β-Ketoacyl-(acyl carrier protein) reductase (FabG), an enzyme from Y. pestis involved in fatty-acid synthesis, was crystallized and X-ray diffraction data were collected to 2.3 Å resolution.
Keywords: β-ketoacyl-ACP reductase, FabG, Yersina pestis
Abstract
The type II fatty-acid biosynthesis pathway of bacteria provides enormous potential for antibacterial drug development owing to the structural differences between this and the type I fatty-acid biosynthesis system found in mammals. β-Ketoacyl-ACP reductase (FabG) is responsible for the reduction of the β-ketoacyl group linked to acyl carrier protein (ACP), and is essential for the formation of fatty acids and bacterial survival. Here, the cloning, expression, purification, crystallization and diffraction of FabG from Yersinia pestis (ypFabG), the highly virulent causative agent of plague, are reported. Recombinant FabG was expressed, purified to homogeneity and crystallized via the hanging-drop vapour-diffusion technique. Diffraction data were collected at the Australian Synchrotron to 2.30 Å resolution. The crystal displayed P212121 symmetry, with unit-cell parameters a = 68.22, b = 98.68, c = 169.84 Å, and four ypFabG molecules in the asymmetric unit.
1. Introduction
The fatty-acid biosynthesis pathway of bacteria provides a number of extremely promising antibacterial drug targets. Most bacteria utilize the type II fatty-acid biosynthesis (FASII) system, in which discrete dissociated enzymes sequentially catalyse the creation of saturated and unsaturated fatty acids of varying lengths from the condensation of malonyl-CoA and acetyl-CoA primers. Such fatty acids form essential components of lipoproteins, phospholipids and lipopolysaccharides (Parsons & Rock, 2013 ▶). In contrast, the type I fatty-acid system (FASI) of mammalian cells is a multifunction protein complex comprised of a homodimer of two 270 kDa polypeptides encoded by a single gene, with the various domains of the complex catalysing the acyltransferase, condensation, reduction and dehydration reactions of the pathway (Asturias et al., 2005 ▶). Whilst the individual reactions in both FASI and FASII are similar, the structural differences between the FASI complex and the dissociated enzymes of the FASII system provide the potential to target an area essential to bacterial survival with minimal adverse effects to the host (Leibundgut et al., 2008 ▶; White et al., 2005 ▶).
FabG catalyses the reduction of the ACP (acyl carrier protein)-bound β-ketoacyl group to a β-hydroxyacyl group and is necessary for the formation of both saturated and unsaturated fatty acids. It is expressed in most bacteria as a single isozyme of approximately 25–26 kDa, forming either a homodimer or homotetramer in solution. The overall structure including the Ser, Tyr and Lys catalytic triad of the active site (Dutta et al., 2012 ▶; Price et al., 2001 ▶) appears to be well conserved in both Gram-negative and Gram-positive bacteria, with an r.m.s.d. of 1.5 Å between Staphylococcus aureus FabG (PDB entry 3osu; Center for Structural Genomics of Infectious Diseases, unpublished work) and Escherichia coli FabG (PDB entry 1i01; Price et al., 2001 ▶) (calculated using DaliLite; Holm & Park, 2000 ▶). FabG also appears to be essential for bacterial survival (Lai & Cronan, 2004 ▶; Subramanian et al., 2011 ▶; Tang et al., 2006 ▶). One notable exception to this is bacteria of the Mycobacterium genus, in which five putative isozymes have been identified; however, the roles of these isozymes have yet to be characterized (Gurvitz, 2009 ▶; Parish et al., 2007 ▶).
This paper describes the cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of FabG from Yersinia pestis, the causative agent of bubonic, pneumonic and septicaemic plague. If left untreated Y. pestis infections are usually fatal, resulting in death in as little as 24 h after exposure (Bertherat et al., 2011 ▶). Additionally, bubonic and septicaemic plague can advance to pneumonic plague (infection in the lungs), a highly contagious manifestation in which human-to-human transmission can occur through respiratory droplets (Bertherat et al., 2011 ▶; Zhou & Yang, 2009 ▶). In recent years, multidrug-resistant isolates of Y. pestis have been observed (Galimand et al., 2006 ▶), indicating a need to develop new antimicrobials to treat infection.
