A feast/famine regulatory protein (Rv2779c) from M. tuberculosis H37Rv was cloned, overexpressed, purified and crystallized. The crystals diffracted to a resolution of 2.8 Å and belonged to space group P21212.
Keywords: feast/famine regulatory proteins, Rv2779c, Mycobacterium tuberculosis
Abstract
Rv2779c from Mycobacterium tuberculosis is a feast/famine regulatory protein. This class of proteins are also known as the leucine-responsive regulatory protein/asparagine synthase C family (Lrp/AsnC) of transcriptional regulators and are known to be involved in various metabolic processes in bacteria and fungi. They contain a RAM (regulator of amino-acid metabolism) domain that is rarely found in humans and acts as the oligomerization domain. Since the oligomeric status is often linked to the particular functional role in these proteins, binding of ligands to the domain can elicit specific functional responses. Full-length Rv2779c corresponding to a molecular mass of 19.8 kDa and 179 residues was cloned and purified to homogeneity following transformation into Escherichia coli C41 (DE3) cells. Crystals were grown by vapour diffusion using the hanging-drop method. Diffraction data extending to 2.8 Å resolution were collected from a single crystal that belonged to space group P21212, with unit-cell parameters a = 99.6, b = 146.0, c = 49.9 Å. Matthews coefficient (V M) calculations suggest that four molecules are present in the asymmetric unit, corresponding to a solvent content of ∼46%. Molecular-replacement calculations using the crystal structure of a homologue, Rv3291c, as the search model gave an unambiguous solution corresponding to four subunits in the asymmetric unit.
1. Introduction
Mycobacterium tuberculosis, which is one of the most successful intracellular pathogens, has the ability to endure in the macrophage and monocytes in a dormant phase (Flynn & Chan, 2003 ▶). The sequencing of its genome, amongst other things, revealed that the pathogen possesses diverse classes of regulatory proteins (Cole et al., 1998 ▶). Nutrient-starved in vitro tuberculosis models designed to mimic persistence/latency have resulted in the identification of several regulatory proteins that are up-regulated in these models (Betts et al., 2002 ▶). Among these, we have focused on a class of regulators called the feast/famine regulatory proteins (FFRPs), also known as leucine-responsive regulatory protein/asparagine synthase C (Lrp/AsnC)-type regulators (Calvo & Matthews, 1994 ▶). This family of transcriptional regulators are widespread among prokaryotes and are involved in the regulation of amino-acid metabolism and related cellular processes (Brinkman et al., 2003 ▶). Additionally, electron-microscopy experiments suggest that they are involved in DNA packaging and form nucleoprotein complexes analogous to the nucleosome (Beloin et al., 2003 ▶). They are distinguished by the presence of a RAM (regulator of amino-acid metabolism) domain, Pfam 01842; these domains are also known as ACT domains after their identification in asparagine synthase C (Ettema et al., 2002 ▶). Since the latter domains are mostly restricted to prokaryotes, targeting them for the development of new inhibitors against pathogenic bacteria is expected to lead to reduced unwanted toxic effects in humans. FFRPs consist of a DNA-binding domain that contains a ‘helix–turn–helix’ motif and a RAM domain. The latter domain contains a conserved Gly that is important in ligand binding and adopts a distinctive αβ sandwich fold (βαββαβ). The two domains are joined by a long linker of about 20 amino-acid residues. The functional unit, as suggested by structural and other studies (Brinkman et al., 2003 ▶), is a dimer. The proteins adopt higher oligomeric states that are multiples of the basic dimeric unit, including tetramers, octamers etc. (Koike et al., 2004 ▶; Shrivastava & Ramachandran, 2007 ▶). Binding of amino acids can trigger specific oligomeric transitions, as observed in another FFRP, Rv3291c (Shrivastava & Ramachandran, 2007 ▶). Crystal structures have been reported for FFRPs from Pyrococcus sp. OT3, Bacillus subtilis, Escherichia coli, Neisseria meningitides, M. tuberculosis and Sulfolobus tokodaii (Leonard et al., 2001 ▶; Koike et al., 2004 ▶; Thaw et al., 2006 ▶; Ren et al., 2007 ▶; Shrivastava & Ramachandran, 2007 ▶; Shrivastava et al., 2009 ▶; Nakano et al., 2007 ▶).
