ABSTRACT
Hemoprotozoan infections often cause serious production losses in livestock. In the present study, we conducted a PCR-based survey of Babesia bovis, Babesia bigemina, Theileria annulata, Theileria orientalis, Trypanosoma evansi and Trypanosoma theileri, using 423 DNA samples extracted from blood samples of cattle (n=202), water buffaloes (n=43), sheep (n=51) and goats (n=127) bred in the Hue and Hanoi provinces of Vietnam. With the exception of T. annulata and T. evansi, all other parasite species (B. bovis, B. bigemina, T. orientalis and T. theileri) were detected in the cattle populations with B. bovis being the most common among them. Additionally, four water buffaloes and a single goat were infected with B. bovis and B. bigemina, respectively. The Hue province had more hemoprotozoan-positive animals than those from the Hanoi region. In the phylogenetic analyses, B. bovis-MSA-2b, B. bigemina-AMA-1 and T. theileri-CATL gene sequences were dispersed across four, one and three different clades in the respective phylograms. This is the first study in which the presence of Babesia, Theileria and Trypanosoma parasites was simultaneously investigated by PCR in Vietnam. The findings suggest that hemoprotozoan parasites, some of which are genetically diverse, continue to be a threat to the livestock industry in this country.
Keywords: epidemiology, hemoprotozoa, livestock, PCR, Vietnam
The livestock industry often suffers enormous economic losses from infectious diseases caused by hemoprotozoan pathogens. Different species of Babesia, Theileria and Trypanosoma parasites that infect farm ruminants, such as cattle, water buffaloes and sheep, afflict the livestock production systems across the world. Bovine babesiosis is caused by Babesia bovis, B. bigemina, B. divergens, B. ovata and B. major [6]. While B. bovis and B. bigemina infect both cattle and water buffaloes, B. divergens, B. ovata and B. major usually infect the cattle [6]. Among these parasites, B. bovis and B. bigemina are known as the most economically important pathogens in many tropical countries [6]. In addition to causing hemolytic anemia, B. bovis induces neurological damage and respiratory syndrome in infected cattle [11, 27]. If untreated, both B. bovis- and B. bigemina-related babesiosis can prove fatal [8].
Theileria parva and T. annulata are highly virulent lymphoproliferative parasites that also damage the health of cattle populations [20]. The causative agent of tropical theileriosis, T. annulata, which infects cattle and yaks, is common in North Africa, Southern Europe and Asia. In contrast, T. parva, which causes East Coast fever in cattle, is mainly found in eastern, central and southern Africa [25]. In addition, a relatively benign group of Theileria parasites, consisting of T. orientalis, T. sergenti and T. buffeli, has a wide host range, including several ruminants and worldwide distribution [3, 19].
Among Trypanosoma species, T. congolense, T. vivax and T. brucei are the most virulent species in Africa; however, T. evansi causes a potentially fatal severe wasting disease amongst susceptible animals, such as cattle, buffaloes, horses, camels and other wildlife in Africa, Asia and South America [9]. Although T. theileri has been described as a benign parasite of cattle, buffaloes, sheep, goats and antelopes, T. theileri co-infections with other hemoprotozoan parasites can worsen the clinical syndrome and cause livestock production losses in the affected ruminants [18, 26].
Vietnam is an agriculturally rich country, and many people are engaged in agricultural and/or livestock farming related activities. The presence of several hemoprotozoan species has been noted previously in Vietnamese livestock populations. However, most of these studies were either based on microscopic or serological testing [13, 24]. Microscopy is not an effective technique for epidemiological surveys, because it lacks sensitivity and specificity [7]. In contrast, although ELISA can be highly sensitive at detecting carrier-stage animals, it only indicates whether an animal has had previous exposure to the parasite, rather than identifying those with active infections. In comparison, well-developed PCR assays have high sensitivity and specificity for detecting the species or strain of interest, thus enabling them to identify current infections in animals [1]. In Vietnam, B. bovis and B. bigemina infections in cattle have already been demonstrated by PCR [14]. Recently, we have also reported the results of an epidemiological survey of T. orientalis conducted among different livestock species using PCR [16]. However, infections of Babesia, Theileria and Trypanosoma parasites were not simultaneously investigated in any of these studies. Therefore, in the present study, we performed a PCR-based cross-sectional epidemiological survey to detect the various hemoprotozoan parasites distributed among Vietnamese cattle and buffalo populations. We also screened sheep and goat populations to find out whether hemoprotozoan pathogens could also infect these host species in Vietnam.
