | 0/6 | |
| Bordetella pertussis | Whooping cough | 0/3 | |
| Borrelia burgdorferi | Lyme disease | 0/4 | |
| Brucella abortus, B. canis, B. melitensis, B. suis | Brucellosis | 0/14 | |
| Campylobacter jejuni | Acute enteritis | 10/11 (ref) | II-C |
| Chlamydia trachomatis | Nongonococcal urethritis, lymphogranuloma venereum, trachoma | 0/21 | |
| Chlamydophila psittaci, C. pneumoniae | Psittacosis | 0/11 | |
| Clostridium botulinum | Botulism | 14/14 | III-B |
| Clostridium difficile | Pseudomembranous colitis | 9/9 | I-B |
| Clostridium perfringens | Gas gangrene, acute food poisoning | 0/3 | |
| Clostridium tetani | Tetanus | 1/1 | I-A/Bc |
| Corynebacterium diphtheriae | Diphtheria | 13/13 | II-C, I-E |
| Enterococcus faecalis | Nosocomial infections | 4/5 | II-A |
| Escherichia coli (enterohemorrhagic (EHEC)) | diarrhea, hemorrhagic colitis, hemolytic-uremic syndrome | 14/15 | I-E |
| Francisella tularensis | Tularemia | 1/14 | II-B |
| Gardnerella vaginalis | Bacterial vaginosis | 2/3 | I-E |
| Haemophilus influenzae | Bacterial meningitis, upper respiratory tract infections, pneumonia, bronchitis | 0/9 | |
| Helicobacter pylori | Peptic ulcer, risk factor for gastric carcinoma | 17/49 | -c |
| Legionella pneumophila | Legionnaire’s Disease | 3/10 (ref) | II-B |
| Leptospira interrogans | Leptospirosis | 2/3 | -c |
| Listeria monocytogenes | Listeriosis | 19/27 | I-B, II-A |
| Mycobacterium leprae | Leprosy (Hansen’s disease) | 0/2 | |
| Mycobacterium tuberculosis | Tuberculosis | 19/19 | III-A |
| Mycoplasma pneumoniae | Mycoplasma pneumonia | 0/3 | |
| Neisseria gonorrhoeae | Gonorrhea | 1/3 | I-C |
| Neisseria meningitidis | Meningococcal disease including meningitis | 12/14 | II-C |
| Pseudomonas aeruginosa | Pseudomonas infection (localized to eye, ear, skin, urinary, respiratory or gastrointestinal tract or CNS, or systemic with bacteremia) | 5/8 | I-F |
| Rickettsia rickettsii | Rocky mountain spotted fever | 0/8 | |
| Salmonella typhi | Typhoid fever type salmonellosis (dysentery, colitis) | 3/4 | I-E |
| Salmonella typhimurium | Salmonellosis with gastroenteritis and enterocolitis | 8/9 | I-E |
| Shigella sonnei | Bacillary dysentery, Shigellosis | 2/3 | I-E |
| Staphylococcus aureus | Coagulase-positive staphylococcal infections (skin infections, acute infective endocarditis, septicemia, necrotizing pneumonia, toxic shock syndrome) | 2/39 | III-A |
| Staphylococcus epidermidis | Infections of implanted prostheses, e.g. heart valves and catheters | 1/2 (ref) | III-A |
| Staphylococcus saprophyticus | Cystitis in women | 0/1 | |
| Streptococcus agalactiae | Meningitis and septicemia in neonates, endometritis in postpartum women | 4/9 (ref) | II-A, I-C |
| Streptococcus pneumoniae | Acute bacterial pneumonia and meningitis in adults, otitis media and sinusitis in children | 0/20 | |
| Streptococcus pyogenes | Streptococcal pharyngitis, scarlet fever, rheumatic fever, impetigo, necrotizing fasciitis | 11/18 (ref) | II-A, I-C |
| Treponema pallidum | Syphilis | 0/8 | |
| Vibrio cholerae | Cholera | 1/7 | I-E |
| Yersinia pestis | Bubonic and pneumonic plague | 12/12 | I-F |
| Yersinia pseudotuberculosis | Gastroenteritis | 4/4 | I-F |
The number of CRISPR-containing strains of the total number of strains with complete genomes is shown as reported by CRISPRdb (http://crispr.u-psud.fr/crispr/). A CRISPR-containing strain is defined as one with a CRISPR locus that harbors at least two spacers. Numbers were corroborated and corrected where necessary using the distribution of CRISPR-Cas loci reported by Makarova et al. (ref).
