Abstract
Significance: It is a well-established scientific observation that mammalian cells contain fidelity or watchdog proteins that maintain the correct function of cellular organelles. Recent Advances: Over the past several years, the Sirtuin deacetylase family protein Sirt3 has emerged as a mitochondrial fidelity protein that directs energy generation and regulates reactive oxygen species (ROS) scavenging proteins. Loss of function or genetic mutation of these fidelity proteins has been shown to create a cellular environment that is permissive for the development of cellular damage associated with processes such as aging and carcinogenesis. Critical Issues: Mitochondria are the primary organelles that direct oxidative metabolism for the production of ATP; however, this is also a significant source of ROS. Thus, it is reasonable to propose that mitochondria should contain proteins that would signal downstream target molecules and/or ROS scavenger enzymes to maintain mitochondrial and cellular homeostatic poise. It is also reasonable to hypothesize that the mitochondria contain fidelity proteins similar to those found in the nucleus and cytoplasm. We discuss a new role of Sirt3 in the direction of the primary superoxide scavenger protein, manganese superoxide dismutase (MnSOD), and how the acetylation or deacetylation of several specific lysines appears to direct MnSOD enzymatic dismutase activity. Future Directions: Aberrant downstream regulation of MnSOD by Sirt3 may be a potential source of cellular damage that accumulates with aging to create a tumor-permissive phenotype. Future studies can explore the role of MnSOD in age-related illness using this new mechanism of enzymatic regulation. Antioxid. Redox Signal. 20, 1646–1654
Introduction
A connection between aging and human cancers
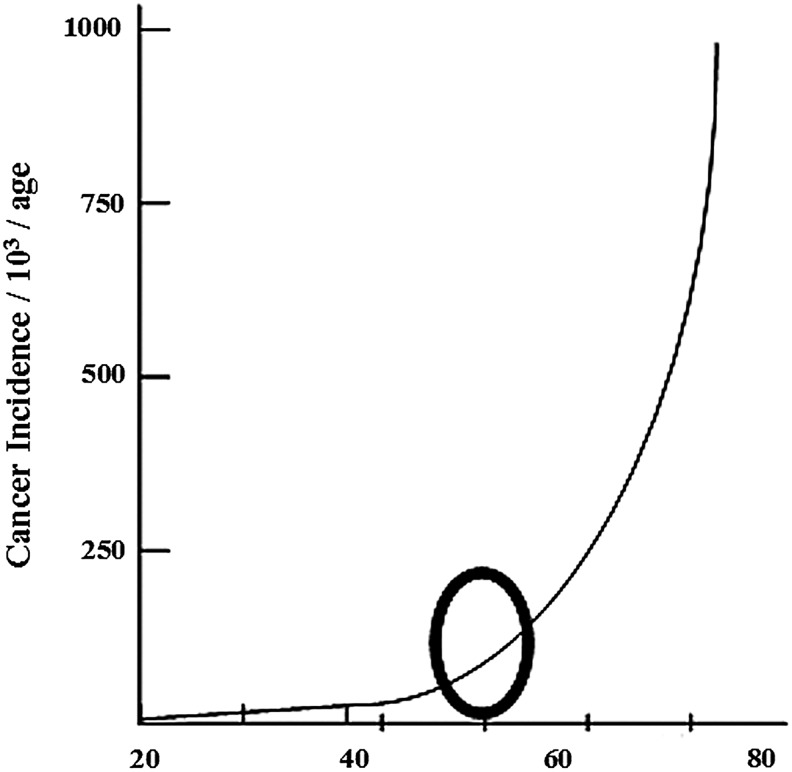
Over the last 20 years, it has become clear that solid tumors in humans have a very strong connection to aging. In fact, increasing age is the most significant statistical variable associated with carcinogenesis for solid tumors arising from somatic cells (14). The incidence of human cancers as a function of age is shown in Figure 1. The curve maintains a relatively flat initial slope (Fig. 1, arrow 1), followed by an inflection point around the age of 50 (Fig. 1, back circle), from which the incidence of cancers begins to rise. This is then followed by a steep increase in incidence after age 50 (Fig. 1, arrow 2). Based on these observations, it seems reasonable to suggest that there are significant changes in certain critical biological processes and/or cellular reparative pathways that occur at this inflection point resulting in a new organismal phenotype that is permissive for the development of events favoring carcinogenesis. As such, our laboratory and many others are interested in the changes that occur in the cell at this critical inflection point that mark the transition to the tumor-permissive phenotype.
FIG. 1.
Cancer incidence rises with age. The incidence of solid tumor cancers derived from somatic cells increases exponentially with age. The circle marks the inflection point at the transition between the flat slope and sharp rise.