Genetic diversity in hemoprotozoan parasites is considered a stumbling block for the development of immune control strategies against them [5]. For example, the merozoite surface antigens (MSAs) in B. bovis were considered as vaccine candidates [5]. However, the genetic diversity and antigenic differences observed within MSAs of different isolates suggested that a mixture of antigens from different genotypes should be used in sub-unit vaccines [2, 5]. Therefore, genetic diversity of B. bovis MSAs in the endemic countries must be analyzed to identify the genotypes. Furthermore, as the past studies on T. orientalis have indicated that the virulence in the hemoprotozoan parasites might be genotype-dependent, investigation on the genetic variations might enable the researchers to predict the clinical significance of parasites [10]. Therefore, in the present study, we analyzed the genetic diversity of the parasites detected in Vietnam.
MATERIALS AND METHODS
Blood samples: This survey was conducted in 2 different regions, the central and northern parts of Vietnam, namely Hue (February 2010) [16] and Hanoi (July 2011) provinces, respectively. While the blood samples were collected from randomly selected cattle, sheep and goats that were reared in several integrated farms from both sampling locations [16], water buffaloes were sampled only in Hue province. All the animals surveyed in the present study were clinically normal. Approximately 2 ml of whole blood was collected from each animal into a Vacutainer tube containing anti-coagulant (EDTA) (NIPRO, Osaka, Japan) and then stored at −20°C.
DNA extraction: DNA was extracted from each blood sample as described previously [23]. Briefly, a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) was employed to extract DNA from 200 µl of each blood sample. After measuring their concentrations, the DNA samples were stored at −20°C.
Polymerase chain reaction (PCR): The target genes for the PCR assays and the sequences of the primers used in the present study are listed in Table 1. Previously described, parasite-specific, single-step PCR assays were employed to detect B. bigemina, T. annulata, T. orientalis, T. evansi and T. theileri [4, 17, 21,22,23], while a nested PCR assay was used for B. bovis [2]. The B. bovis- and T. orientalis-specific PCR assays were performed according to previously described methods [2, 21]. However, PCR assays for detection of B. bigemina, T. annulata, T. evansi and T. theileri were conducted with minor modifications. Briefly, 1 µl of each DNA sample was added to a 9 µl reaction mixture that consisted of 200 µM dNTPs (Applied Biosystems, Branchburg, NJ, U.S.A.), 1 µl 10 × buffer (Applied Biosystems), 0.5 µM forward and reverse primers, 0.5 unit of Taq polymerase (Applied Biosystems) and double distilled water; PCR cycling conditions were as described previously [4, 17, 22, 23]. For the B. bigemina-, T. annulata-, T. evansi- and T. theileri-specific PCR assays, the number of amplification cycles was increased to 45.
Table 1. Parasite species-specific PCR primers.
| Parasite | Target gene | Primers sequence (5'− 3') |
Amplicon size (bp) | Reference | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| B. bovis − 1st step | Spherical body protein 2 | CTGGAAGTGGATCTCATGCAACC | TCACGAGCACTCTACGGCTTTGCAG | 1,236 | [2] |
| − 2nd step | TGAATCTAGGCATATAAGGCATTTCG | AACCCCTCCTAAGGTTGGCTAC | 584 | ||
| B. bigemina | Apical membrane antigen − 1 | TACTGTGACGAGGACGGATC | CCTCAAAAGCAGATTCGAGT | 211 | [23] |
| T. annulata | Merozoite-piroplasm surface antigen | ATGCTGCAAATGAGGAT | GGACTGATGAGAAGACGATGAG | 768 | [17] |
| T. orientalis | Major piroplasm surface protein | CTTTGCCTAGGATACTTCCT | ACGGCAAGTGGTGAGAACT | 776 | [21] |
| T. evansi | Minicircle DNA | CAACGACAAAGAGTCAGT | ACGTGTTTTGTGTATGGT | 373 | [4] |
| T. theileri | Cathepsin L-like protein | CGTCTCTGGCTCCGGTCAAAC | TTAAAGCTTCCACGAGTTCTTGATGATCCAGTA | 289 | [22] |
Cloning and sequencing: Amplicons with high band intensities from each species were cloned and sequenced, as described previously [28]. In addition, full-length merozoite surface antigen-2b (MSA-2b) gene sequences were amplified from the DNA samples, which were B. bovis-positive by the above PCR screening assay, using a pair of forward (5′-ATGATCGGGAAAATCTTCTTGTTAA-3′) and reverse (5′-TTAAAATGCAGAGAGAACGAAGTAGC-3′) primers. The composition of the reaction mixture and cycling conditions of the PCR assay to amplify MSA-2b were similar to those of the B. bigemina-specific screening PCR, except that the annealing temperature was lowered to 52°C.