The most predominant CRISPR subtype among strains is noted first, followed by the less prevalent. Subtypes were confirmed by indicated references.
A definitive determination cannot be made based on the available information.
Staphylococci
Staphylococci are commensal opportunistic pathogens that reside on the skin and mucous membranes of humans. In recent years, pathogenic staphylococci that are resistant to all known antibiotics have emerged in both hospital and community settings [30] and have become a major threat to public health. The primary means of antibiotic resistance dissemination in staphylococci is through the horizontal transfer of conjugative plasmids [31]. Staphylococcus epidermidis RP62a, a clinical isolate, was found to harbor a Type III CRISPR-Cas system with a spacer that matches all sequenced staphylococcal conjugative plasmids [32]. This observation underscores the potential for this system to prevent the spread of such plasmids and the antibiotic resistance genes they carry. Experimentally, CRISPR immunity in S. epidermidis was shown to limit the uptake of a conjugative plasmid harboring a mupirocin resistance cassette [26]. Considering the enormous selective pressure applied by antibiotics upon staphylococci, this finding suggested that anti-plasmid CRISPR immunity could be detrimental for the evolution of these bacteria. In other words, CRISPR immunity could prevent the dissemination of antibiotic resistance mediated by conjugative plasmids that is so common between staphylococci. Alternatively, CRISPR-Cas systems could have a mechanism to distinguish “bad” DNA (such as an invading phage) form the “good”, antibiotic resistance-carrying, plasmid DNA. To address this possibility, a recent study analyzed the few CRISPR-escape cells that are able to acquire the plasmids that confer mupirocin resistance in spite of the presence of a targeting CRISPR-Cas system in the population of recipient cells [33]. All of the “escapers” analyzed (111) contained mutations in the CRISPR-Cas locus: 50 % lost the compete CRISPR-Cas region, 21 % contained transposon insertions in the cas genes, 16 % harbored single-nucleotide deletions or substitutions that abrogated Cas function, and 13 % of the cells lost the spacer sequence that matches the conjugative plasmid. These results indicate that the CRISPR system and its target cannot coexist in the cell; the only bacteria that survive the antibiotic selection have lost a functional CRISPR-Cas system. Therefore it can be speculated that the absence of CRISPR loci in most S. aureus strains (Table 1) is a consequence of the importance of plasmids, prophages and other mobile genetic elements for the virulence of this pathogen [34,35] and the ability of CRISPR-Cas systems to attack these elements.
Pneumococci
A similar scenario could explain the lack of CRISPR-Cas loci in Streptococcus pneumoniae, a causative agent of pneumonia. Pathogenic pneumococci are covered by a polysaccharide capsule that allows the bacterium to escape the immune system. The importance of the capsule for virulence led Griffith to the discovery of natural transformation [36] and allowed Avery to determine that DNA is the molecule that carries the genetic information of the cell [37]. In both cases, experiments relied on the acquisition of the capsule synthesis genes from heat-killed encapsulated cells by non-encapsulated pneumococci. In Griffith’s experiment, while mice injected with either heat-killed encapsulated cells or live non-encapsulated cells survive infection, co-injection leads to infection due to transformation and expression of capsule synthesis genes by non-encapsulated pneumococci. Avery’s experiment measures this transformation in vitro by using antiserum raised against non-encapsulated cells that prevents their planktonic growth but allows the growth of the encapsulated transformants. Capsular transformation, also known as capsule switching, is very common in nature and is the source of the more than 90 different pneumococcal serotypes. The importance of the capsule in pathogenesis has been exploited for vaccine development. Current pneumococcal vaccines contain several different types of capsular polysaccharides and therefore S. pneumoniae undergoes a rapid evolution in response to the selective pressure posed by the introduction of the vaccines [38].