The longevity curve for humans relating to cancer incidence
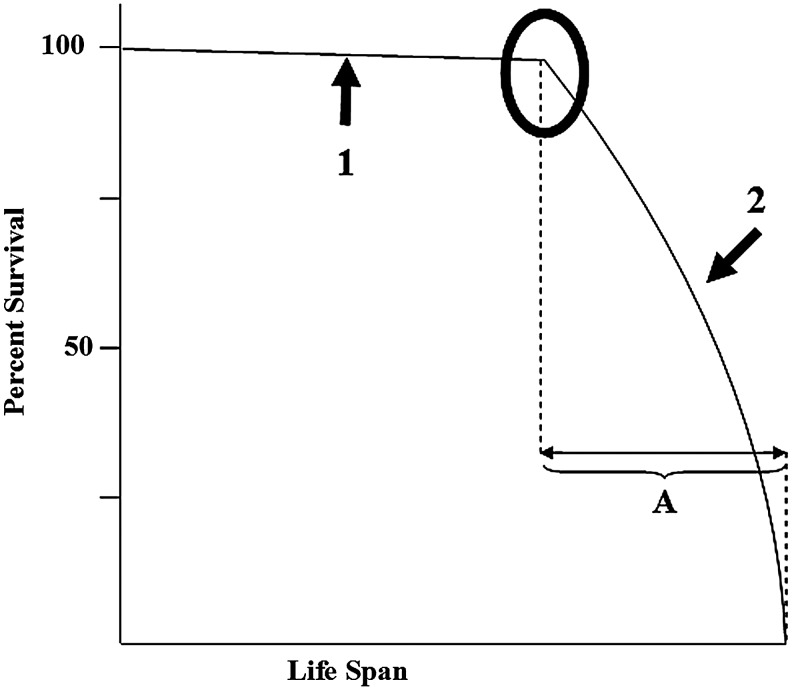
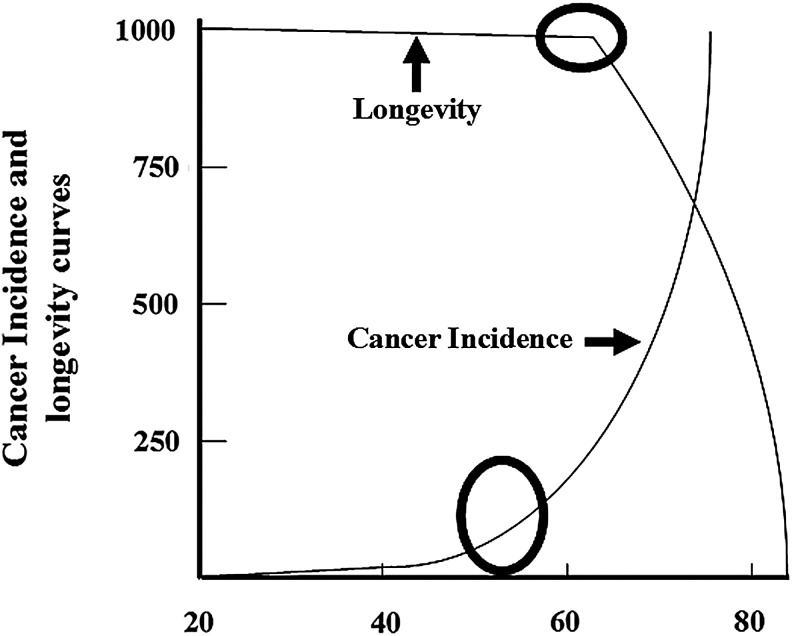
Aging is a universal process characterized by a decline in an organism's fertility and probability of survival. Although it occurs in every organism, the means to quantitatively measure its underlying biologic processes remains elusive. Whereas not the only metric, quantifying patterns of longevity is one basic method to critically investigate the processes of aging (27). Longevity is the statistical probability that an organism survives over time and can be represented by curves illustrating the probability of survival across different life spans. Although the maximum life span between species varies enormously, a common pattern emerges from longevity curves across various species ranging from Caenorhabditis elegans to humans (25, 49–51). This curve, similar to the curve in Figure 1, also has a relatively flat initial slope (Fig. 2, arrow 1), representing the high chance of survival at birth and early life. This is followed by an inflection point (Fig. 2, black circle) after which the slope of the curve changes rapidly, representing a steep decline in the probability of survival before the maximal longevity of a species reaches its limit (Fig. 2, arrow 2). The overall pattern of the inflection point followed by the second slope (Fig. 2, shown as region A, where A represents the time from the inflection point to maximum longevity) appears to be present in most species (25, 51). Whereas the curves in Figures 1 and 2 are inverted, the same pattern is evident: both start with a flat slope followed by a sharp transition to a steep slope marked by an inflection point (Fig. 3, black circle). This raises a rather interesting question: do the inherent cellular reparative biological processes that direct longevity play a role in the observed increased incidence of solid tumors and are these alterations occurring at the inflection point?
FIG. 2.
Example longevity curve. The longevity curve shows the probability of survival over the course of a life span. The circle marks the inflection point. Arrow 1 points to the flat slope and arrow 2 points to the fast decrease in survival probability at the maximum life span. The region labeled “A” shows a pattern observed across different organisms.
FIG. 3.
Cancer incidence and longevity curves superimposed to show their common patterns. Although the curves are inverted, they both exhibit a flat region followed by the inflection point (marked by the circle) and a rapid rise/fall.
Sirtuins as a gene family that plays a role in aging
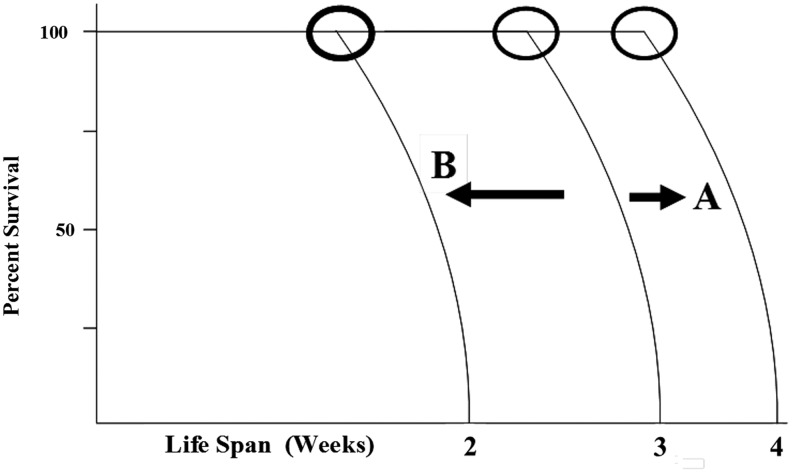
Sirtuins are present in almost all species and catalyze the deacetylation of a variety of substrates ranging from transcription factors to metabolic enzymes (39, 41, 47, 83). Over the past decade, they have come to particular attention as the human and murine homologs of the Saccharomyces cerevisiae Sir2 gene have been shown to regulate both the replicative and overall lifespan in model organisms, including C. elegans and Drosophila melanogaster (18, 25, 47, 66, 90). In an elegant set of experiments performed by many laboratories, including the Guarente group, (6, 25, 26), it appears that the sirtuin genes play a role, at least in part, in directing the longevity of C. elegans. The data summarized demonstrate that the overexpression of sirtuin genes increases the longevity curve in a very specific manner by moving the inflection point and extending the length of the initial flat slope of the survival curve (Fig. 4, Arrow A). In addition, deletion of the sirtuin genes in C. elegans has the expected and opposite effect by decreasing the length of the initial flat slope of the survival curve, while the steep portion following the inflection point remains roughly the same (Fig. 4, arrow B). Interestingly, the length of survival from the inflection point to the point of maximal longevity (as shown in Fig. 2, designated as A) is unchanged. It has also been proposed that the levels of mammalian sirtuin activity will decrease with increasing age. Whereas this has not yet been published, we have preliminary data with specific SIRT3 target anti-lysine-acetyl antibodies suggesting that downstream SIRT3 targets exhibit an increase in lysine acetylation with increasing age (data not shown). Based on these observations, it is reasonable to suggest that the family of sirtuin genes plays a role in directing the longevity time span at the species inflection point, as do many other proteins important to aging (25, 50, 62).