For each PCR amplicon, a single sequence was determined as previously described [2]. Briefly, PCR products were extracted from agarose gels (Alquick Gel Extraction Kit, Qiagen), ligated to a plasmid vector (pCR2.1-TOPO, Invitrogen, Carlsbad, CA, U.S.A.), transferred to Escherichia coli competent cells (TOP 10, Invitrogen) and then cultured in LB broth (Invitrogen). Thereafter, plasmids were extracted (QIAprep Spin Miniprep kit, Qiagen) from the cultures, and the nucleotide sequences of the inserts were determined using an ABI PRISM 3100 genetic analyzer (Applied Biosystems).
Sequence identity estimates and phylogenetic analyses: After initial analyses using GENTYX 7.0 software (GENTYX, Tokyo, Japan) and the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi), the sequence identity values among the gene sequences determined in the present study and those already published in GenBank were calculated using the EMBOSS-needle online program (http://emboss.bioinformatics.nl/cgi-bin/emboss/needle).
The sequences determined in the present study were phylogenetically analyzed, together with those available in GenBank. The previously described MAFFT software program was used to construct the phylogenetic trees [15].
RESULTS
PCR detection of hemoprotozoan parasites: B. bovis, B. bigemina, T. orientalis and T. theileri were all detected among the animals surveyed in Vietnam (Table 2). B. bovis was the most common blood parasite in cattle (8.9%), and Hue province had a higher percentage of B. bovis-positive cattle (15.6%) than Hanoi province (2.8%). In addition, B. bigemina was identified in 3.5% of the cattle, while 5% of the cattle had T. theileri. In the present study, only the animals from Hanoi were surveyed for T. orientalis, because we have previously reported on the prevalence of this pathogen in Hue province [16]. In Hanoi, a single DNA sample from the cattle was positive for T. orientalis (0.9%). In water buffaloes, B. bovis was the only parasite detected in the present study (9.3%) (Table 2). While all the sheep samples were parasite-negative, a single goat bred in Hanoi was positive for B. bigemina. In addition, while 8 cattle bred in the Hue province were infected with more than one parasite, none of the animals surveyed in Hanoi was infected with multiple parasites (data not shown). Among the 8 cattle that had mixed infections, one animal was infected with 3 parasite species (B. bovis, T. orientalis [16] and T. theileri), while the remaining had duel infections.
Table 2. Blood sample collection and PCR detection of various hemoprotozoan parasites from Hue and Hanoi, Vietnam.
| Animaltype | Location | Sample N. | PCR detection of parasite DNA (%) |
|||||
|---|---|---|---|---|---|---|---|---|
| B. bovis | B. bigemina | T. annulata | T. orientalis | T. evansi | T. theileri | |||
| Cattle | Hue | 96 | 15 (15.6) | 5 (5.2) | 0 | ND | 0 | 10 (10.4) |
| Hanoi | 106 | 3 (2.8) | 2 (1.9) | 0 | 1 (0.9) | 0 | 0 | |
| (Total) | 202 | 18 (8.9) | 7 (3.5) | 0 | 0 | 10 (5.0) | ||
| Buffaloa | Hue | 43 | 4 (9.3) | 0 | 0 | ND | 0 | 0 |
| Sheep | Hue | 21 | 0 | 0 | 0 | ND | 0 | 0 |
| Hanoi | 30 | 0 | 0 | 0 | 0 | 0 | 0 | |
| (Total) | 51 | 0 | 0 | 0 | 0 | 0 | ||
| Goat | Hue | 21 | 0 | 0 | 0 | ND | 0 | 0 |
| Hanoi | 106 | 0 | 1 (0.9) | 0 | 0 | 0 | 0 | |
| (Total) | 127 | 0 | 1 (0.8) | 0 | 0 | 0 | ||
ND: Not done in this study. a) Water buffaloes were sampled only in the Hue region.