None of the 26 pneumococcal strains with completed genomes contain CRISPR sequences [29]. To understand the impact that CRISPR immunity could have in capsule switching, a recent study introduced the type II CRISPR-Cas system of Streptococcus pyogenes into S. pneumoniae R6, a non-encapsulated strain, and engineered an anti-capsule gene spacer [39]. By performing the Griffith and Avery experiments with this strain, investigators were able to demonstrate that the attack of transforming DNA by the CRISPR-Cas machinery could prevent pneumococcal capsule-switching, both in vitro and in vivo. Interestingly, “escaper” strains arise during mice infections that were able to acquire a capsule and cause disease. Similar to the case of antibiotic-resistant staphylococci CRISPR “escapers” (see above), these cells contained mutations in the cas genes that abrogated CRISPR function. Because of the artificial set up of these experiments, it is difficult to evaluate the effect of CRISPR loci in the evolution of pneumococci from their results. Nevertheless, considering the importance of natural transformation and capsule-switching for the emergence of new virulent strains of S. pneumoniae, it is tempting to speculate that the potential to acquire spacers targeting capsule synthesis or other virulence genes could have contributed to the selection against CRISPR-Cas loci in pneumococci.
Enterococci
Enterococcus faecalis and Enterococcus faecium, while natural inhabitants of the human digestive tract, are among the most common causes of antibiotic resistant hospital-acquired infections [40,41]. These organisms exhibit a propensity to incorporate mobile genetic elements such as independently replicating plasmids, prophages, and pathogenicity islands, all of which can constitute up to 25% of the genome [42]. These mobile genetic elements are rife with antibiotic resistance genes and virulence factors [43,44]. Noting the preponderance of horizontally acquired elements in these organisms and the function of CRISPR-Cas system of targeting these elements, studies explored the co-distribution of CRISPR loci and the presence of antibiotic resistance genes in E. faecalis and E. faecium strains [42,45]. While many strains harbor Type II CRISPR systems, there is a correlation between the absence of CRISPR loci and the presence of drug resistance genes. Many of the strains containing CRISPR loci were isolated prior to the widespread use of antibiotics, supporting the notion that CRISPR-Cas loss of function can occur in response to antibiotic use [42].
Group A Streptococci
S. pyogenes, or Group A Streptococci (GAS), is a major human pathogen that causes a variety of infections including pharyngitis, sepsis, and necrotizing fasciitis [46]. S. pyogenes strains are unique in that they have acquired a plethora of toxins and virulence factors by means of multiple prophage integrations [47]. The prophage content of S. pyogenes is believed to be a major determinant of the type of disease caused by specific strains and that the exchange and acquisition of prophages over time results in periodic shifts of disease manifestation and increased virulence [48]. Group A streptococci usually carry two CRISPR-Cas loci in their genomes, one type II-B, which has been shown to be functional [15], and another type I-C. A bioinformatics analysis examining the 13 available GAS genome sequences revealed that eight contained at least one CRISPR-Cas systems [49]. Interestingly, the strains that lack CRISPR sequences have the highest number of prophages (Table 2). More importantly, analysis of the CRISPR targets shows a mutually exclusive relationship between CRISPR spacer sequences and their prophage targets (Table 2 and [49]). This suggests that CRISPR function antagonizes prophage insertion and therefore influences the evolution as well as the type of disease caused by this pathogen.
Table 2.