2. Experimental methods
2.1. Cloning
The gene encoding ypFabG (accession No. NP_669075.1) was amplified from genomic DNA by polymerase chain reaction (PCR) using HotStarTaq PCR Master Mix (Qiagen). The PCR product was inserted into the expression vector pMCSG21 via ligation-independent cloning as described previously (Eschenfeldt et al., 2009 ▶), with the following modifications: T4 treatment was conducted using 80 ng of PCR product and 1.5 µl (∼4 U) of T4 polymerase. T4-treated products were annealed using a 3:1 ratio of insert to vector before being transformed into chemically competent E. coli TOP10 cells (Invitrogen). The final construct encoded full-length ypFabG fused to an N-terminal hexahistidine tag with a Tobacco etch virus (TEV) protease cleavage site for tag removal.
2.2. Expression and purification
Protein expression and purification were performed similarly to that described by Forwood et al. (2007 ▶). Plasmid containing recombinant ypFabG was transformed into E. coli BL21 (DE3) pLysS competent cells. A single colony was used to inoculate a 5 ml Luria–Bertani (LB) starter culture supplemented with spectinomycin (100 µg ml−1) and chloramphenicol (34 µg ml−1), and was then used to inoculate auto-induction medium (Studier, 2005 ▶) containing spectinomycin (100 µg ml−1) and chloramphenicol (34 µg ml−1), which was incubated at 310 K until an OD600 of 6–10 was reached. Cells were harvested by centrifugation at 7459g at 277 K before being resuspended in buffer A (50 mM phosphate buffer pH 8.0, 300 mM NaCl, 20 mM imidazole) to a final volume of 35 ml and frozen at 253 K. Cell pellets were thawed and incubated with 1 ml 20 mg ml−1 lysozyme and 10 µl 50 mg ml−1 DNAse on ice for 30 min. The cell lysate was cleared via centrifugation and the recombinant protein was purified using affinity chromatography (HisTrap HP, GE Healthcare) equilibrated with buffer A. Protein was eluted using an increasing concentration of buffer B (50 mM phosphate buffer pH 8.0, 300 mM NaCl, 500 mM imidazole). Following elution, pooled fractions (14 ml) were incubated with 200 µl TEV protease (∼5 mg ml−1) for 14 h at 277 K (additional residues following tag removal: SNA–) before being further purified by size-exclusion chromatography (HiLoad 26/60 S200 column, GE Healthcare). Fractions containing ypFabG were eluted in buffer C (50 mM Tris pH 8.0, 125 mM NaCl) and concentrated to 70 mg ml−1 using an Amicon ultracentrifugal device (Millipore) with a 10 kDa molecular-weight cutoff. The concentrated protein was assessed by SDS–PAGE to be >95% pure, and was aliquoted and stored at 253 K.
2.3. Crystallization
Screening for initial crystallization conditions was conducted using the following commercially available screens: Crystal Screen, Crystal Screen 2, PEG/Ion and PEG/Ion 2 (Hampton Research). Screening was performed in VDX 48-well plates (Hampton Research) via the hanging-drop vapour-diffusion technique, using 1.5 µl recombinant ypFabG solution combined with 1.5 µl reservoir solution suspended over 300 µl reservoir solution and incubated at 296 K. Further optimization was conducted by varying salt and additive concentrations, and varying the concentration of protein using neat and 1:2 dilutions of recombinant ypFabG solution (diluted in buffer C). Large diffraction-quality crystals were obtained in 300 mM ammonium phosphate monobasic, 30%(w/v) glycerol, using a protein concentration of 35 mg ml−1.
