Abstract
CYP3A4 metabolizes more than 50% of the drugs on the market. The large inter-individual differences of CYP3A4 expression may contribute to the variability of human drug responses. Post-transcriptional regulation of CYP3A4 is poorly understood, whereas transcriptional regulation has been studied much more thoroughly. In this study, we used multiple software programs to predict miRNAs that might bind to CYP3A4 and identified 112 potentially functional miRNAs. Then a luciferase reporter system was used to assess the effect of the overexpression of each potentially functional miRNA in HEK 293T cells. Fourteen miRNAs that significantly decreased reporter activity were measured in human liver samples (N = 27) as candidate miRNAs. To establish a more effective way to analyze in vivo data for miRNA candidates, the relationship between functional miRNA and target mRNA was modeled mathematically. Taking advantage of this model, we found that hsa-miR-577, hsa-miR-1, hsa-miR-532-3p and hsa-miR-627 could significantly downregulate the translation efficiency of CYP3A4 mRNA in liver. This study used in silico, in vitro and in vivo methods to progressively screen functional miRNAs for CYP3A4 and to enhance our understanding of molecular events underlying the large inter-individual differences of CYP3A4 expression in human populations.
The cytochrome P450 superfamily (CYP) is a group of monooxygenases that catalyze the oxidation of organic substances. The substrates of CYP enzymes include metabolic intermediates, such as lipids and steroidal hormones, as well as xenobiotic substances, including drugs and other toxic chemicals. CYPs are the major enzymes involved in drug metabolism and bioactivation, and they account for approximately 75% of the total number of different metabolic reactions. Fifty-seven human genes that code for various cytochrome P450 enzymes have been identified.
Cytochrome P450 3A4 (CYP3A4) is a member of the CYPs. The expression level of the CYP3A4 gene is highest in the human liver, followed by the prostate, breast, gut, colon, small intestine and brain1,2,3,4,5,6. The CYP3A4 protein is involved in the metabolism of more than 50% of the current prescription drugs7,8,9, and it has the greatest range of substrates among all the CYPs. As a result, CYP3A4 is the predominant isozyme of all the CYPs expressed in adult human liver.
A large inter-individual difference (approximately 50-fold or more) has been reported for CYP3A4 levels in the general population10 and contributes significantly to the striking individual differences in the therapeutic and toxic effects of drugs. It has been suggested that approximately 90% of the inter-individual differences in hepatic CYP3A4 activity could be accounted for by genetics10. However, the coding variants of the CYP3A4 gene may contribute to but are not likely to be the major cause of inter-individual differences in CYP3A4 activity because of the low allele frequencies and limited alterations in enzyme expression and catalytic function11. Therefore, the variability in CYP3A4 levels might be explained by the dynamic facets of functional genomics related to gene expression, including transcriptional and post-transcriptional processes. Recently, transcriptional regulation of CYP3A4 has been well studied12, but post-transcriptional regulation remains elusive, especially downregulation by micro-RNAs (miRNAs)13.
miRNAs are a family of short, noncoding RNAs of approximately 22 nucleotides14. Binding to complementary regions of transcripts, miRNAs play important roles in the regulation of target genes, either by repressing the translation of mRNAs (frequently in animals) or triggering their degradation (mainly in plants). At present, more than 1900 miRNAs have been identified in humans15,16, and miRNAs are predicted to control approximately 60% of the genes in the human genome17,18,19. The regulatory roles of miRNAs have received a great deal of attention, especially in the cancer field, but are still hardly known in the field of pharmacology.
Only two studies have thus far shown the involvement of miRNAs in the regulation of CYP3A4 expression. Takagi et al. reported that CYP3A4 can be indirectly regulated by miRNA via microRNA-148a (miR-148a)-controlled regulation of the human Pregnane X Receptor (PXR)20. Pan et al. showed that miR-27b may negatively regulate CYP3A4 protein expression in human embryonic kidney 293 cells21. These studies provided a useful clue for the mechanism of CYP3A4 regulation, although a single gene can usually be regulated by multiple miRNAs22, and it is very likely that other miRNAs are also involved in the regulation of CYP3A4 expression. A complete understanding of the role of miRNAs in the regulation of CYP3A4 expression using large-scale screening experiments will provide new insights into CYP3A4-specific expression patterns and inter-individual variability, which have so far been unexplained. Furthermore, most investigations of CYP3A4 regulation are based on studies of cell lines. These cell lines do not completely mimic the regulation of gene expression observed in normal hepatocytes because the cell lines are isolated from their natural environment, and the interactions with other liver cell types and the extracellular matrix are missing. Therefore, it is necessary to know how these regulatory pathways operate in humans.