CRISPR and prophage content of GAS genomes.
| Strain(a) | Serotype | CRISPR(b) | Prophages in the genome(c) | Prophages targeted by CRISPR(d) | Ref. | |
|---|---|---|---|---|---|---|
| SF370 | M1 | 9 | 370.1 – 4 | 10270.1, 2 315.2, 3, 4, 5 SPsP2, 3, 4, 5 10750.1, 2, 3 10394.3, 4, 5 |
9429.2 8232.2, 3, 5 6180.2 NZ131.3 |
[61] |
| MGAS9429 | M12 | 9 | 9429.1 – 3 | 5005.1, 2 315.1, 2 SPsP5, 6 Man.1, 4 |
8232.1, 2,
5 6180.1 NZ131.3 |
[62] |
| NZ131 | M49 | 9 | NZ131.1 - 3 | 370.1,
3 5005.2 10270.3 315.3, 4 SPsP3, 4 10750.3 |
Man.1, 2 10394.4, 5, 6 2096.1 8232.2, 4, 5 6180.2 |
[63] |
| MGAS2096 | M12 | 8 | 2096.1 – 2 | 5005.1 315.1, 2 SPsP5 Man.1, 4 |
8232.1, 2,
5 6180.1 NZ131.3 |
[62] |
| MGAS5005 | M1 | 7 | 5005.1 – 3 | 370.1 10270.1, 2, 3 315.2, 4, 5 SPsP2, 4, 5 10750.1, 2 |
Man.1, 3,
4 10394.6 9429.1 8232.2, 5 NZ131.3 |
[64] |
| MGAS10270 | M2 | 6 | 10270.1 – 5 | 370.1 315.1, 2, 3 SPsP3, 4, 5 10750.3 |
Man.2, 3, 4 10394.3, 5 8232.1, 2, 3 6180.1 |
[62] |
| MGAS6180 | M28 | 5 | 6180.1 – 4 | 370.2, 3 5005.2 315.3, 5 SPsP2, 4, 5 |
10750.3 10394.4, 5 9424.2 8232.3, 4, 5 |
[65] |
| MGAS10750 | M4 | 5 | 10750.1 – 4 | 370.3 315.5 SPsP2 |
Man.1 8232.4 |
[62] |
| MGAS315 | M3 | - | 315.1 – 6 | - | [66] | |
| SSI-1 | M3 | - | SPsP1 – 6 | - | [67] | |
| Manfredo | M5 | - | Man.1 – 5 | - | [68] | |
| MGAS10394 | M6 | - | 10394-1 – 8 | - | [69] | |
| MGAS8232 | M18 | - | 8232.1 – 5 | - | [70] | |
Strains are ordered from highest to lowest total number of CRISPR spacers.
Number of spacers is indicated.
Prophages and prophage remnants are indicated.
Prophages carry the name of the host strain followed by a serial number. Note that there is a mutually exclusive relationship between CRISPR-Cas systems and prophage content. For example, the CRISPR spacers present in strain SF370 match prophages present in other strains, but none of the 370.1-4 endogenous prophages.
Pseudomonas
Pseudomonas aeruginosa is an opportunistic human pathogen that colonizes immune-compromised patients such as those with cystic fibrosis, cancer, or AIDS. Their pathology relies upon the formation of biofilms, surface-attached bacterial communities that can become more resistant to antibiotics than their planktonic counterparts by up to three orders of magnitude [50]. A recent study showed that about 37% of 122 P. aeruginosa clinical isolates harbored Type I CRISPR-Cas systems [51], that are functional against many clinical and environmental bacteriophage isolates [52]. The temperate phage DMS3, however, is able to lysogenize its Pseudomonas host despite the presence of a functional CRISPR-Cas system that targets it [53]. It was later demonstrated that DMS3 escape from CRISPR immunity occurs as a result of imperfect complementarity between the CRISPR spacer and the targeted region on the phage genome [52]. This phage is important because its lysogenization inhibits biofilm formation and swarming motility the P. aeruginosa PA14 clinical isolate. Strikingly, this inhibition requires an intact CRISPR-Cas system. While the mechanism underlying this CRISPR-Cas mediated biofilm elimination is still being investigated, the available data indicates that the biofilm defect relies upon an imperfect match between a CRISPR spacer and the DMS3-42 gene in the phage genome [53,54]. The DMS3-42 protein is not required for inhibition of biofilms formation, suggesting that the interaction between the CRISPR-Cas machinery and the DMS3-42 target affects the expression of the prophage genes that mediate the inhibition. This study highlights the potential for CRISPR-Cas systems to modulate bacterial physiology in unexpected ways that might mitigate their pathogenesis.