2.4. Data collection and molecular replacement
ypFabG crystals were flash-cooled in liquid nitrogen at 100 K on day 12. Diffraction data were collected at the Australian Synchrotron on the MX2 microcrystallography beamline. A total of 200° of data with 0.5° oscillations were collected at a wavelength of 0.9537 Å and a crystal-to-detector distance of 240 mm. Collected data were indexed, integrated and scaled using iMosflm (Battye et al., 2011 ▶) and SCALA (Evans, 2006 ▶) from the CCP4 suite (Potterton et al., 2003 ▶; Winn et al., 2011 ▶). Molecular replacement was performed by Phaser (McCoy et al., 2007 ▶) using a monomer of E. coli FabG as the search model (PDB entry 1i01; Price et al., 2001 ▶).
3. Results and discussion
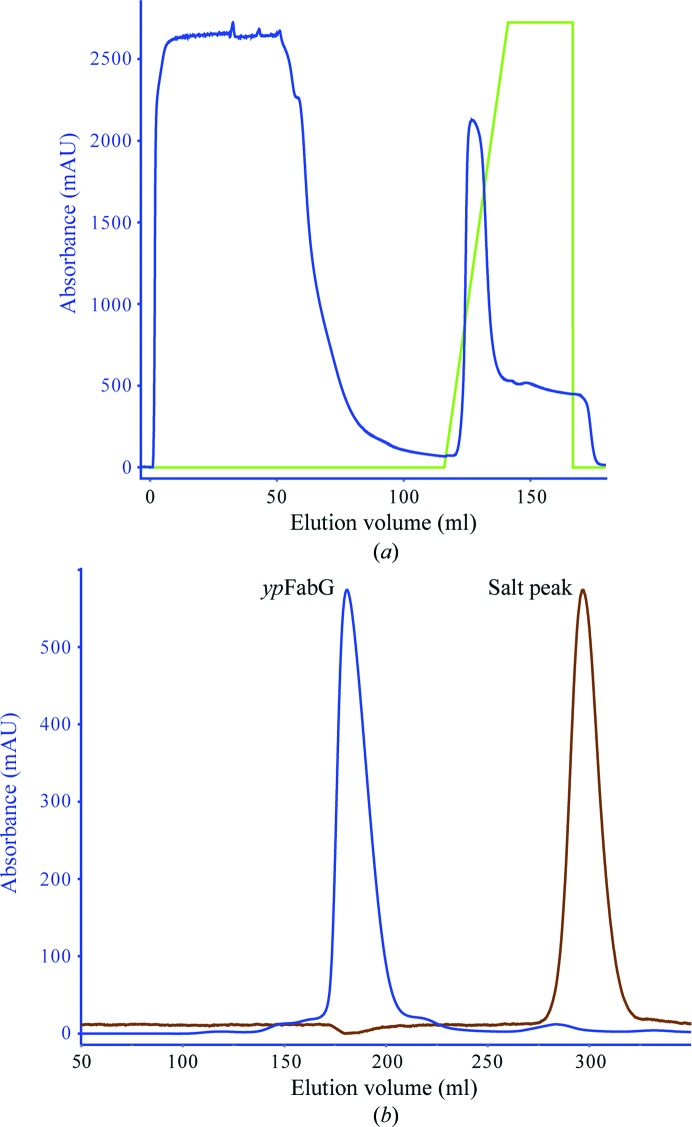
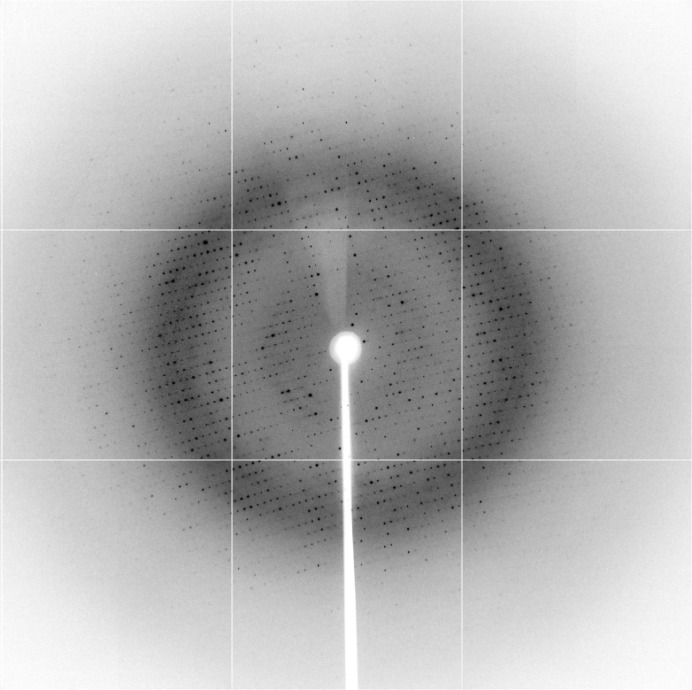
Recombinant ypFabG protein was solubly overexpressed in E. coli at levels of ∼50 mg per litre of culture. Purification by Ni2+-affinity chromatography (as described in §2) resulted in greater than 95% purity, and subsequent purification by size-exclusion chromatography (Fig. 1 ▶) yielded a highly pure, monodisperse protein of approximately 100 kDa, indicative of a tetramer. Initial crystallization conditions were obtained by screening against the commercial kits Crystal Screen, Crystal Screen 2, PEG/Ion and PEG/Ion 2. Preliminary crystals, obtained in condition No. 3 of Crystal Screen (400 mM ammonium phosphate monobasic), were optimized around salt/glycerol mixtures