Sequencing and phylogenetic analyses: Sequence analyses confirmed that the PCR assays were specific for the target protozoan species. In the present study, 9 (AB742543 − AB742551), 5 (AB742552 − AB742556), 1 (AB742557), 5 (AB742558 − AB742562) and 5 (AB786920 − AB786924) sequences were successfully determined for B. bovis spherical body 2 (SBP-2), B. bigemina apical membrane antigen 1 (AMA-1), T. orientalis major piroplasm surface protein (MPSP), T. theileri cathepsin L-like protein (CATL) and B. bovis MSA-2b genes, respectively. While a single SBP-2 (AB742548) and MSA-2b (AB786923) gene sequences were derived from buffalo DNA samples, an AMA-1(AB742556) sequence was isolated from a goat. All of the remaining sequences were determined using DNA samples of infected cattle blood.
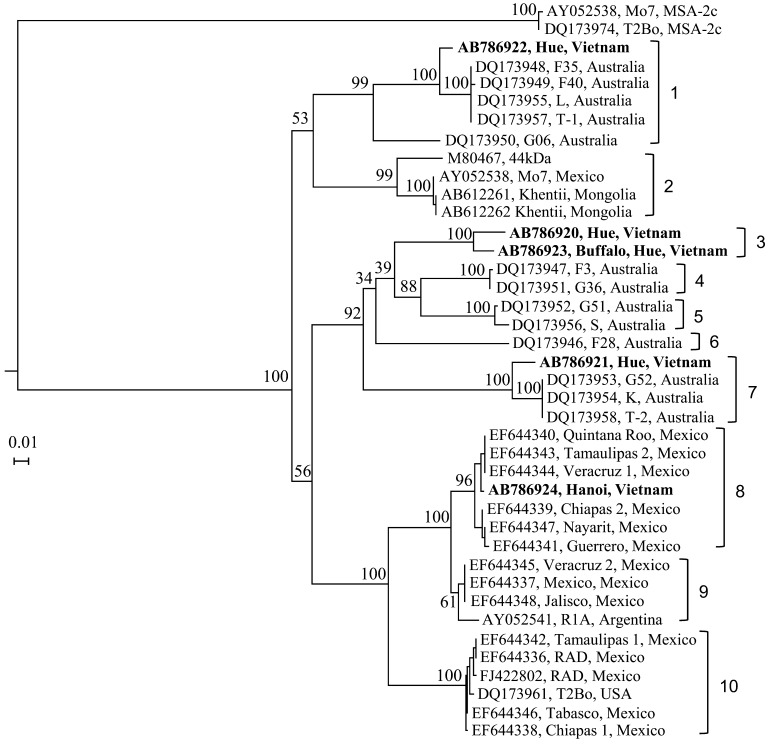
B. bovis SBP-2 gene sequences (AB742549, AB742550 and AB742551) amplified from the Hanoi samples were closest to those from Thailand (JN974305) with sequence identities of 95.0–95.9%. Out of 6 SBP-2 sequences determined for the Hue samples, 2 (AB742546 and AB742548) were closest to those from Thailand (JN974305) (95.4% identity), but others (AB742543, AB742544, AB742545 and AB742547) had higher sequence identities to the American (T2Bo strain) gene sequences (XM_001611639 and XM_001611732) (94.0–97.8%) than those from Thailand (88.2–92.7%). In the B. bovis-MSA-2b-based phylogram (Fig. 1), which divided the gene sequences into 10 clades (designated as 1–10), the Vietnamese sequences (n=5) (AB786920 − AB786924) were observed in 4 different clades (1, 3, 7 and 8). In particular, clade 3, which is closest to the Australian clades (4 and 5), was formed by Vietnamese MSA-2b sequences (AB786920 and AB786923) only. Two other Vietnamese sequences (AB786922 and AB786921) also clustered together with the Australian sequences (clades 1 and 7, respectively), while another Vietnamese sequence (AB786924) appears with the Mexican isolates in clade 8.
Fig. 1.
Phylogenetic analysis of B. bovis-MSA-2b gene sequences. Sequences generated in the present study (n=5) are indicated by boldface-type letters. Note that the Vietnamese sequences fall into four different clades.
A T. orientalis MPSP gene sequence (AB742557) determined in the present study was closer to sequences from Brazil (AB581616), Mongolia (AB571919) and Thailand (AB562534) (99.1% identity) and was classified as MPSP-type 3 in the phylogenetic analysis (data not shown).