Francisella
Francisella tularensis, the causative agent of tularemia, is an intracellular pathogen known for its lethality to humans and potential for use as a biological weapon [55]. The less-virulent subspecies novicida is thought to be an opportunistic pathogen, and has been found to harbor a Type II CRISPR-Cas system [56]. This CRISPR-Cas system has been shown to directly enhance F. novicida virulence by chromosomal gene regulation and influence the outcome of a murine model of infection [57,58]. Critical for innate immunity against F. novicida are eukaryotic pattern-recognition receptors, which recognize bacterial lipoproteins and initiate a pro-inflammatory host response. The Type II CRISPR-Cas system in F. novicida was shown to use a unique small CRISPR-Cas associated RNA (scaRNA), distinct from the tracrRNA and crRNA, to repress an endogenous transcript encoding a bacterial lipoprotein. While the tracrRNA harbors the sequence complementarity to the lipoprotein transcript that mediates the down-regulation of this gene, the scaRNA has homology to tracrRNA sequences and is required for repression. These RNA-RNA interactions, mediated by Cas9, result in a reduction of the lipoprotein transcript, leading to less expression of this antigen and a decrease in the host pro-inflammatory response. F. novicida strains harboring mutations in the CRISPR-Cas system or the scaRNA were attenuated during murine infection, and mice vaccinated with these attenuated strains were protected from future infection. This seminal study establishes a direct role for CRISPR-Cas in regulating the interaction between the bacteria with its eukaryotic host.
Conclusions
The specific cases described in this review reveal a love-hate relationship between bacterial pathogens and their CRISPR-Cas systems. On the one hand, the ability of CRISPR immunity to prevent the transfer of antibiotic resistance and virulence genes into pathogens can reduce the evolvability of the pathogen. On the other, CRISPR-Cas systems can be repurposed to regulate gene expression and enhance pathogenesis. The application of novel sequencing technologies to epidemiological studies will allow us to understand the full effect of CRISPR-Cas loci on the evolution of important human pathogens such as S. pneumoniae, S. aureus and E. fecaelis. The role of CRISPR-Cas systems in the modulation of virulence is certainly a new and exciting area of research. In addition to the case of F. novicida, a number of recent observations support the influence of CRISPR immunity on host-pathogen interactions, although the mechanisms remain obscure. For example, deletion of cas9 from the Type II CRISPR-Cas system in Nisseria meningitidis resulted in its reduced ability to adhere to, invade, and replicate within human epithelial cells, leading to a defect in survival of the pathogen [57]. Similarly, Cas9 was shown to be critical for host interactions in Campylobacter jejuni [59]. These effects are most likely mediated by a scaRNA, however the gene(s) down-regulated are unknown. Also Cas2, a component of the Type II CRISPR-Cas adaptation machinery in Legionella pneumophilia, was shown to facilitate infection of its amoeba host, a parasitic interaction that is required for L. pneumophilia proliferation in aqueous environments and transmission to humans [60]. Taken together, these examples demonstrate the remarkable versatility of CRISPR-Cas systems and warrant future investigations focused on the roles of CRISPR-Cas that extend beyond the canonical interference against foreign nucleic acids.
Highlights.
CRISPR-Cas systems protect bacteria from bacteriophages and other mobile genetic elements.