as described in §2.3 (Fig. 2 ▶). Crystals appearing after 2 d were flash-cooled in liquid nitrogen on day 12 and X-ray diffraction data were collected on the MX2 microcrystallography beamline at the Australian Synchrotron (Fig. 3 ▶). Images were indexed, integrated and scaled to a resolution of 2.30 Å (full data-collection statistics are listed in Table 1 ▶). The crystal was orthorhombic with P212121 symmetry, with unit-cell parameters a = 68.22, b = 98.68, c = 169.84 Å. Four ypFabG molecules were expected in the asymmetric unit, giving a Matthews coefficient (V M) of 2.60 Å3 Da−1 and a solvent content of 52.78% (Kantardjieff & Rupp, 2003 ▶; Matthews, 1968 ▶). This was confirmed by the location of a tetramer in the asymmetric unit by molecular replacement. Following iterative cycles of model building with WinCoot (Emsley et al., 2010 ▶) and structure refinement with REFMAC5 (Murshudov et al., 2011 ▶), the R and R free values were 0.26060 and 0.28938, respectively. The structure will be fully refined, with a detailed structural and functional analysis to be published in a separate paper. Structural analysis of ypFabG will provide new insights into the biochemical properties of ypFabG and of FabG proteins of other species from the Yersinia genus.
Figure 1.
Recombinant ypFabG was purified using (a) affinity chromatography (HisTrap HP, GE Healthcare) and (b) size-exclusion chromatography. Protein expression and purity were assessed by SDS–PAGE (not shown).
Figure 2.