In this study, we used in silico, in vitro and in vivo methods to systematically investigate the effects of miRNAs on CYP3A4 expression and found that hsa-miR-577, hsa-miR-1, hsa-miR-532-3p and hsa-miR-627 repress the translation of CYP3A4 mRNA.
Results
Bioinformatic prediction of miRNA binding
A total of 61, 28 and 77 miRNAs were predicted to bind CYP3A4 mRNA by TargetScan, miRbase and DIANA, respectively. In total, 65, 40 and 7 miRNAs were predicted by one, two and all three of the software programs, respectively. Among the 112 predicted miRNAs, 105 were considered potentially functional miRNAs and were examined by luciferase reporter expression in this study. The other 7 miRNAs were not included in our miRNA expression library (unpublished data). Please see Table S1 for the list of predicted miRNAs.
Functional examination in vitro
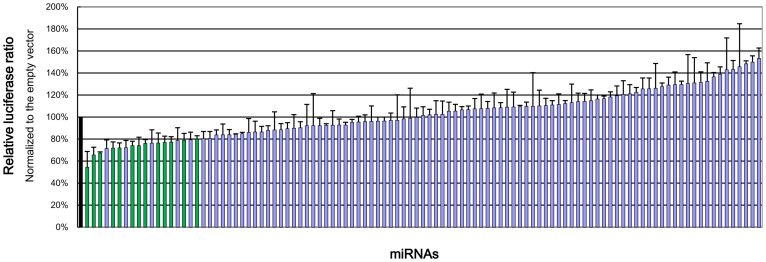
From a miRNA expression library, 105 potentially functional miRNA expression vectors were chosen for a large-scale experimental validation using dual luciferase assays. Expression of the psiCHECK2-CYP3A4 3′UTR vector characterized by the ratio of Renilla activity/firefly activity was compared between the empty vector group (control) and each miRNA group. Candidate miRNAs were chosen according to the following criteria: (1) the relative luciferase ratios were lower than 0.8, normalized to the empty vector; (2) the relative luciferase ratios were significantly different from that of the empty vector, with p values lower than 0.025; (3) both criteria were reproducible. Among the miRNAs assayed (Figure 1), 14 candidate miRNAs were found (Table 1) and investigated in liver samples. Please see Table S1 for the relative luciferase ratios of all predicted miRNAs.
Figure 1. Relative luciferase ratio of each measured miRNA normalized to the empty vector.
Black: empty vector group (control). Green: candidate miRNAs with significant (p < 0.025) reduction of relative luciferase activity (ratio < 0.8). Table S1 shows the data for each individual miRNA.
Table 1. Candidate miRNAs that significantly reduced luciferase reporter expression.
| miRNA | Relative luciferase ratio (Normalized to the empty vector) | p value |
|---|---|---|
| hsa-mir-27b | 0.544 ± 0.143 | 0.018 |
| hsa-mir-627 | 0.656 ± 0.070 | 0.016 |
| hsa-mir-653 | 0.672 ± 0.012 | 0.006 |
| hsa-miR-660 | 0.684 ± 0.058 | 0.006 |
| hsa-miR-361-3p | 0.718 ± 0.056 | 0.014 |
| hsa-miR-613 | 0.719 ± 0.046 | 0.008 |
| hsa-miR-526b | 0.739 ± 0.041 | 0.020 |
| hsa-miR-569 | 0.740 ± 0.077 | 9E-04 |
| hsa-miR-548a-3p | 0.761 ± 0.034 | 0.019 |
| hsa-miR-1 | 0.764 ± 0.091 | 0.013 |
| hsa-miR-629 | 0.769 ± 0.058 | 0.008 |
| hsa-miR-532-3p | 0.773 ± 0.049 | 0.010 |
| hsa-miR-559 | 0.790 ± 0.062 | 0.006 |
| hsa-miR-577 | 0.799 ± 0.030 | 0.006 |
Data are expressed as the mean ± SD.