Rv2779c is homologous to transcriptional regulators of the FFRP family. It is up-regulated threefold to fourfold in nutrient-deprived models of M. tuberculosis. Consequently, it has been thought to play an important role in the latent/persistence stage of the pathogen (Betts et al., 2002 ▶). It is present in the upstream region of the ald gene that encodes alanine dehydrogenase (ALD). Incidentally, the latter is listed in one study as among the top three targets against persistence (Hasan et al., 2006 ▶). Recently, it has been reported that in a hypoxic model of M. smegmatis the expression of alanine dehydrogenase increases and is regulated by aldR (Jeong et al., 2013 ▶), a gene whose product is homologous to the current protein. We have purified and crystallized the protein in order to understand the molecular mechanisms that are fundamental to its regulatory activity.
2. Materials and methods
2.1. Cloning
The Rv2779c gene was PCR-amplified from M. tuberculosis H37Rv genomic DNA. The sense primer, 5′-CGC ACC ATG GTA ATT CTT TTT CGA GGC C-3′, contains an NcoI site (shown in bold), while the antisense primer, 5′-GTT AAG CTT GCT TGC TCT GGA TGG GCG CCG C- 3′, contains a HindIII site (shown in bold). The 540 bp product was digested with NcoI and HindIII and further cloned into pET-21d (Novagen, USA), such that a hexahistidine tag was added at the C-terminus. The integrity of the construct was verified by sequencing.
2.2. Overexpression
The recombinant plasmid was transformed into chemically competent E. coli C41 (DE3) cells. Transformed cells were cultured in LB medium supplemented with 100 µg ml−1 carbenicillin at 303 K. Expression of recombinant Rv2779c-His6 was induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when an OD600 of 0.6 was reached. After induction, the cells were further grown for 12–14 h at 303 K, harvested by centrifugation at 8000g for 10 min at 270 K and resuspended in 40 ml ice-cold sonication buffer A (50 mM HEPES pH 7.0, 1.5 M NaCl, 10 mM imidazole) supplemented with 12% glycerol. The cells were frozen, thawed and then lysed by sonication using a Vibra-Cell (Sonics & Materials, USA) instrument using a medium-size probe at 20% output power, 50% duty cycle with a pulse time of 40 s. Before sonication, 1000 µM phenylmethylsulfonyl fluoride (PMSF), a protease inhibitor, was added to the thawed culture. The cell lysate was then centrifuged at 21 365g (13 000 rev min−1) in a Heraeus Multifuge X3R for 30 min at 270 K to remove cell debris.
2.3. Purification
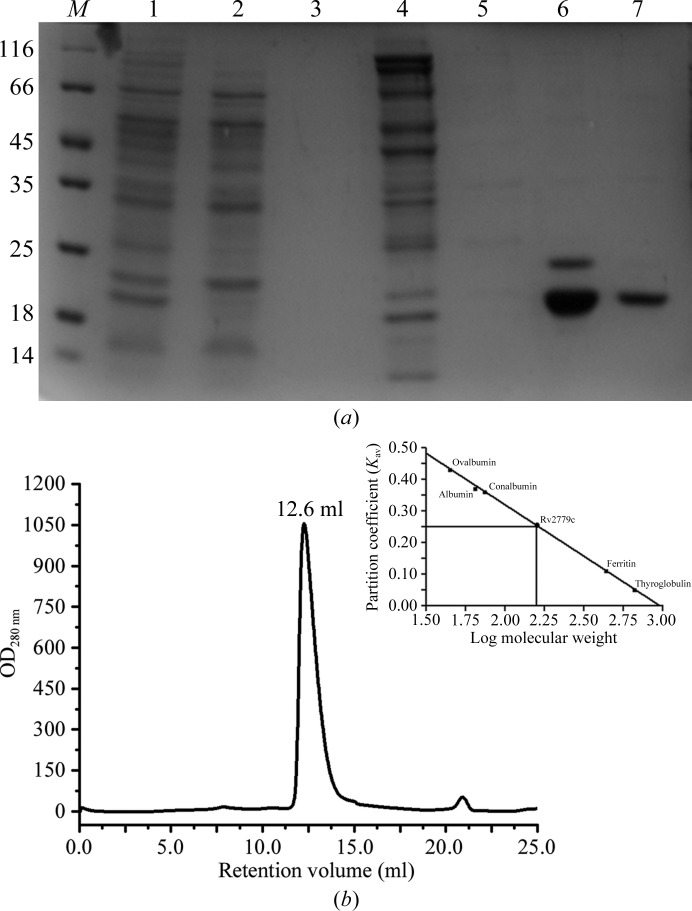
The clear supernatant from the above step was loaded onto an Ni2+–IDA column (GE Healthcare) pre-equilibrated with buffer A. The column was subsequently washed with five column volumes of buffer A and then with five column volumes of wash buffer B (50 mM HEPES pH 7.0, 500 mM NaCl, 80 mM imidazole), and finally with five column volumes of wash buffer C (50 mM HEPES pH 7.0, 500 mM NaCl, 125 mM imidazole). The bound protein was then eluted using a linear gradient of 130–600 mM imidazole in buffer C. Purity was monitored by running the samples on a 12% SDS–PAGE gel (Fig. 1 ▶ a). The gel was stained with Coomassie Brilliant Blue R-250 (CBB). Protein fractions that