The B. bigemina AMA-1 gene sequences from cattle and goats in Hanoi (AB742555 and AB742556) were identical to each other and to those from Sri Lanka (AB690862), Turkey (JN572801), Argentina (AB481200) and Italy (HM543729), while the AMA-1 sequences from Hue province (AB742552, AB742553 and AB742554) shared 98.6–99.1% identity with each other and 99.1–99.5% with the above named sequences from other countries (Fig. 2). B. bigemina AMA-1 sequences, which are relatively conserved among different isolates, were dispersed across 2 different clades (designated as 1 and 2) in the phylogenetic tree (Fig. 2). All the Vietnamese AMA-1 gene sequences, including that from a goat, were found in clade 2.
Fig. 2.
Phylogenetic analysis of B. bigemina-AMA-1 gene sequences. Sequences generated in the present study (n=5) are indicated by boldface-type letters. Note that all the Vietnamese sequences are in clade 2.
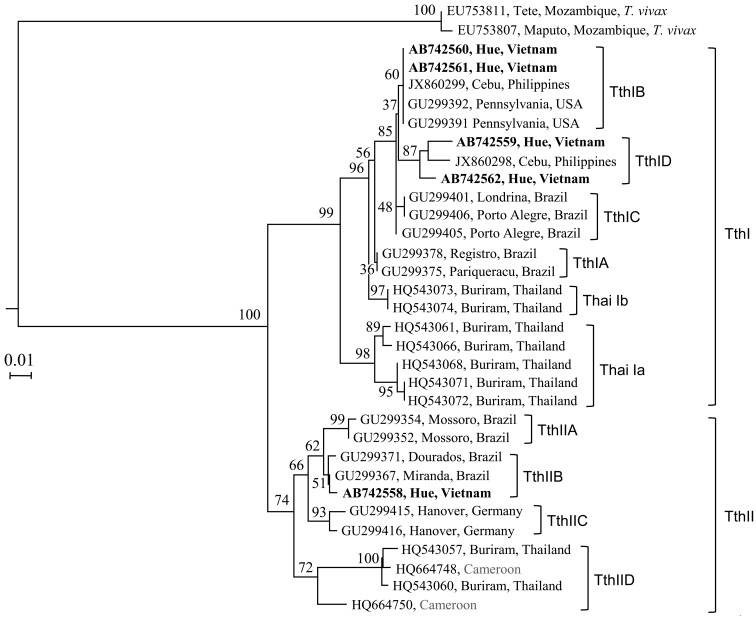
Two T. theileri CATL gene sequences (AB742558 and AB742562) shared high sequence identities (99.6 and 98.2%, respectively) with their Brazilian counterparts (GU299367 and GU299405, respectively), while 2 other sequences (AB742560 and AB742561) were identical to sequences from the United States (GU299391 and GU299392) and Philippines (JX860299) (Fig. 3). In addition, a CATL sequence from the Philippines (JX860299) shared high sequence identity (97.8%) with a Vietnamese sequence (AB742558). Based on a previously described classification method for the phylogenetic positions of T. theileri-CATL gene sequences [12], the gene sequences can be grouped into 2 major clades, TthI and TthII, which can be further divided into several sub-clades (Fig. 3). In the present study, the CATL sequences from Vietnam were found in three different sub-clades. Within the major clade TthI, 2 Vietnamese sequences (AB742560 and AB742561) clustered together with sequences from the United States and the Philippines to form TthIB, while 2 other Vietnamese sequences (AB742559 and AB742562) together with a Philippine sequence (JX860298) formed a new sub-clade, which was named TthID in the present study. Within TthII, a single Vietnamese sequence (AB742558) was observed in the TthIIB sub-clade together with those from Brazil (GU299371 and GU299367).
Fig. 3.
Phylogenetic analysis of T. theileri-CATL gene sequences. Sequences determined in the present study (n=5) are indicated by boldface-type letters. Note that the Vietnamese sequences fall into three different sub-clades. Because the two Cameroon sequences (HQ664748 and HQ664750) were found in previously described ThaiIIa [12], this sub-clade has been renamed TthIID in the present study.
DISCUSSION
Our findings show that the Vietnamese cattle populations investigated here were infected with B. bovis, B. bigemina, T. orientalis or T. theileri. Detection of B. bovis as the most common bovine hemoprotozoan species identified in the cattle is an important finding, because this parasite is often fatal to cattle [6]. While the prevalence of B. bovis in Hanoi (2.8%) was slightly lower than that previously reported (4.2%) [14], the combined prevalence in the 2 regions was higher (8.9%) than that estimated in earlier surveys (1.5–4.2%) [13, 14]. Because SBP-2 is a relatively conserved multi-copy gene, it is unlikely to become a good marker for phylogenetic analysis. Therefore, in addition to SBP-2, we used MSA-2b to analyze genetic variation among B. bovis isolates in Vietnam. The findings have shown that MSA-2b gene sequences among B. bovis in Vietnam are genetically diverse. Therefore, the possible immune control strategies against B. bovis in Vietnam must be designed in light of the genetic diversity of this parasite.