A mathematical model for miRNA function
The mathematical model we established is explained with details in the Supplemental Results. In brief, our model showed that a functional miRNA should theoretically have the following relationship with its target:
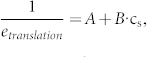 |
where A and B are constants, cs is the concentration of miRNA and etranslation, the “translation efficiency”, is the ratio of protein concentration to mRNA concentration of the target.
To our knowledge, there has been no gold standard to correlate a miRNA with its potential target. The most common method is the linear correlation of miRNA concentration with either mRNA or protein concentration. A better method was proposed by Takagi et al.20, in which translation efficiency was used empirically. Our model refined their method and provided a theoretical foundation. In addition, the linear correlation in our model is convenient for researchers to evaluate statistical significance. Therefore, we took advantage of this model to screen functional miRNAs from candidates in vivo.
Clinical investigation
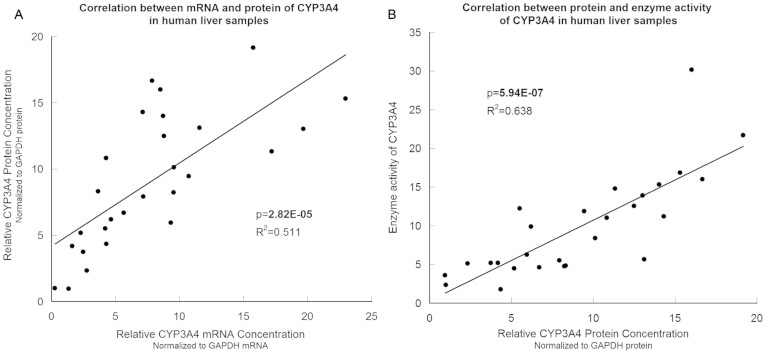
Enzymatic activity, relative protein concentration, relative mRNA concentration of CYP3A4 and relative concentration of 14 candidate miRNAs were measured in 27 liver samples. All of these data were normally distributed, as verified by the one-sample Kolmogorov-Smirnov test (p > 0.05). As shown in Figure 2, the enzymatic activity was significantly correlated with the relative protein concentration (p = 5.97E-07, R2 = 0.638), and the latter was also significantly correlated with the relative mRNA concentration (p = 2.82E-05, R2 = 0.511).
Figure 2. The relationship between mRNA concentration, protein concentration and enzymatic activity of CYP3A4 in liver samples.
(A), The CYP3A4 mRNA level was significantly correlated with the CYP3A4 protein level. (B), The CYP3A4 enzymatic activity was significantly correlated with the CYP3A4 mRNA level. N = 27.
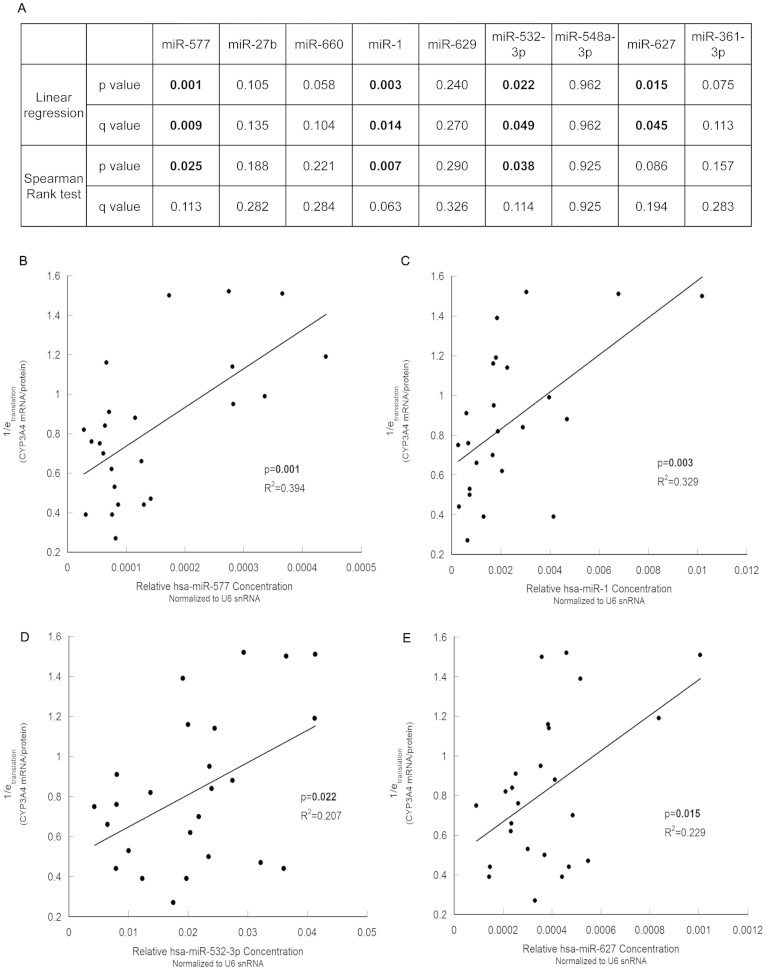
Among 14 candidate functional miRNAs, hsa-miR-577, hsa-miR-1, hsa-miR-532-3p and hsa-miR-627 showed significant linear correlations (p < 0.05) with the reciprocal of translation efficiency (Figure 3). The concentrations of hsa-miR-569, hsa-miR-559, hsa-miR-613, hsa-miR-653 and hsa-miR-526b were too low to be analyzed. Moreover, hsa-miR-577 and hsa-miR-1 also correlated significantly with translation efficiency using Spearman's rank method.