FIG. 4.
The effect of sirtuin gene expression on life span. This is the graphical summary of data obtained from increased or decreased sirtuin expression in Caenorhabditis elegans. Overexpression of sirtuin genes results in increases in the life span (arrow A), whereas underexpression of these genes shortened the life span (arrow B). The time of the inflection point (circle) is shifted, but the general shape of the survival curve remains unchanged.
Finally, when the longevity data presented in Figures 2 and 4 are compared to the cancer incidence curve (Figs. 1 and 3), it prompts us to hypothesize that the biological cell processes directing the length of the initial longevity curve at the inflection point might also direct the inflection point for cancer incidence. As such, we asked whether mammalian sirtuins play a role in the cellular genetic and biochemical events that prevent a pro-aging phenotype and the establishment of a tumor-permissive phenotype in mammals (42–44, 63, 64, 84, 87). Ten years ago, we constructed our first murine sirtuin knockout mouse and performed a series of experiments to test this hypothesis. Based on the results from this murine model, we present evidence to support our conclusion that sirtuins do play an important role in linking aging and carcinogenesis.
Sirt3 is a mitochondrial metabolism fidelity protein
Sirtuins are present in almost all species and catalyze the deacetylation of a variety of substrates ranging from transcription factors to metabolic enzymes (39, 41, 47, 83). In humans, the sirtuin homologs include seven NAD+-dependent, type III histone and protein deacetylases (39, 57). These proteins share a common 275-amino acid catalytic domain and are localized to the nucleus (SIRT1, 6, and 7), mitochondria (SIRT3, 4, and 5), and cytoplasm (SIRT2) (29). Whereas these genes have not been shown to determine longevity in mammals, they regulate critical signaling networks, and mice lacking several of the sirtuin genes develop illnesses that closely mimic human diseases that have strong statistical, genetic, and biochemical connections to increasing age (13, 24, 54, 90). The proteins encoded by these genes require NAD+ as a cofactor and are induced by caloric restriction and fasting, suggesting that they function as cellular metabolic sensors and subsequently induce signaling cascades that adjust processes governing metabolic homeostasis. Therefore, it has been proposed that the mammalian sirtuins play a role, at least in part, in directing the acetylome signaling network that is critical to regulating cellular processes mechanistically connected to aging (63, 64, 76).
The sirtuin family of proteins appears to monitor and direct the integrity of cellular metabolism and respond to stressful conditions or perhaps, better said, these proteins respond to nutrient stress by activating compensatory pathways (55, 85, 89). Interestingly, these proteins have also been shown to play a role, at least in part, in the cellular process of aging (22–26). It has been proposed that loss of function and/or genetic mutation of these fidelity proteins create a cellular environment that is permissive for the development of disease, which can lead to human illnesses connected to aging such as carcinogenesis (40, 41, 83). As discussed above, it is well known that the risk of developing many human cancers increases exponentially with age. These observations suggest that sirtuin fidelity proteins may function as tumor suppressors (TS) in the context of aging and metabolism (12, 38, 68, 69), providing a mechanistic link between the processes critical to aging, metabolism, and carcinogenesis. As it is unlikely that mammalian cells are under direct evolutionary pressure to develop proteins specifically against carcinogenesis, sirtuins are more likely guardian proteins that have evolved over time to sense specific damage induced by cellular and/or nutrient stress and activate reparative signaling pathways involving the cell acetylome.
Among the different sirtuins, Sirt3 is particularly interesting as it is localized to the mitochondria, where many cellular metabolic and energy production processes take place. As a consequence, mitochondria are also a major source of cellular reactive oxygen species (ROS). Sirt3 is the primary mitochondrial deacetylase (53) and has been demonstrated to function as a mitochondrial TS both in vitro and in vivo (43, 84). The in vitro data can be summarized with the experiments comparing Sirt3 knockout mouse embryonic fibroblasts (MEFs) to their wild-type or heterozygous counterparts. Sirt3 knockout MEFs were shown to exhibit aberrant glucose metabolism (3, 16, 43) and generate increased levels of ROS when exposed to various types of environmental and/or cellular stress. Exposure to oxidative stress, genotoxic stress, and ionizing radiation resulted in a loss of contact inhibition in cells lacking Sirt3 (43). In addition, the Sirt3 knockout MEFs were immortalized and transformed by infection with a single oncogene, suggesting that Sirt3 is an in vitro TS. These results were extended in vivo, whereby mice lacking Sirt3 were found to spontaneously develop estrogen receptor (ER)-positive mammary tumors (43). An interesting correlation is that ER-positive breast cancers in humans also tend to occur in older women. Interrogation of human breast cancer samples as measured by (1) staining for protein levels; (2) RT-PCR for RNA levels; and (3) genomic analysis all showed that SIRT3 levels were statistically significantly decreased in tumor cells as compared to normal tissue controls. Given these results, we proposed that SIRT3 is a genomically expressed, mitochondrially localized TS protein (43). A key question in SIRT3 biology was then to determine the downstream targets that are aberrantly regulated when Sirt3 is deleted, resulting in the observed tumor-permissive cellular phenotype. We proposed that MnSOD may be one such protein.
MnSOD is a critical enzyme in the defense against oxidative stress
Almost every complex cell utilizes mitochondrial aerobic respiration as an energy source. However, as a result of using oxygen to generate ATP, mitochondria generate ROS as a by-product. One such extremely damaging ROS molecule is the superoxide radical (O2−), which can react with DNA, lipids, and other biomolecules to induce mutagenesis, senescence, and ultimately cause cell injury or cell death (17, 20, 75). Hence, mitochondria have evolved mechanisms to process ROS into less toxic substances and maintain metabolic homeostasis (19). The primary mitochondrial mechanism to eliminate or maintain the correct levels of O2− involves the evolutionarily conserved manganese superoxide dismutase (MnSOD) protein (61, 80, 81). MnSOD, also known as SOD2, is found across organisms and localized to mitochondria in eukaryotes (60, 61).