were obtained following affinity purification were pooled and concentrated using an Amicon Ultra-10 concentrator (Millipore, USA). The concentrated sample was then subjected to size-exclusion chromatography using a Superdex S-200 HR10/300 column pre-equilibrated with buffer consisting of 50 mM HEPES pH 7.0, 250 mM NaCl, 5 mM EDTA, 10% glycerol and mounted on an ÄKTA FPLC system (GE Healthcare). The protein eluted at 12.6 ml (Fig. 1 ▶ b) elution volume. The purity was confirmed by running a 12% SDS–PAGE gel. The column was calibrated using standard molecular-weight protein markers for gel filtration procured from GE Healthcare. The standard proteins were dissolved in 1 ml of the same buffer and loaded onto the same column. The partition coefficient (K av) was then plotted as a function of log M r of the standard protein according to
where V e is the elution volume of the protein, V o is the void volume of the column, V t is the total volume of the column and M r is the molecular weight of the particular protein.
Figure 1.
(a) Coomassie Brilliant Blue R-250-stained 12% SDS–PAGE gel of expressed Rv2779c. Lane M, molecular-weight markers (labelled in kDa); lane 1, total lysate after sonication; lane 2, flowthrough collected after loading; lane 3, sample washed with buffer A; lane 4, sample washed with buffer B; lane 5, sample washed with buffer C; lane 6, Ni2+–IDA-purified protein; lane 7, purified protein after size-exclusion chromatography. (b) Gel-filtration profile of Rv2779c from a Superdex S-200 HR10/300 column. The total column volume is 25 ml, while the void volume of the column is 8.43 ml. The protein eluted at 12.6 ml, consistent with an octameric association in solution.
2.4. Crystallization and data collection
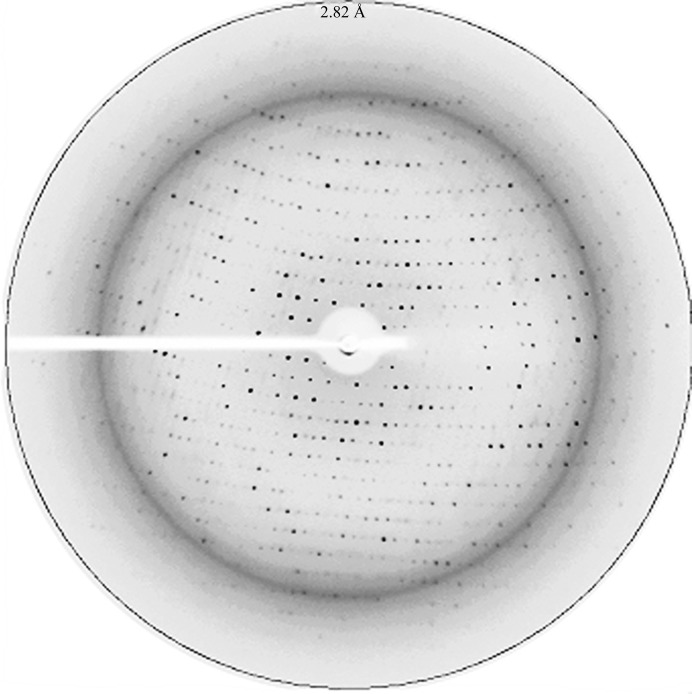
For crystallization, the protein was concentrated to about 7 mg ml−1; the concentration was determined by the Bradford method (Bradford, 1976 ▶). Crystallization trials were initiated by the hanging-drop vapour-diffusion method at 295 K using the commercially available Crystal Screen, Crystal Screen 2 and Index screens from Hampton Research. Crystallization experiments were set up using a variety of protein concentrations (5, 6 and 7 mg ml−1). In the experiments, 3 µl protein solution was mixed with 1.5 µl reservoir solution and equilibrated against 1000 µl reservoir solution. Initial hits were optimized by grid screening. The crystals used in the diffraction experiments appeared in 2–3 d and grew to full size within a week (Fig. 2 ▶). For X-ray data collection, crystals were mounted on CryoLoops (Hampton Research), rinsed with cryoprotectant solution [25%(v/v) glycerol in reservoir solution] and flash-cooled directly in a nitrogen stream at 100 K (Oxford Cryosystems). Diffraction data to 2.8 Å resolution (Fig. 3 ▶) were collected in-house using a Rigaku MicroMax-007 HF rotating-anode X-ray generator operated at 40 kV and 30 mA and a MAR345 detector. The crystal-to-detector distance was set to 275 mm. A total range of 160° was covered with 1° oscillation per frame. The reflections were processed and scaled using the HKL-2000 program package (Otwinowski & Minor, 1997 ▶). Diffraction data statistics are given in Table 1 ▶.