Figure 3. The correlation between each miRNA and 1/etranslation (mRNA/protein) of CYP3A4 in human liver samples.
(A), The significance of linear regression and Spearman's rank test between candidate miRNAs and 1/etranslation. Significant p values or q values are in boldface. (B), (C), (D) and (E), the 1/etranslation of CYP3A4 was significantly correlated with the relative concentration of hsa-miR-577 (B), hsa-miR-1 (C), hsa-miR-532-3p (D) and hsa-miR-627 (E). N = 27.
Rescue validation
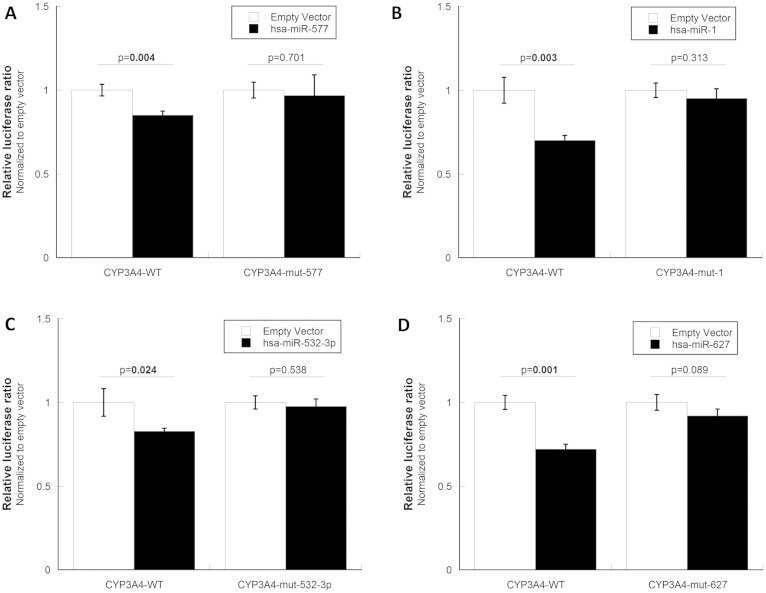
To verify the direct targeting, rather than an indirect mechanism, of the four regulatory miRNAs identified above, we designed CYP3A4 3′UTR vectors with mutated miRNA binding sites to test the rescuing effect of each miRNA in vitro. Multiple binding sites were mutated for hsa-miR-532-3p because it has three predicted targets. At each binding site, six to twelve base pairs were mutated to complementary sequences using QuikChange II XL Site-Directed Mutagenesis Kits (Agilent Technologies, Santa Clara, CA) so that the corresponding miRNAs could not target the mutant mRNAs. The success of the mutagenesis was confirmed by sequencing. Co-transfection of the mutated CYP3A4 3′UTR vectors with the corresponding miRNA expression vectors showed that all four functional miRNAs identified in this study interacted with the CYP3A4 3′UTR to perform their post-transcriptional regulation (Figure 4).
Figure 4. Rescue validation for four functional miRNAs.
The CYP3A4 3′UTR vector was mutated to produce several rescue vectors that did not have the corresponding miRNA binding sites. After mutagenesis, the expression of the CYP3A4 3′UTR vectors was no longer significantly affected by hsa-miR-577 (A), hsa-miR-1 (B), hsa-miR-532-3p (C) or hsa-miR-627 (D).