MnSOD has long been regarded as a simple scavenging enzyme with its activity thought to be only stoichiometrically dependent on levels of superoxide in the mitochondria (36, 37, 48). However, it now appears that the MnSOD activity is regulated by a variety of diverse cellular mechanisms. Depending on the intracellular signals or environmental triggers, the MnSOD activity may undergo transcriptional, translational, and, perhaps most importantly, post-translational regulation. Various inflammatory cytokines such as TNF-alpha, interleukin (IL)-1, and lipopolysaccharide have been shown to activate transcription of MnSOD (88). RNA-binding proteins have also been shown to increase translation of MnSOD mRNA (9, 11). Because post-translational modification of proteins is the most established mechanism to direct and regulate enzymatic activity, it seems reasonable to suggest that this may also be the case with MnSOD.
The mitochondrial acetylome directs MnSOD enzymatic activity
A fundamental paradigm in biology is that intracellular sensing proteins recognize specific cellular conditions and initiate post-translational signaling cascades (5, 19, 21, 28). The most common example of this is the cytoplasmic activation of sensing kinases that phosphorylate a series of downstream targets in response to different environmental conditions, thereby minimizing any potential permanent and detrimental cellular effects (31, 46). In this regard, lysine acetylation has recently emerged as an important and perhaps primary post-translational modification mechanism employed to regulate mitochondrial proteins (8, 45, 46, 53). Thus, we proposed that mitochondria also rely upon sensing proteins, such as sirtuins, to maintain mitochondrial homeostasis through changes in downstream target protein acetylation (33, 44). As acetylation appears to play such a significant role in the regulation of mitochondrial protein function, we hypothesized that acetylation may regulate the MnSOD enzymatic activity as well.
On this topic, three manuscripts have been published over the course of 6 months, all of which demonstrated that MnSOD contains several reversible acetyl lysines and that acetylation alters its enzymatic function (7, 65, 84). These groups also showed using somewhat different models that loss of Sirt3 resulted in increased intracellular and mitochondrial superoxide levels, whereas overexpression of Sirt3 decreased cellular ROS and mitochondrial superoxide levels. In addition, it was uniformly shown that MnSOD was directly deacetylated by SIRT3 in cell-free, in vitro, and in vivo (murine) model systems. The reintroduction of the wild-type, but not the deacetylation-null, Sirt3 gene decreased MnSOD acetylation and increased MnSOD activity. Although superoxide levels were higher in cells lacking Sirt3, there was no decrease in the total MnSOD protein levels. Finally, Sirt3 and MnSOD have also been shown to physically interact within the mitochondria. The totality of these results clearly demonstrates that the functional enzymatic activity of MnSOD is determined by acetylation. Sirt3 plays a critical role in driving this process, and, similar to other proteins, MnSOD is also regulated by post-translational modifications.
The acetylation status of MnSOD lysines 68 and 122 directs enzymatic activity
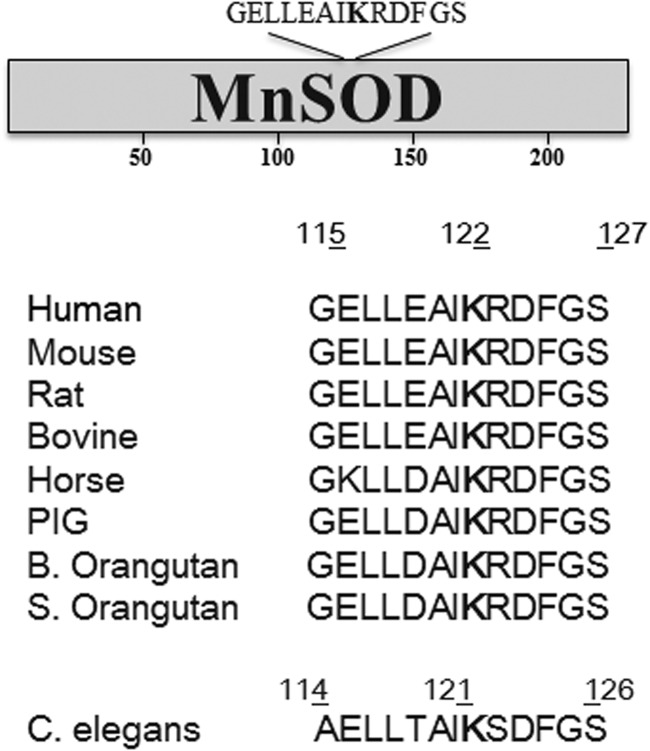
We then hypothesized that MnSOD may contain specific lysine residues, which could be targets of acetylation/deacetylation. In this regard, we found that MnSOD contains lysine residues that are reversibly acetylated and demonstrated that MnSOD acetylation was increased in the livers of Sirt3 knockout mice, while the total MnSOD protein levels remained the same compared to Sirt3 wild-type mice (84). Additionally, we found that a specific 13 amino acid motif (GELLEAIK*RDFGS) containing an evolutionarily conserved lysine residue at amino acid position 122 in the enzyme was especially conserved among multiple mammalian and nonmammalian species (Fig. 5).
FIG. 5.
Evolutionary conservation of the MnSOD target with a central lysine. A 13 amino acid motif containing an evolutionarily conserved lysine at amino acid 122 is shown.
Examining the protein structure of MnSOD demonstrated that lysine 122 is located on the outside of the tetramer complex, where it can be easily targeted for deacetylation (Fig. 6) (84). Using site-directed mutagenesis, we created MnSOD mutants at lysine 122, replacing it with an arginine residue to mimic the positively charged deacetylated form of the enzyme. This increased the MnSOD enzymatic activity and decreased intracellular superoxide levels. When lysine 122 was replaced with a glutamine residue that carries a neutral charge, mimicking the acetylated form of the enzyme, the MnSOD activity decreased and increased superoxide levels were found in the cell. Given that the lysine residue is located near the entrance to the active site of MnSOD, we proposed that the mechanism of increased enzymatic activity is due to the attraction of the negatively charged superoxide anion toward the positively charged lysine residues (93). The deacetylated lysine residues create an electrostatic funnel propelling superoxide into the catalytic core of the enzyme. This mechanism of electrostatic facilitation was initially proposed in 1983 by Irwin Fridovich and colleagues (4, 93) who were the pioneers of research on MnSOD, and can now be explained by acetylation/deacetylation as the mechanism at the direct substrate level (93).