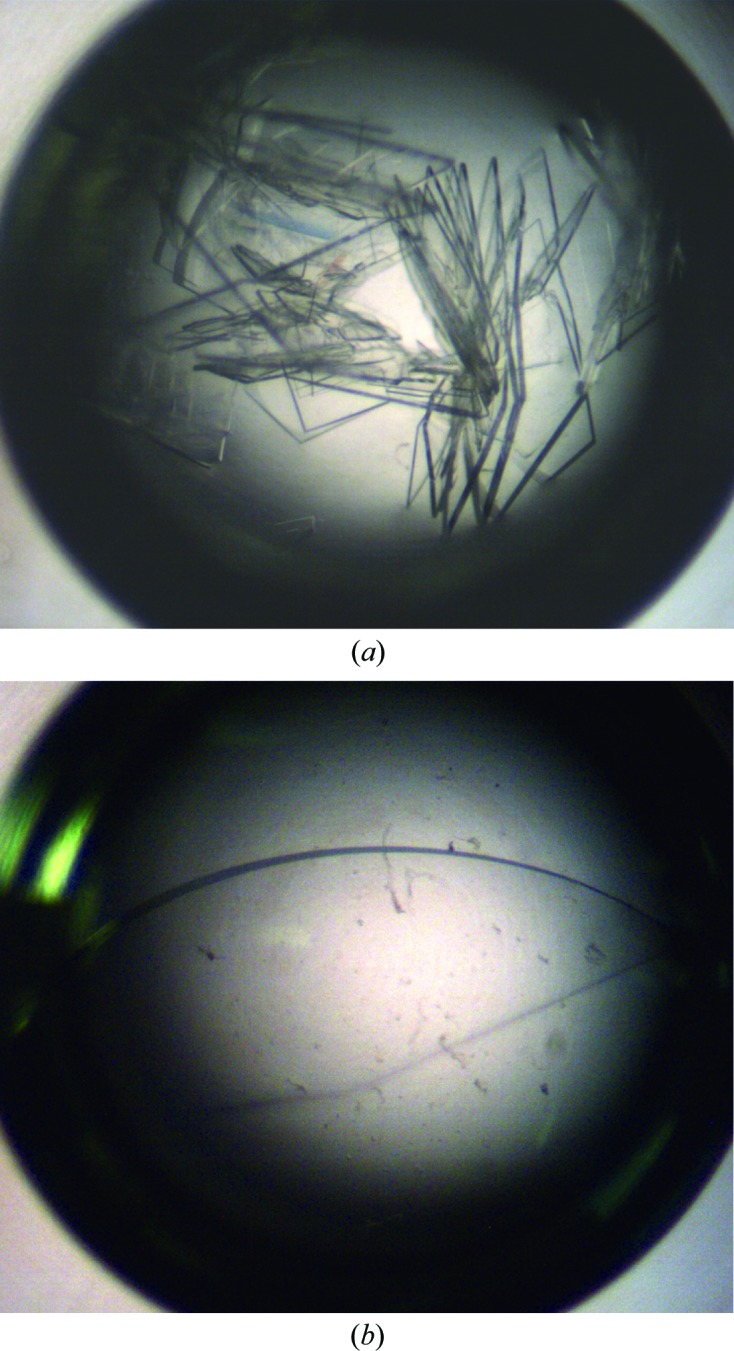
ypFabG crystals at day 7. (a) Crystals obtained using Hampton Research Crystal Screen condition No. 3 (400 mM ammonium phosphate monobasic) and (b) optimized crystal obtained using 300 mM ammonium phosphate monobasic and 30%(w/v) glycerol.
Figure 3.
A diffraction pattern of the ypFabG data set collected at the Australian Synchrotron after 180° of data collection.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the outermost resolution shell.
| Crystal system | Orthorhombic |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 68.22, b = 98.68, c = 169.84 |
| Wavelength (Å) | 0.9537 |
| Oscillation angle per frame (°) | 0.5 |
| Exposure time per frame (s) | 1.0 |
| Resolution (Å) | 34.11–2.30 (2.37–2.30) |
| Total reflections | 319791 (24239) |
| Unique reflections | 50945 (4329) |
| Mean I/σ(I) | 11.8 (3.5) |
| Completeness (%) | 98.8 (97.9) |
| Multiplicity | 6.3 (5.6) |
| R meas | 0.119 (0.562) |
| R merge † | 0.103 (0.480) |
| V M (Å3 Da−1) | 2.60 |
| Solvent content (%) | 52.78 |
| Wilson B factor (Å2) | 33.6 |
R
merge = 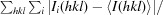
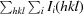 , where Ii(hkl) is the intensity of an individual measurement of the reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity of that reflection.