Discussion
There is a huge inter-individual variability of CYP3A4 levels in human populations. Such variability may be caused post-transcriptionally by miRNAs. In the present study, we aimed to clarify whether and which miRNAs influenced human CYP3A4 gene expression. We began the study using a computational prediction of miRNA binding to CYP3A4 mRNA, which revealed 112 potentially functional miRNAs. The luciferase reporter cloned with the human CYP3A4 3′-UTR fragment was then used to evaluate the effect of each potentially functional miRNA on gene expression in HEK 293T cells. A total of 14 miRNAs with significant repression effects was chosen as the candidates for further investigation. Using our mathematical model, four candidate miRNAs (hsa-miR-577, hsa-miR-1, hsa-miR-532-3p and hsa-miR-627) were found to significantly influence the translation efficiency of CYP3A4 mRNA in human liver samples (Figure S1).
Although the roles of miRNAs in the regulation of the cytochrome P450s have only begun to become clear, many studies have reported interesting findings. The first study revealing the regulation of P450 by miRNA23 found that hsa-miR-27b downregulated human CYP1B124. In addition, hsa-miR-27b was reported to decrease human CYP3A4 expression using the luciferase reporter21. This result is consistent with our in vitro result but not in our liver samples. Human CYP2E1 and CYP24A1 were found to be repressed by hsa-miR-37825 and hsa-miR-125b26, respectively. In addition, miR-126* was reported to regulate CYP2A4 in rats27. All of the studies mentioned indicated that miRNAs participate in the regulation of CYPs.
All of the miRNAs we found to affect CYP3A4 expression, especially hsa-miR-1, have been reported to be involved to some extent in various cancers. Both hsa-miR-577 and hsa-miR-1 were found to be significantly associated with chemosensitivity after chemotherapy for gastric cancer28, which may be partly explained by their influence on the cytochrome P450 enzymes.
In our simplified mathematical model, there are many issues that need to be addressed cautiously. (1) Because this model aimed to evaluate whether and how much each miRNA regulated the expression of a certain gene in a population, it was assumed that the miRNA level was the unique variable. Therefore, individual differences of transcription efficiency and of decay rates were neglected. (2) In metazoans, including humans, miRNAs function with non-perfect target matches primarily by blocking translation, whereas miRNAs in plants function with perfect or near-perfect matches and cause mRNA degradation. Our model only considered the function of miRNAs in translation repression because the subjects are human, and it is not applicable to the study of miRNAs in plants. (3) We also neglected the one-to-multiple relationships between mRNA and miRNA because a one-to-one relationship has not been well understood. (4) Feedback widely exists in biological events but is not considered in this model. All these simplifications were designed to reveal a linear correlation to assess the significance of the influence of a miRNA that would be practically useful for clinical investigations. However, such practical applicability sacrificed the accuracy of the mathematical model. For more accurate but complicated models, please see the literature29,30. Better models for practical applications are urgently needed because in vivo miRNA studies are beginning to multiply.
The concept of “translation efficiency” was proposed by Takagi et al20 to minimize the transcriptional differences of evaluating the post-transcriptional regulation by miRNAs. In our model, we found that the reciprocal of the translation efficiency was theoretically linearly correlated with miRNA level, which is the reason we used linear regression in the data analysis. In addition, we also performed Spearman's rank test to correlate ranks of translation efficiency and those of miRNAs in the population, as used in Takagi's study20, which revealed less significant but similar findings.
Although using a single reference gene is still a common practice, it may introduce a bias into qPCR results. To minimize such a possibility, we also measured other widely used reference genes in the liver samples. As shown in Figure S2, there were significant correlations of expression between two reference mRNAs (GAPDH and ACTB) and between two reference small RNAs (U6 and SNORD38B) in liver samples, which suggested that only a limited bias was caused by the arbitrary reference genes in this study. Future studies with more reference genes and better normalization, e.g., geNorm31, in bigger sample size are warranted to confirm the functional miRNAs we identified in the current study.
To minimize the possibility of losing any functional miRNAs for subsequent in vivo investigations, we did not use any correction in our luciferase reporter experiments in vitro. This strategy is reasonable because any false-positive results from in vitro examination can be verified by in vivo investigations. Moreover, triplicates in each group are not suitable for correction in high-throughput in vitro examination. Arbitrarily, we set the p value threshold to 0.025 to limit the number of candidate miRNAs.