FIG. 6.
Protein structure of MnSOD showing the location of lysine 122 residues positioned on the outside of the tetramer complex. The position of the evolutionarily conserved lysine allows it to be a feasible target for SIRT3 deacetylation.
Similar results were shown for MnSOD lysine 68, which was also deacetylated in a series of cell-free, in vitro, and in vivo techniques (7, 65). These results showed that SOD2 is acetylated at lysine 68 and that this acetylation decreases the SOD2 activity. SIRT3 was again shown to deacetylate MnSOD resulting in an increase in the SOD2 enzymatic activity. In addition, it was demonstrated that agents or conditions that increased intracellular ROS levels also stimulated SIRT3 transcription. The results of these three manuscripts present SIRT3-dependent deacetylation of SOD2 as a novel post-translational regulation in response to oxidative stress (7, 65, 84).
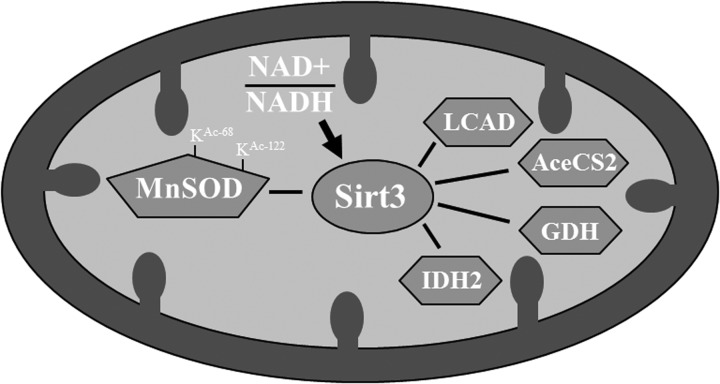
Other groups have demonstrated that Sirt3 is the primary mitochondrial protein deacetylase catalyzing the removal of lysine acetyl groups using NAD+ as a cofactor (52). Sirt3 has also been shown to upregulate mitochondrial enzymes that are critical to energy production through deacetylation of specific lysines on these proteins (Fig. 7). Acetyl-coenzyme A synthetase, long-chain acyl-coenzyme A dehydrogenase (LCAD), and isocitrate dehydrogenase (IDH) are the three of many examples of enzymes regulated by Sirt3 in response to the nutrient status of the cell (32, 34, 35, 58, 59, 71, 72, 77). IDH is a component of the mitochondrial citric acid cycle and, interestingly, Sirt3 has been show to activate IDH during caloric restriction (70, 77). Activation of IDH increases NADH generation and is proposed to increase the ratio of reduced-to-oxidized glutathione, which serves as a form of protection against oxidative stress (77). The activation of IDH by Sirt3 under conditions of caloric restriction is, thus, another example of how Sirt3 may protect the cell from oxidative stress, while sensing the nutrient status of the cell.
FIG. 7.
SIRT3 mitochondrial deacetylation targets. Data suggest that the MnSOD enzymatic activity is directed by acetylation of lysine 122 following fasting or exposure to radiation through the activation of Sirt3. It has also been proposed that the SIRT3 activity is directed and subsequently regulated by changes in cellular NAD+/NADH ratios and this alters the SIRT3 enzymatic activity. After activation, SIRT3 has also been shown to regulate the activity of other mitochondrial proteins, including acetyl-CoA synthetase (AceCS2), glutamate dehydrogenase (GDH), long-chain acyl-CoA dehydrogenase (LCAD), and isocitrate dehydrogenase (IDH2).
It has also been suggested that the SIRT3 activity is directed, at least in some part, by changes in the cellular ratio of NAD+/NADH (18, 25, 47). Whereas it is likely that other mechanisms will be identified that direct and regulate the SIRT3 activity, it is intriguing to propose that a potential feedback loop involving factors that determine NAD+ and NADH levels that may monitor the cellular energy needs and, subsequently, respond by altering the enzymatic activity to match mitochondrial energy production to cellular ATP consumption. If one extends this idea, it could be proposed that several potential mechanisms might direct SIRT3 post-translational modifications that increase the enzymatic activity and mediate the activation of MnSOD, analogous to how activation of IDH then affects the NAD+/NADH ratio. In this regard, these mitochondrial signaling processes should subsequently activate a series of downstream targets that alter the cellular phenotype resulting in a adaptive, reparative, and/or protective cellular phenotype.
Aberrant regulation of MnSOD activity leads to a damage-permissive phenotype
MnSOD is a primary mitochondrial antioxidant in a diverse network of detoxification enzymes that neutralizes mitochondrial ROS through the conversion of superoxide to hydrogen peroxide and ultimately to H2O by catalase (78). ROS, in order of sequential reduction from O2 include superoxide (O2•−), hydrogen peroxide (H2O2), the hydroxyl radical (•OH), and organic peroxides, which are normally produced during oxidative metabolism. Whereas low or physiologically normal amounts of these ROS molecules are well tolerated by the cell, abnormally high levels of ROS from any number of possible sources induce oxidative stress and can cause damage by peroxidizing lipids and disrupting proteins and nucleic acids (74, 79). Therefore, it seems logical to propose that the cell contains specific mitochondrial processes to maintain a fine balance between the mitochondrial antioxidant detoxification system and ROS production (81).