Although B. bigemina is less pathogenic to cattle than B. bovis, babesiosis caused by B. bigemina can also be fatal if not properly treated [8]. In addition to B. bigemina infections in cattle, this species was also detected in a goat in the present study. To our knowledge, this is the first report on the infection of B. bigemina in goats. In contrast to B. bovis-MSA-2b (Fig. 1), B. bigemina-AMA-1 sequences from Vietnam were observed in a single clade, suggesting that these gene sequences are highly conserved in the Vietnamese isolates (Fig. 2). However, additional genes should be identified and developed for use as markers to obtain a better picture of genetic diversity in B. bigemina populations.
In the present study, a single T. orientalis-positive sample was detected in a cattle population from Hanoi. However, when all the Hue samples used in the present study were tested for T. orientalis in a recent investigation, 13 (13.8%), 11 (25.6%) and 1 (4.8%) samples were positive for this parasite among cattle, water buffaloes and sheep, respectively [16]. While the sequence obtained in the present study can be classified as MPSP-type 3, seven different genotypes, 1, 3, 5, 7, N1, N2 and N3 of T. orientalis were detected in the previous investigation [16]. Further studies using T. orientalis outbreak isolates might shed additional light on the clinical relevance of different genotypes in Vietnam.
T. theileri co-infections with other bovine blood pathogens or where the animals are malnourished or under stress sometimes make them more susceptible to illness, which can lead to economic losses in cattle-farming operations [18, 26]. In the present study, T. theileri was associated with coinfections in 4 of 8 cattle that had mixed infections. Therefore, the presence of this parasite in Vietnam might be of clinical significance. T. theileri-CATL sequences from Vietnam were observed in three different clusters in the phylogram (Fig. 3), suggesting that T. theileri in Vietnam is genetically diverse. The future studies must focus on the functional relevance of the genetic diversity in T. theileri, because it is not very clear whether the genetic variations within this parasite are related to any differences in antigenicity or virulence among isolates.
Hemoprotozoan parasites were more commonly detected in Hue than Hanoi. Large herds of animals are reared under an extensive system in Hue, while animals in the Hanoi area are maintained by an intensive system, under which vector control methods are regularly practiced. Therefore, the animals in Hue province might have been more frequently exposed to parasite-infected vectors than those in Hanoi. Additionally, differences in the distribution of the transmission vectors in the study areas could also account for the differences observed.
This is the first study in which Babesia, Theileria and Trypanosoma parasites were simultaneously surveyed by PCR in Vietnam. The findings suggest that hemoprotozoan parasites, some of which are genetically diverse, continue to be a threat to the livestock industry in this country.
ACKNOWLEDGMENTS
We thank all of the staff from the farms that were surveyed and the veterinarians in Vietnam who participated in the present study. In addition, we thank Ms. Hiroko Yamamoto (National Research Center for Protozoan Diseases, Obihiro University of Agriculture, Obihiro, Japan) for her excellent technical assistance. This study was supported by a grant from the Global COE Program from the Japanese Ministry of Education, Culture, Sports, Science and Technology and Grants-in-Aid for Scientific Research from the Japan Society for Promotion of Science (JSPS), Japan.