In clinical investigations, the FDR (false discovery rate) correction was used to avoid false-positive conclusions caused by multiple comparisons. As shown in Figure 3A, all of the hsa-miR-577, hsa-miR-1, hsa-miR-532-3p and hsa-miR-627 results were still significant after the FDR correction (q < 0.05) of the linear regression analysis. However, significance for miRNAs decreased after the FDR correction in Spearman's rank test, which suggested that our mathematical model may analyze miRNA function better than traditional methods.
This study systematically investigated miRNAs in silico, in vitro and in vivo, considering all known miRNAs as targets and examining them in a high-throughput system. Moreover, the use of liver samples consolidated our conclusions because miRNAs exhibit unique expression patterns in specific cells and/or tissues at various developmental stages, and the liver is the principal site where CYP3A4 metabolizes drugs. These miRNAs involved in the regulation of human liver CYP3A4 expression are potential biomarkers that can be used to guide personalized medicine. In addition, our simplified mathematical model provides a theoretical basis for the empirical use of translational efficiency in analyses of miRNA functions.
However, the sample size of liver samples is still small, although similar to those of other studies. This increases the risk of type II errors (false-negative errors), as observed by the power = 0.61 with a medium size effect (f2 = 0.2). In addition, we could not rule out the possibility of indirect regulation of miRNAs by other proteins, which becomes a serious issue for miRNA studies. Finally, future studies with appropriate animal models may assist in confirming the regulation of CYP3A4 expression by miRNAs.
In this systematic study, we first found 112 miRNAs that potentially regulate CYP3A4 from miRNA databases in silico. Then, we used an overexpression method with a luciferase reporter system to identify 14 candidate miRNAs in vitro. Finally, four miRNAs were found to correlate significantly with the reciprocal of the translation efficiency of CYP3A4 in human liver samples in vivo, following our mathematical model. We concluded that human CYP3A4 is post-transcriptionally regulated by hsa-miR-577, hsa-miR-1, hsa-miR-532-3p and hsa-miR-627 in liver. This study enhanced our understanding of the molecular events underlying the large inter-individual differences of CYP3A4 expression in human populations, which would be a potent risk factor for drug response variability.
Methods
Ethics statement
The protocol complied with the Declaration of Helsinki and its subsequent revisions and was approved by the Ethics Committees of Zhengzhou University.
Bioinformatics prediction
The miRNA binding was predicted using three online software programs: TargetScan (v6.2; http://www.targetscan.org/)32, miRanda in combination with mirSVR (August 2010 release; http://www.microrna.org/microrna/getGeneForm.do)33 and DIANA (v3.0; http://diana.cslab.ece.ntua.gr/microT/)34 with default thresholds. The full-length sequence of the human CYP3A4 3′-UTR was input into the prediction, if available.
Luciferase reporter activity assay
Human CYP3A4 3′-UTR fragments, corresponding to nucleotides +26057 to +27229 in the CYP3A4 gene (NG_008421), were PCR-amplified from genomic DNA and cloned into the psiCHECK2 vector (Promega, Madison, WI) downstream of the Renilla luciferase reporter gene. The psiCHECK2 vector also includes an independent firefly luciferase reporter gene, which functions as a control for transfection efficiency. This psiCHECK2-CYP3A4 vector was transiently co-transfected with each miRNA expression vector or empty pLL3.7 vector (control) into HEK 293T cells, using Lipofectamine 2000 (Life Technologies, Carlsbad, CA). The miRNA expression library was constructed based on the miRBase reference database (August 2010 Release). The Renilla and firefly luciferase activities in cell lysates were measured 48 hours after transfection using the dual-luciferase reporter assay system (Promega), and the results were expressed as the ratio of Renilla activity/firefly activity in the lysate. Each experiment included at least three replicates. The difference between the miRNA group and the control was evaluated using a t-test.
Tissue samples
Human liver samples from 27 Chinese Han donors were obtained from the First Affiliated Hospital of Zhengzhou University after informed consent. Liver samples were stored in liquid nitrogen immediately after resection. Total RNAs were extracted from approximately 0.1 g of liver using the standard TRIzol method (Life Technologies) followed by agarose gel electrophoresis for quality control. Randomly selected RNA samples were also examined using an Agilent Bioanalyzer (Agilent Technologies) for further quality control (RIN value > 7). RNA concentration was measured using a NanoDrop 2000c (Thermo Fisher Scientific, Waltham, MA). Total proteins were extracted from approximately 0.3 g of liver using RIPA lysis buffer containing the protease inhibitor PMSF followed by quantification using the BCA method (Pierce, Rockford, Illinois).