What follows from this logic is that aberrant direction of MnSOD should result in cellular phenotypes permissive for the development of pathologic conditions, including human illnesses, some of which have a strong connection to increasing age. We have demonstrated that the tumor-permissive phenotype in Sirt3−/− MEFs could be reversed when the deacetylated MnSOD was overexpressed in the cells, suggesting that the lack of MnSOD regulation by Sirt3 was a key step, at least in part, in permitting transformation (17, 44). There is also in vivo evidence supporting the critical role of MnSOD in pathogenesis, as mice lacking MnSOD cannot live beyond a few weeks of life, while the MnSOD+/− mice display higher oxidative damage and a higher incidence of tumor formation compared to their wild-type counterparts (86, 92). Additionally, an excessive amount of superoxide is believed to shorten the life span (73) and induce age-associated pathological conditions such as carcinogenesis and cardiovascular diseases (2, 56). In humans, mutations in SOD2 are also associated with age-related disorders, suggesting that upstream signaling proteins that regulate MnSOD may also play a role in these pathologies. We would suggest that SIRT3, with its direction of the mitochondrial acetylome, is one such protein.
These observations, and the results demonstrating that MnSOD is regulated by the acetylation status of specific lysines (7, 65, 84), raise several intriguing questions. Does the aberrant dysregulation of mitochondrial proteins through increased acetylation, which may naturally occur with increasing age, result in a damage-permissive phenotype? Is MnSOD just one of those that are likely to be many such mitochondrial proteins? This idea would not only suggest a critical role for both the mitochondrial and cellular acetylome in age-related illness, but also imply that increased protein acetylation results in a physiological phenotype favoring the accumulation of cell damage. Caloric restriction, on the other hand, appears to activate sirtuins favoring deacetylation and has been shown to increase the mammalian life span, while decreasing the incidence of age-related human illnesses, such as neurodegeneration, insulin resistance, cardiovascular disease, and cancer (14, 15, 54, 67). Given these findings, identifying mitochondrial proteins that are hyperacetylated may lead to the discovery of other proteins critical to the damage-permissive phenotype.
Finally, additional questions raised by these findings include as to which evolutionary tradeoffs lead to the acetylation/downregulation of critical stress–response proteins, and why this seems to be a consequence of aging. A fundamental paradox in aging biology is why aging even occurs given that natural selection should continue to drive longer lifespans and delayed senescence. If hyperacetylation favors a damage-permissive phenotype, and deacetylation favors a protective phenotype, then natural selection should theoretically drive the persistent deacetylation of mitochondrial repair proteins. Part of the answer to this paradox is that the eukaryotic cell and mitochondria are under numerous different selective pressures, and limitations exist to maintaining repair enzymes in the deacetylated/constitutively active state. As discussed above, we hypothesize that post-translational modification in the form of acetylation/deacetylation is a quick way for the cell to respond to stressful conditions when needed.
Conclusions
The results summarized above suggest that loss of a single mitochondrial protein and the aberrant regulation of the mitochondrial acetylome signaling network places the entire cell at risk for damage and may initiate the development of many human illnesses. In this regard, MnSOD, as well as several other recently identified mitochondrial proteins such as acetyl-CoA synthetase (30, 71), glutamate dehydrogenase (30, 70), LCAD(71), succinate dehydrogenase (10), Ku70 (82), mitochondrial ribosome subunit MRPL10 (91), and isocitrate dehydrogenase (77), appear to be potential downstream SIRT3 deacetylation candidates that may play a role in age-related illness. Development of anti-acetyl-lysine antibodies that recognize specific lysines in a hyperacetylated protein may identify potential molecular biomarkers that predict the risk of human illnesses. Moreover, it would be interesting to explore why aging is connected to hyperacetylation of important mitochondrial proteins and how these proteins affect the inflection points on both the aging and cancer incidence graphs. Additional understanding of the evolutionary tradeoffs resulting in age-driven acetylation of mitochondrial repair proteins could offer insight into mechanisms other than caloric restriction to delay the effects of aging. Further characterization of these seemingly esoteric mitochondrial proteins can thus lead to better understanding of human disease with potential to impact multiple areas of medicine.
Abbreviations Used
- ER
estrogen receptor
- GDH
glutamate dehydrogenase
- IDH
isocitrate dehydrogenase
- LCAD
long-chain acyl-coenzyme A dehydrogenase
- MEFs
mouse embryonic fibroblasts
- MnSOD
manganese superoxide dismutase
- ROS
reactive oxygen species
- TS
tumor suppressors
Acknowledgments
DG is supported by NCI-1R01CA152601-01, 1R01CA152799-01A1, 1R01CA168292-01A1, 1R01CA16383801A1, and BC093803 from the DOD and the Hirshberg Foundation for Pancreatic Cancer Research Seed Grant Award.
References
- 1.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, and Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A 105: 14447–14452, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban RS, Nemoto S, and Finkel T. Mitochondria, oxidants, and aging. Cell 120: 483–495, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Bell EL, Emerling BM, Ricoult SJ, and Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene 30: 2986–2996, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benovic J, Tillman T, Cudd A, and Fridovich I. Electrostatic facilitation of the reaction catalyzed by the manganese-containing and the iron-containing superoxide dismutases. Arch Biochem Biophys 221: 329–332, 1983 [DOI] [PubMed] [Google Scholar]
- 5.Bisht KS, Bradbury CM, Mattson D, Kaushal A, Sowers A, Markovina S, Ortiz KL, Sieck LK, Isaacs JS, Brechbiel MW, Mitchell JB, Neckers LM, and Gius D. Geldanamycin and 17-allylamino-17-demethoxygeldanamycin potentiate the in vitro and in vivo radiation response of cervical tumor cells via the heat shock protein 90-mediated intracellular signaling and cytotoxicity. Cancer Res 63: 8984–8995, 2003 [PubMed] [Google Scholar]
- 6.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, and Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol 4: e31, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, and Xiong Y. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep 12: 534–541, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, and Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840, 2009 [DOI] [PubMed] [Google Scholar]
- 9.Chung DJ, Wright AE, and Clerch LB. The 3′ untranslated region of manganese superoxide dismutase RNA contains a translational enhancer element. Biochemistry 37: 16298–16306, 1998 [DOI] [PubMed] [Google Scholar]