CRISPR-Cas systems can be barriers to horizontal gene transfer, reducing the evolvability of pathogens.
CRISPR-Cas systems can increase virulence by modulating gene expression.
These different functions have opposite effects on the survival of different pathogens.
As a result, CRISPR-Cas systems are not universal in bacterial pathogens.
Acknowledgments
L.A.M is supported by the Searle Scholars Program, the Rita Allen Scholars Program, an Irma T. Hirschl Award, a Sinsheimer Foundation Award and a NIH Director’s New Innovator Award (1DP2AI104556-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brussow H, Hendrix RW. Phage genomics: small is beautiful. Cell. 2002;108:13–16. doi: 10.1016/s0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 2.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 3.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 5.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 6**.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. Seminal paper demosntrating the function of CRISPR sequences as an adaptive immune system of bacteria. [DOI] [PubMed] [Google Scholar]
- 7*.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. This work (along with reference # 8, elucidated the importance of partial matches between existing spacers and invading elements for the acquisition process. [DOI] [PubMed] [Google Scholar]
- 8.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodt A, Lurie-Weinberger MN, Gophna U. CRISPR loci reveal networks of gene exchange in archaea. Biology direct. 2011;6:65. doi: 10.1186/1745-6150-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. Seminal paper that small crRNAs within a ribonucleoprotein complex mediate CRISPR immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. Discovery of the crRNA biogenesis mechanism for type II CRISPR-Cas systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci US A. 2011;108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 19*.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. Demonstration that the CMR complex can cleave RNA molecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23**.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. Important paper that unified the classification and nomenclature of the very diverse CRISPR-Cas systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koonin EV, Makarova KS. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013;10:679–686. doi: 10.4161/rna.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. This work demonstrated that CRISPR-Cas systems can prevent plasmid conjugation by targeting DNA but not RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatoum-Aslan A, Samai P, Maniv I, Jiang W, Marraffini LA. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem. 2013;288:27888–27897. doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuya EY, Lowy FD. Antimicrobial-resistant bacteria in the community setting. Nat Rev Microbiol. 2006;4:36–45. doi: 10.1038/nrmicro1325. [DOI] [PubMed] [Google Scholar]
- 31.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 32.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. Dealing with the Evolutionary Downside of CRISPR Immunity: Bacteria and Beneficial Plasmids. PLoS Genet. 2013;9:e1003844. doi: 10.1371/journal.pgen.1003844. This paper explores the consequences of CRISPR immunity against beneficial plasmids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 36.Griffith F. The significance of pneumococcal types. J Hyg. 1928;27:113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Top J, Willems R, Bonten M. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS immunology and medical microbiology. 2008;52:297–308. doi: 10.1111/j.1574-695X.2008.00383.x. [DOI] [PubMed] [Google Scholar]
- 41.Isnard C, Malbruny B, Leclercq R, Cattoir V. Genetic Basis for In Vitro and In Vivo Resistance to Lincosamides, Streptogramins A, and Pleuromutilins (LSAP Phenotype) in Enterococcus faecium. Antimicrobial agents and chemotherapy. 2013;57:4463–4469. doi: 10.1128/AAC.01030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. mBio. 2010;1:e00227–00210. doi: 10.1128/mBio.00227-10. This work revealed a negative correlation between the CRISPR and antibiotic resistance gene content in enterococci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 44.Shankar N, Baghdayan AS, Gilmore MS. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature. 2002;417:746–750. doi: 10.1038/nature00802. [DOI] [PubMed] [Google Scholar]
- 45.Burley KM, Sedgley CM. CRISPR-Cas, a prokaryotic adaptive immune system, in endodontic, oral, and multidrug-resistant hospital-acquired Enterococcus faecalis. J Endod. 2012;38:1511–1515. doi: 10.1016/j.joen.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Bisno AL. Group A streptococcal infections and acute rheumatic fever. The New England journal of medicine. 1991;325:783–793. doi: 10.1056/NEJM199109123251106. [DOI] [PubMed] [Google Scholar]