REFERENCES
- 1.Almeria S., Castella J., Ferrer D., Ortuno A., Estrada-Peña A., Gutierrez J. F. 2001. Bovine piroplasms in Minorca (Balearic Islands, Spain): a comparison of PCR-based and light microscopy detection. Vet. Parasitol. 99: 249–259. doi: 10.1016/S0304-4017(01)00464-2 [DOI] [PubMed] [Google Scholar]
- 2.Altangerel K., Sivakumar T., Battsetseg B., Battur B., Ueno A., Igarashi I., Yokoyama N. 2012. Phylogenetic relationships of Mongolian Babesia bovis isolates based on the merozoite surface antigen (MSA)-1, MSA-2b, and MSA-2c genes. Vet. Parasitol. 184: 309–316. doi: 10.1016/j.vetpar.2011.09.021 [DOI] [PubMed] [Google Scholar]
- 3.Altangerel K., Sivakumar T., Inpankaew T., Jittapalapong S., Terkawi M. A., Ueno A., Xuan X., Igarashi I., Yokoyama N. 2011. Molecular prevalence of different genotypes of Theileria orientalis detected from cattle and water buffaloes in Thailand. J. Parasitol. 97: 1075–1079. doi: 10.1645/GE-2846.1 [DOI] [PubMed] [Google Scholar]
- 4.Artama W. T., Agey M. W., Donelson J. E. 1992. DNA comparisons of Trypanosoma evansi (Indonesia) and Trypanosoma brucei spp. Parasitology 104: 67–74. doi: 10.1017/S0031182000060819 [DOI] [PubMed] [Google Scholar]
- 5.Berens S. J., Brayton K. A., Molloy J. B., Bock R. E., Lew A. E., McElwain T. F. 2005. Merozoite surface antigen 2 proteins of Babesia bovis vaccine breakthrough isolates contain a unique hypervariable region composed of degenerate repeats. Infect. Immun. 73: 7180–7189. doi: 10.1128/IAI.73.11.7180-7189.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bock R., Jackson L., de Vos A., Jorgensen W. 2004. Babesiosis of cattle. Parasitology 129:(Suppl): S247–S269. doi: 10.1017/S0031182004005190 [DOI] [PubMed] [Google Scholar]
- 7.Böse R., Jorgensen W. K., Dalgliesh R. J., Friedhoff K. T., de Vos A. J. 1995. Current state and future trends in the diagnosis of babesiosis. Vet. Parasitol. 57: 61–74. doi: 10.1016/0304-4017(94)03111-9 [DOI] [PubMed] [Google Scholar]
- 8.Brown W. C., Norimine J., Goff W. L., Suarez C. E., McElwain T. F. 2006. Prospects for recombinant vaccines against Babesia bovis and related parasites. Parasite Immunol. 28: 315–327. doi: 10.1111/j.1365-3024.2006.00849.x [DOI] [PubMed] [Google Scholar]
- 9.Brun R., Hecker H., Lun Z. 1998. Trypanosoma evansi and T. equiperdum: distribution, biology, treatment and phylogenetic relationship (a review). Vet. Parasitol. 79: 95–107. doi: 10.1016/S0304-4017(98)00146-0 [DOI] [PubMed] [Google Scholar]
- 10.Eamens G. J., Gonsalves J. R., Jenkins C., Collins D., Bailey G. 2013. Theileria orientalis MPSP types in Australian cattle herds associated with outbreaks of clinical disease and their association with clinical pathology findings. Vet. Parasitol. 191: 209–217. doi: 10.1016/j.vetpar.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 11.Everitt J. I., Shadduck J. A., Steinkamp C., Clabaugh G. 1986. Experimental Babesia bovis infection in Holstein calves. Vet. Pathol. 23: 556–562 [DOI] [PubMed] [Google Scholar]
- 12.Garcia H. A., Kamyingkird K., Rodrigues A. C., Jittapalapong S., Teixeira M. M. G., Desquesnes M. 2011. High genetic diversity in field isolates of Trypanosoma theileri assessed by analysis of cathepsin L-like sequences disclosed multiple and new genotypes infecting cattle in Thailand. Vet. Parasitol. 180: 363–367. doi: 10.1016/j.vetpar.2011.03.017 [DOI] [PubMed] [Google Scholar]
- 13.Hanh H. T., Khai N. D., Lang P. S., Lan P. D., Tan N. D., Lam H. M. 1997. Blood parasites of cattle in Daklak province, Vietnam. Khoa. Hoc. Ky. Thuat. Thu. Y 4: 50–53 [Google Scholar]
- 14.Hau N. V., Thu N. V., Hanh H. T., Sat L. M. 1999. A preliminary study on application of polymerase chain reaction in diagnosis of haemosporidiosis in cattle. Khoa. Hoc. Ky. Thuat. Thu. Y 6: 48–52 [Google Scholar]