- 10.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, and Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry 49: 304–311, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clerch LB. Post-transcriptional regulation of lung antioxidant enzyme gene expression. Ann N Y Acad Sci 899: 103–111, 2000 [DOI] [PubMed] [Google Scholar]
- 12.Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci 5: 147–152, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donmez G. and Guarente L. Aging and disease: connections to sirtuins. Aging Cell 9: 285–290, 2010 [DOI] [PubMed] [Google Scholar]
- 14.Ershler WB. and Longo DL. Aging and cancer: issues of basic and clinical science. J Natl Cancer Inst 89: 1489–1497, 1997 [DOI] [PubMed] [Google Scholar]
- 15.Ershler WB. and Longo DL. The biology of aging: the current research agenda. Cancer 80: 1284–1293, 1997 [PubMed] [Google Scholar]
- 16.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, and Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell 19: 416–428, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freeman BA. and Crapo JD. Biology of disease: free radicals and tissue injury. Lab Invest 47: 412–426, 1982 [PubMed] [Google Scholar]
- 18.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273: 793–798, 2000 [DOI] [PubMed] [Google Scholar]
- 19.Gius D, Botero A, Shah S, and Curry HA. Intracellular oxidation/reduction status in the regulation of transcription factors NF-kappaB and AP-1. Toxicol Lett 106: 93–106, 1999 [DOI] [PubMed] [Google Scholar]
- 20.Gius D. and Spitz DR. Redox signaling in cancer biology. Antioxid Redox Signal 8: 1249–1252, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Gius DR, Ezhevsky SA, Becker-Hapak M, Nagahara H, Wei MC, and Dowdy SF. Transduced p16INK4a peptides inhibit hypophosphorylation of the retinoblastoma protein and cell cycle progression prior to activation of Cdk2 complexes in late G1. Cancer Res 59: 2577–2580, 1999 [PubMed] [Google Scholar]
- 22.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev 14: 1021–1026, 2000 [PubMed] [Google Scholar]
- 23.Guarente L. Ageless quest: one scientist's search for genes that prolong youth. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2003, p. 154 [Google Scholar]
- 24.Guarente L. Mitochondria—a nexus for aging, calorie restriction, and sirtuins? Cell 132: 171–176, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guarente L. and Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature 408: 255–262, 2000 [DOI] [PubMed] [Google Scholar]
- 26.Guarente L, Partridge L, and Wallace DC. Molecular biology of aging. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Pres, 2008 [Google Scholar]
- 27.Guarente L. and Picard F. Calorie restriction—the SIR2 connection. Cell 120: 473–482, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Hallahan DE, Gius D, Kuchibhotla J, Sukhatme V, Kufe DW, and Weichselbaum RR. Radiation signaling mediated by Jun activation following dissociation from a cell type-specific repressor. J Biol Chem 268: 4903–4907, 1993 [PubMed] [Google Scholar]
- 29.Hallows WC, Albaugh BN, and Denu JM. Where in the cell is SIRT3?—functional localization of an NAD+-dependent protein deacetylase. Biochem J 411: e11–e13, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallows WC, Lee S, and Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A 103: 10230–10235, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill CS. and Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell 80: 199–211, 1995 [DOI] [PubMed] [Google Scholar]
- 32.Hirschey MD, Shimazu T, Capra JA, Pollard KS, and Verdin E. SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2. Aging (Albany NY) 3: 635–642, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr., Alt FW, Kahn CR, and Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464: 121–125, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirschey MD, Shimazu T, Huang JY, and Verdin E. Acetylation of mitochondrial proteins. Methods Enzymol 457: 137–147, 2009 [DOI] [PubMed] [Google Scholar]
- 35.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr., Kahn CR, and Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell 44: 177–190, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hitchler MJ, Oberley LW, and Domann FE. Epigenetic silencing of SOD2 by histone modifications in human breast cancer cells. Free Radic Biol Med 45: 1573–1580, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y, He T, and Domann FE. Decreased expression of manganese superoxide dismutase in transformed cells is associated with increased cytosine methylation of the SOD2 gene. DNA Cell Biol 18: 643–652, 1999 [DOI] [PubMed] [Google Scholar]
- 38.Hunter T. Oncoprotein networks. Cell 88: 333–346, 1997 [DOI] [PubMed] [Google Scholar]
- 39.Imai S, Armstrong CM, Kaeberlein M, and Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800, 2000 [DOI] [PubMed] [Google Scholar]
- 40.Kaeberlein M, Jegalian B, and McVey M. AGEID: a database of aging genes and interventions. Mech Ageing Dev 123: 1115–1119, 2002 [DOI] [PubMed] [Google Scholar]
- 41.Kaeberlein M, McVey M, and Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13: 2570–2580, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, Sack MN, Jing E, Kahn CR, Friedman JE, and Jonscher KR. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J 433: 505–514, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, and Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17: 41–52, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, Ji J, Wang XW, Park SH, Cha YI, Gius D, and Deng CX. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20: 487–499, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, and Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23: 607–618, 2006 [DOI] [PubMed] [Google Scholar]
- 46.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? Embo J 19: 1176–1179, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, and Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A 97: 5807–5811, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li M, Chiu JF, Mossman BT, and Fukagawa NK. Down-regulation of manganese-superoxide dismutase through phosphorylation of FOXO3a by Akt in explanted vascular smooth muscle cells from old rats. J Biol Chem 281: 40429–40439, 2006 [DOI] [PubMed] [Google Scholar]
- 49.Libina N, Berman JR, and Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115: 489–502, 2003 [DOI] [PubMed] [Google Scholar]
- 50.Lin K, Dorman JB, Rodan A, and Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319–1322, 1997 [DOI] [PubMed] [Google Scholar]
- 51.Lin K, Hsin H, Libina N, and Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28: 139–145, 2001 [DOI] [PubMed] [Google Scholar]
- 52.Lombard DB. Sirtuins at the breaking point: SIRT6 in DNA repair. Aging (Albany NY) 1: 12–16, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr., Weissman S, Verdin E, and Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27: 8807–8814, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Longo VD. and Kennedy BK. Sirtuins in aging and age-related disease. Cell 126: 257–268, 2006 [DOI] [PubMed] [Google Scholar]