Immunoblot analysis
Total protein (20 μg) was input in 10% SDS-PAGE gels and transferred electrophoretically to nitrocellulose membranes. The membranes were blocked with 5% (wt/vol) powdered/dry milk in Tris-buffered saline with 0.1% Tween-20. After incubation with a primary rabbit anti-CYP3A4 antibody (ab3572; Abcam, MA), the membranes were washed and incubated with a secondary antibody. The membranes were then developed using the Supersignal west pico-chemiluminescent substrate (Pierce). Anti-GAPDH was used as the endogenous control. Quantification of immunoreactive bands was performed using Quantity One software (Bio-Rad, Hercules, California).
CYP3A4 enzymatic activity
Liver microsomes were extracted from tissue using the CaCl2 method. The BCA method was used to measure total protein concentration (Pierce). Midazolam (0.25–75 μmol/L) was incubated with liver microsomes at 37°C for 10 minutes. The 1′-OH midazolam concentration was measured using an Agilent 1200 HPLC (Agilent Technologies). Michaelis constants (Km, μM) were estimated to quantify enzymatic activity with five replicates.
qPCR for mRNA and miRNA
Total RNA (500 ng) was reverse transcribed to cDNA using PrimeScript RT Reagent kits (Takara Bio, Dalian, China). The relative expression of CYP3A4 mRNA was measured by qPCR using the appropriate primers (forward, 5′-CCAAGCTATGCTCTTCACCG-3′; reverse, 5′-TCAGGCTCCACTTACGGTGC-3′) and the FastStart Universal SYBR Green Master (Roche, Germany) according to the manufacturer's protocol. GAPDH mRNA was quantified as the endogenous control using the appropriate primers (forward, 5′-ATCACCATCTTCCAGGAGCGA-3′; reverse, 5′-GCTTCACCACCTTCTTGATGT-3′). For both sets of qPCR primers, the forward primer and the reverse primer were designed to target different exons to avoid the possibility of amplifying genomic DNA. The specificity of the qPCR primers was verified by the presence of a single peak in the melting curves. For miRNA quantification, reverse transcription and qPCR were performed using the miRCURY LNA™ Universal RT microRNA PCR system (Exiqon, Denmark) following the manufacturer's protocol. The U6 snRNA was measured as the endogenous control. All qPCRs were run on a ViiA 7 instrument (Life Technologies) with three replicates. The Cq values were determined automatically by the ViiA 7 RUO Software v1.1 (Life Technologies). All NTCs' Cq values were higher than 40 and were labeled as “undetermined” by the ViiA 7 RUO Software v1.1.
Statistics
Linear regressions and Spearman's rank correlations were determined using SPSS (Statistical Package for the Social Sciences) for Windows, version 11.0 (IBM, NY). All tests were two-tailed, and statistical significance was assumed at p < 0.05.
Author Contributions
Z.W., Y.Z., C.M., Y.L. and S.Q. performed the RNA experiments. S.J. and X.P. performed the prediction and performed the experiments with the cell line. X.W., F.S. and L.Z. provided the tissue samples and performed the enzyme experiments. Y.Z. and H.W. performed the immunoblot experiments. Z.W. and L.G. performed the mathematical modeling. L.H., L.Z. and Q.X. supervised the studies. Z.W. and Q.X. designed the study and wrote the paper.
Supplementary Material
Supplemental Information
Acknowledgments
This work was supported by the National Natural Science Foundation of China [grants 30971582, 61240031 and 81173127] and the 973 Program [2010CB529600]. Those funding the study had no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript. We are deeply grateful to all the participants as well as to the clinicians working on this project.