- 47.Beres SB, Musser JM. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One. 2007;2:e800. doi: 10.1371/journal.pone.0000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banks DJ, Beres SB, Musser JM. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 2002;10:515–521. doi: 10.1016/s0966-842x(02)02461-7. [DOI] [PubMed] [Google Scholar]
- 49.Nozawa T, Furukawa N, Aikawa C, Watanabe T, Haobam B, Kurokawa K, Maruyama F, Nakagawa I. CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS One. 2011;6:e19543. doi: 10.1371/journal.pone.0019543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 51.Cady KC, White AS, Hammond JH, Abendroth MD, Karthikeyan RS, Lalitha P, Zegans ME, O’Toole GA. Prevalence, conservation and functional analysis of Yersinia and Escherichia CRISPR regions in clinical Pseudomonas aeruginosa isolates. Microbiology. 2011;157:430–437. doi: 10.1099/mic.0.045732-0. [DOI] [PubMed] [Google Scholar]
- 52.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O’Toole GA. The CRISPR/Cas Adaptive Immune System of Pseudomonas aeruginosa Mediates Resistance to Naturally Occurring and Engineered Phages. J Bacteriol. 2012;194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O’Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54*.Cady KC, O’Toole GA. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol. 2011;193:3433–3445. doi: 10.1128/JB.01411-10. This works shows how mismatches between the crRNA and its target can prevent DNA cleavage but affect gene expression and virulence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, et al. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 56.Schunder E, Rydzewski K, Grunow R, Heuner K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol. 2013;303:51–60. doi: 10.1016/j.ijmm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 57**.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. Seminal work showing the repurposing of CRISPR-Cas systems for the regulation of gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. Corrigendum: A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;501:262. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Louwen R, Horst-Kreft D, de Boer AG, van der Graaf L, de Knegt G, Hamersma M, Heikema AP, Timms AR, Jacobs BC, Wagenaar JA, et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. Eur J Clin Microbiol Infect Dis. 2013;32:207–226. doi: 10.1007/s10096-012-1733-4. [DOI] [PubMed] [Google Scholar]
- 60.Gunderson FF, Cianciotto NP. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. mBio. 2013;4:e00074–00013. doi: 10.1128/mBio.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci US A. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McShan WM, Ferretti JJ, Karasawa T, Suvorov AN, Lin S, Qin B, Jia H, Kenton S, Najar F, Wu H, et al. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. Journal of bacteriology. 2008;190:7773–7785. doi: 10.1128/JB.00672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. The Journal of infectious diseases. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 65.Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, LeFebvre RB, Musser JM. Genome sequence of a serotype M28 strain of group a streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. The Journal of infectious diseases. 2005;192:760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- 66.Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, Liu MY, Smoot JC, Porcella SF, Parkins LD, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakagawa I, Kurokawa K, Yamashita A, Nakata M, Tomiyasu Y, Okahashi N, Kawabata S, Yamazaki K, Shiba T, Yasunaga T, et al. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome research. 2003;13:1042–1055. doi: 10.1101/gr.1096703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holden MT, Scott A, Cherevach I, Chillingworth T, Churcher C, Cronin A, Dowd L, Feltwell T, Hamlin N, Holroyd S, et al. Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain manfredo. Journal of bacteriology. 2007;189:1473–1477. doi: 10.1128/JB.01227-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Banks DJ, Porcella SF, Barbian KD, Beres SB, Philips LE, Voyich JM, DeLeo FR, Martin JM, Somerville GA, Musser JM. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. The Journal of infectious diseases. 2004;190:727–738. doi: 10.1086/422697. [DOI] [PubMed] [Google Scholar]
- 70.Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, Sylva GL, Sturdevant DE, Ricklefs SM, Porcella SF, Parkins LD, et al. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4668–4673. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]