- 15.Katoh K., Misawa K., Kuma K., Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066. doi: 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khukhuu A., Lan D. T., Long P. T., Ueno A., Li Y., Luo Y., Macedo A. C., Matsumoto K., Inokuma H., Kawazu S. I., Igarashi I., Xuan X., Yokoyama N. 2011. Molecular epidemiological survey of Theileria orientalis in Thua Thien Hue. J. Vet. Med. Sci. 73: 701–705. doi: 10.1292/jvms.10-0472 [DOI] [PubMed] [Google Scholar]
- 17.Kirvar E., Ilhan T., Katzer F., Hooshmand-Rad P., Zweygarth E., Gerstenberg C., Phipps P., Brown C. G. D. 2000. Detection of Theileria annulata in cattle and vector ticks by PCR using the Tams1 gene sequences. Parasitology 120: 245–254. doi: 10.1017/S0031182099005466 [DOI] [PubMed] [Google Scholar]
- 18.Mansfield J. M. 1977. Nonpathogenic trypanosomes of mammals. pp. 297–327. In: Parasitic Protozoa, 1 (Kreier, J. P. ed.), Academic Press, London. [Google Scholar]
- 19.McFadden A. M., Rawdon T. G., Meyer J., Makin J., Clough R. R., Tham K., Mullner P., Geysen D. 2011. An outbreak of haemolytic anaemia associated with infection of Theileria orientalis in naive cattle. N. Z. Vet. J. 59: 79–85. doi: 10.1080/00480169.2011.552857 [DOI] [PubMed] [Google Scholar]
- 20.McKeever D. J. 2009. Bovine immunity —a driver for diversity in Theileria parasites? Trends Parasitol. 25: 269–276. doi: 10.1016/j.pt.2009.03.005 [DOI] [PubMed] [Google Scholar]
- 21.Ota N., Mizuno D., Kuboki N., Igarashi I., Nakamura Y., Yamashina H., Hanzaike T., Fujii K., Onoe S., Hata H., Kondo S., Matsui S., Koga M., Matsumoto K., Inokuma H., Yokoyama N. 2009. Epidemiological survey of Theileria orientalis infection in grazing cattle in the Eastern Part of Hokkaido, Japan. J. Vet. Med. Sci. 71: 937–944. doi: 10.1292/jvms.71.937 [DOI] [PubMed] [Google Scholar]
- 22.Rodrigues A. C., Garcia H. A., Ortiz P. A., Cortez A. P., Martinkovic F., Paiva F., Bartista J. S., Minervino A. H., Campaner M., Pral E. M., Alfieri S. C., Teixeira M. M. 2010. Cysteine proteases of Trypanosoma (Megatrypanum) theileri: cathepsin L-like gene sequences as targets for phylogenetic analysis, genotyping diagnosis. Parasitol. Int. 59: 318–325. doi: 10.1016/j.parint.2010.03.002 [DOI] [PubMed] [Google Scholar]
- 23.Sivakumar T., Altangerel K., Battsetseg B., Battur B., AbouLaila M., Munkhjargal T., Yoshinari T., Yokoyama N., Igarashi I. 2012. Genetic detection of Babesia bigemina from Mongolian cattle using apical membrane antigen-1 gene based PCR technique. Vet. Parasitol. 187: 17–22. doi: 10.1016/j.vetpar.2012.01.008 [DOI] [PubMed] [Google Scholar]
- 24.Verloo D., Holland W., My L. N., Thanh N. G., Tam P. T., Goddeeris B., Vercruysse J., Busher P. 2000. Comparison of serological tests for Trypanosoma evansi natural infections in water buffaloes from North Vietnam. Vet. Parasitol. 92: 87–96. doi: 10.1016/S0304-4017(00)00284-3 [DOI] [PubMed] [Google Scholar]
- 25.Weir W., Karagenc T., Baird M., Tait A., Shiels B. R. 2010. Evolution and diversity of secretome genes in the apicomplexan parasite Theileria annulata. BMC Genomics 11: 42. doi: 10.1186/1471-2164-11-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wells E. A. 1976. Subgenus Megatrypanum pp. 257–275. In: Biology of the kinetoplastida (Lumsden, W. H. R. and Evans, D. A. eds.), Academic Press, London. [Google Scholar]
- 27.Wright I. G., Goodger B. V. 1988. Pathogenesis of babesiosis. pp. 99–118. In: Babesiosis of Domestic Animals and Man (Ristic, M. ed.), CRC Press, Boca Raton. [Google Scholar]
- 28.Yokoyama N., Ueno A., Mizuno D., Kuboki N., Khukhuu A., Igarashi I., Miyahara T., Shiraishi T., Kudo R., Oshiro M., Zakimi S., Sugimoto C., Matsumoto K., Inokuma H. 2011. Genotypic Diversity of Theileria orientalis Detected from Cattle Grazing in Kumamoto and Okinawa Prefectures of Japan. J. Vet. Med. Sci. 73: 305–312. doi: 10.1292/jvms.10-0263 [DOI] [PubMed] [Google Scholar]