References
- Guengerich F. P. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol 39, 1–17, 10.1146/annurev.pharmtox.39.1.1 (1999). [DOI] [PubMed] [Google Scholar]
- Watkins P. B., Wrighton S. A., Schuetz E. G., Molowa D. T. & Guzelian P. S. Identification of glucocorticoid-inducible cytochromes P-450 in the intestinal mucosa of rats and man. J Clin Invest 80, 1029–1036, 10.1172/JCI113156 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lown K. S. et al. Grapefruit juice increases felodipine oral availability in humans by decreasing intestinal CYP3A protein expression. J Clin Invest 99, 2545–2553, 10.1172/JCI119439 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolars J. C. et al. CYP3A gene expression in human gut epithelium. Pharmacogenetics 4, 247–259 (1994). [DOI] [PubMed] [Google Scholar]
- Huang Z., Fasco M. J., Figge H. L., Keyomarsi K. & Kaminsky L. S. Expression of cytochromes P450 in human breast tissue and tumors. Drug Metab Dispos 24, 899–905 (1996). [PubMed] [Google Scholar]
- Shimada T., Yamazaki H., Mimura M., Inui Y. & Guengerich F. P. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 270, 414–423 (1994). [PubMed] [Google Scholar]
- Blumberg B. et al. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev 12, 3195–3205 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B., Hodgson E. & Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 56, 1329–1339 (1999). [DOI] [PubMed] [Google Scholar]
- Lehmann J. M. et al. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102, 1016–1023, 10.1172/JCI3703 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir V. et al. Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics 10, 373–388 (2000). [DOI] [PubMed] [Google Scholar]
- Lamba J. K., Lin Y. S., Schuetz E. G. & Thummel K. E. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev 54, 1271–1294 (2002). [DOI] [PubMed] [Google Scholar]
- Martinez-Jimenez C. P., Jover R., Donato M. T., Castell J. V. & Gomez-Lechon M. J. Transcriptional regulation and expression of CYP3A4 in hepatocytes. Curr Drug Metab 8, 185–194 (2007). [DOI] [PubMed] [Google Scholar]
- Koturbash I., Beland F. A. & Pogribny I. P. Role of microRNAs in the regulation of drug metabolizing and transporting genes and the response to environmental toxicants. Expert Opin Drug Metab Toxicol 8, 597–606, 10.1517/17425255.2012.673587 (2012). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Bentwich I. et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37, 766–770, 10.1038/ng1590 (2005). [DOI] [PubMed] [Google Scholar]
- Kozomara A. & Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39, D152–157, 10.1093/nar/gkq1027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B. & Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20, 10.1016/j.cell.2004.12.035 (2005). [DOI] [PubMed] [Google Scholar]
- Xie X. et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434, 338–345, 10.1038/nature03441 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R. C., Farh K. K., Burge C. B. & Bartel D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19, 92–105, 10.1101/gr.082701.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S., Nakajima M., Mohri T. & Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem 283, 9674–9680, 10.1074/jbc.M709382200 (2008). [DOI] [PubMed] [Google Scholar]
- Pan Y. Z., Gao W. & Yu A. M. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos 37, 2112–2117, 10.1124/dmd.109.027680 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene 29, 2302–2308, 10.1038/onc.2010.34 (2010). [DOI] [PubMed] [Google Scholar]
- Nakajima M. & Yokoi T. MicroRNAs from biology to future pharmacotherapy: regulation of cytochrome P450s and nuclear receptors. Pharmacol Ther 131, 330–337, 10.1016/j.pharmthera.2011.04.009 (2011). [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Nakajima M., Takagi S., Taniya T. & Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res 66, 9090–9098, 10.1158/0008-5472.CAN-06-1403 (2006). [DOI] [PubMed] [Google Scholar]
- Mohri T. et al. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol 79, 1045–1052, 10.1016/j.bcp.2009.11.015 (2010). [DOI] [PubMed] [Google Scholar]
- Komagata S. et al. Human CYP24 catalyzing the inactivation of calcitriol is post-transcriptionally regulated by miR-125b. Mol Pharmacol 76, 702–709, 10.1124/mol.109.056986 (2009). [DOI] [PubMed] [Google Scholar]
- Kalscheuer S., Zhang X., Zeng Y. & Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis 29, 2394–2399, 10.1093/carcin/bgn209 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H. et al. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genomics 4, 79, 10.1186/1755-8794-4-79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine E., Ben Jacob E. & Levine H. Target-specific and global effectors in gene regulation by MicroRNA. Biophys J 93, L52–54, 10.1529/biophysj.107.118448 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyev A. et al. Dynamical modeling of microRNA action on the protein translation process. BMC Syst Biol 4, 13, 10.1186/1752-0509-4-13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27, 91–105, 10.1016/j.molcel.2007.06.017 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B. et al. Human MicroRNA targets. PLoS Biol 2, e363, 10.1371/journal.pbio.0020363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M. et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 37, W273–276, 10.1093/nar/gkp292 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information