, where Ii(hkl) is the intensity of an individual measurement of the reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity of that reflection. 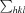 is the sum over all reflections and
is the sum over all reflections and 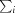 is the sum over i measurements of reflection hkl. Calculated for I > 3σ(I).
is the sum over i measurements of reflection hkl. Calculated for I > 3σ(I).
Acknowledgments
We sincerely thank the beamline scientists and staff of the Australian Synchrotron for their knowledge and assistance. JKF is an Australian Research Council (ARC) Future Fellow. JDN is an Australian Postgraduate Award recipient. Funding has been provided by the Australian government.
References
- Asturias, F. J., Chadick, J. Z., Cheung, I. K., Stark, H., Witkowski, A., Joshi, A. K. & Smith, S. (2005). Nature Struct. Mol. Biol. 12, 225–232. [DOI] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Bertherat, E., Thullier, P., Shako, J. C., England, K., Koné, M.-L., Arntzen, L., Tomaso, H., Koyange, L., Formenty, P., Ekwanzala, F., Crestani, R., Ciglenecki, I. & Rahalison, L. (2011). Emerg. Infect. Dis. 17, 778–784. [DOI] [PMC free article] [PubMed]
- Dutta, D., Bhattacharyya, S. & Das, A. K. (2012). Proteins, 80, 1250–1257. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Eschenfeldt, W. H., Lucy, S., Millard, C. S., Joachimiak, A. & Mark, I. D. (2009). Methods Mol. Biol. 498, 105–115. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Forwood, J. K., Thakur, A. S., Guncar, G., Marfori, M., Mouradov, D., Meng, W., Robinson, J., Huber, T., Kellie, S., Martin, J. L., Hume, D. A. & Kobe, B. (2007). Proc. Natl Acad. Sci. USA, 104, 10382–10387. [DOI] [PMC free article] [PubMed]
- Galimand, M., Carniel, E. & Courvalin, P. (2006). Antimicrob. Agents Chemother. 50, 3233–3236. [DOI] [PMC free article] [PubMed]
- Gurvitz, A. (2009). Mol. Genet. Genomics, 282, 407–416. [DOI] [PMC free article] [PubMed]
- Holm, L. & Park, J. (2000). Bioinformatics, 16, 566–567. [DOI] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Lai, C. Y. & Cronan, J. E. (2004). J. Bacteriol. 186, 1869–1878. [DOI] [PMC free article] [PubMed]
- Leibundgut, M., Maier, T., Jenni, S. & Ban, N. (2008). Curr. Opin. Struct. Biol. 18, 714–725. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Parish, T., Roberts, G., Laval, F., Schaeffer, M., Daffé, M. & Duncan, K. (2007). J. Bacteriol. 189, 3721–3728. [DOI] [PMC free article] [PubMed]
- Parsons, J. B. & Rock, C. O. (2013). Prog. Lipid Res. 52, 249–276. [DOI] [PMC free article] [PubMed]
- Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. (2003). Acta Cryst. D59, 1131–1137. [DOI] [PubMed]
- Price, A. C., Zhang, Y.-M., Rock, C. O. & White, S. W. (2001). Biochemistry, 40, 12772–12781. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Subramanian, S., Abendroth, J., Phan, I. Q. H., Olsen, C., Staker, B. L., Napuli, A., Van Voorhis, W. C., Stacy, R. & Myler, P. J. (2011). Acta Cryst. F67, 1118–1122. [DOI] [PMC free article] [PubMed]
- Tang, Y., Lee, H. Y., Tang, Y., Kim, C.-Y., Mathews, I. & Khosla, C. (2006). Biochemistry, 45, 14085–14093. [DOI] [PubMed]
- White, S. W., Zheng, J., Zhang, Y.-M. & Rock, C. O. (2005). Annu. Rev. Biochem. 74, 791–831. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zhou, D. & Yang, R. (2009). Infect. Immun. 77, 2242–2250. [DOI] [PMC free article] [PubMed]