- 55.Mallakin A, Sugiyama T, Taneja P, Matise LA, Frazier DP, Choudhary M, Hawkins GA, D'Agostino RB, Jr., Willingham MC, and Inoue K. Mutually exclusive inactivation of DMP1 and ARF/p53 in lung cancer. Cancer Cell 12: 381–394, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merry BJ. Oxidative stress and mitochondrial function with aging—the effects of calorie restriction. Aging Cell 3: 7–12, 2004 [DOI] [PubMed] [Google Scholar]
- 57.Michishita E, Park JY, Burneskis JM, Barrett JC, and Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16: 4623–4635, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.North BJ, Marshall BL, Borra MT, Denu JM, and Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11: 437–444, 2003 [DOI] [PubMed] [Google Scholar]
- 59.North BJ. and Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol 5: 224, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oberley LW. and Buettner GR. Role of superoxide dismutase in cancer: a review. Cancer Res 39: 1141–1149, 1979 [PubMed] [Google Scholar]
- 61.Oberley LW. and Oberley TD. Role of antioxidant enzymes in cell immortalization and transformation. Mol Cell Biochem 84: 147–153, 1988 [DOI] [PubMed] [Google Scholar]
- 62.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, and Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999, 1997 [DOI] [PubMed] [Google Scholar]
- 63.Ozden O, Park SH, Kim HS, Jiang H, Coleman MC, Spitz DR, and Gius D. Acetylation of MnSOD directs enzymatic activity responding to cellular nutrient status or oxidative stress. Aging (Albany NY) 3: 102–107, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park SH, Ozden O, Jiang H, Cha YI, Pennington JD, Aykin-Burns N, Spitz DR, Gius D, and Kim HS. Sirt3, Mitochondrial ROS, Ageing, and Carcinogenesis. Int J Mol Sci 12: 6226–6239, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiu X, Brown K, Hirschey MD, Verdin E, and Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 12: 662–667, 2010 [DOI] [PubMed] [Google Scholar]
- 66.Rogina B. and Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A 101: 15998–16003, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MS, Ohlsson R, Longo DL, and Feinberg AP. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science 307: 1976–1978, 2005 [DOI] [PubMed] [Google Scholar]
- 68.Saunders LR. and Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene 26: 5489–5504, 2007 [DOI] [PubMed] [Google Scholar]
- 69.Saunders LR. and Verdin E. Cell biology. Stress response and aging. Science 323: 1021–1022, 2009 [DOI] [PubMed] [Google Scholar]
- 70.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, and Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol 382: 790–801, 2008 [DOI] [PubMed] [Google Scholar]
- 71.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, and Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A 103: 10224–10229, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwer B, North BJ, Frye RA, Ott M, and Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 158: 647–657, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sebastian C. and Mostoslavsky R. SIRT3 in calorie restriction: can you hear me now? Cell 143: 667–668, 2010 [DOI] [PubMed] [Google Scholar]
- 74.Sies H. Oxidative stress: from basic research to clinical application. Am J Med 91: 31S–38S, 1991 [DOI] [PubMed] [Google Scholar]
- 75.Singh KK. Mitochondria damage checkpoint, aging, and cancer. Ann N Y Acad Sci 1067: 182–190, 2006 [DOI] [PubMed] [Google Scholar]
- 76.Smith KT. and Workman JL. Introducing the acetylome. Nat Biotechnol 27: 917–919, 2009 [DOI] [PubMed] [Google Scholar]
- 77.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, and Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143: 802–812, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spitz DR, Azzam EI, Li JJ, and Gius D. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: a unifying concept in stress response biology. Cancer Metastasis Rev 23: 311–322, 2004 [DOI] [PubMed] [Google Scholar]
- 79.Spitz DR. and Li GC. Heat-induced cytotoxicity in H2O2-resistant Chinese hamster fibroblasts. J Cell Physiol 142: 255–260, 1990 [DOI] [PubMed] [Google Scholar]
- 80.St Clair DK. and Oberley LW. Manganese superoxide dismutase expression in human cancer cells: a possible role of mRNA processing. Free Radic Res Commun 12–13Pt 2: 771–778, 1991 [DOI] [PubMed] [Google Scholar]
- 81.St Clair DK, Wan XS, Oberley TD, Muse KE, and St Clair WH. Suppression of radiation-induced neoplastic transformation by overexpression of mitochondrial superoxide dismutase. Mol Carcinog 6: 238–242, 1992 [DOI] [PubMed] [Google Scholar]
- 82.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, and Gupta MP. SIRT3 is a stress responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku-70. Mol Cell Biol 28: 6384–6401, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanny JC, Dowd GJ, Huang J, Hilz H, and Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99: 735–745, 1999 [DOI] [PubMed] [Google Scholar]
- 84.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, and Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell 40: 893–904, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, Lagcher W, Sie D, Tanger E, Cox T, Reinders M, Hubbard TJ, Rogers J, Jonkers J, Wessels L, Adams DJ, van Lohuizen M, and Berns A. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell 133: 727–741, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, and Richardson A. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics 16: 29–37, 2003 [DOI] [PubMed] [Google Scholar]
- 87.Vassilopoulos A, Fritz KS, Petersen DR, and Gius D. The human sirtuin family: evolutionary divergences and functions. Hum Genomics 5: 485–496, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Visner GA, Chesrown SE, Monnier J, Ryan US, and Nick HS. Regulation of manganese superoxide dismutase: IL-1 and TNF induction in pulmonary artery and microvascular endothelial cells. Biochem Biophys Res Commun 188: 453–462, 1992 [DOI] [PubMed] [Google Scholar]
- 89.Vousden KH. and Prives C. Blinded by the Light: The growing complexity of p53. Cell 137: 413–431, 2009 [DOI] [PubMed] [Google Scholar]
- 90.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, and Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430: 686–689, 2004 [DOI] [PubMed] [Google Scholar]
- 91.Yang Y, Cimen H, Han MJ, Shi T, Deng JH, Koc H, Palacios OM, Montier L, Bai Y, Tong Q, and Koc EC. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem 285: 7417–7429, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y, Zhang HM, Shi Y, Lustgarten M, Li Y, Qi W, Zhang BX, and Van Remmen H. Loss of manganese superoxide dismutase leads to abnormal growth and signal transduction in mouse embryonic fibroblasts. Free Radic Biol Med 49: 1255–1262, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu Y, Park SH, Ozden O, Kim HS, Jiang H, Vassilopoulos A, Spitz DR, and Gius D. Exploring the electrostatic repulsion model in the role of Sirt3 in directing MnSOD acetylation status and enzymatic activity. Free Radic Biol Med 53: 828–833, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]