Figure 2.
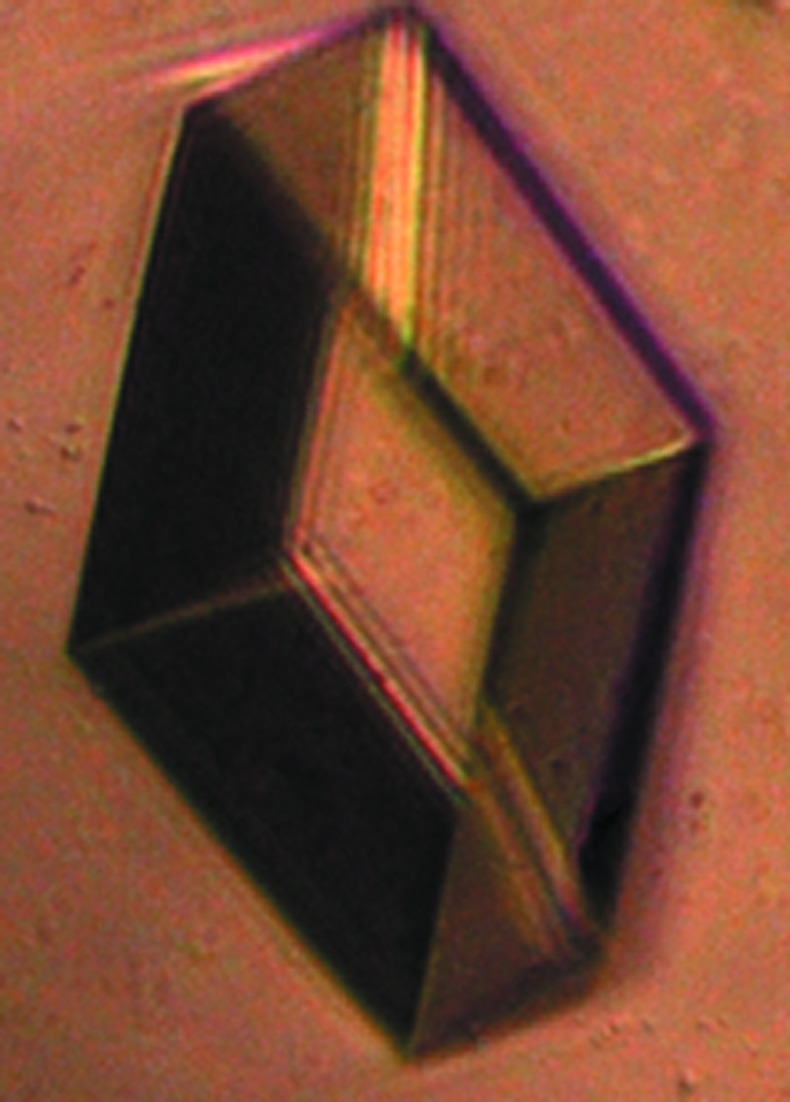
A single crystal of Rv2779c grown by hanging-drop vapour diffusion. The crystals have typical dimensions of 0.13 × 0.1 × 0.07 mm.
Figure 3.
A snapshot of the diffraction pattern of an Rv2779c crystal. The image corresponds to 1° oscillation with 4 min exposure time and a crystal-to-detector distance of 275 mm. The edge of the circle represents 2.82 Å resolution.
Table 1. Data-collection statistics for Rv2779c.
Values in parentheses are for the outermost shell.
| Wavelength (Å) | 1.5418 |
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 99.6, b = 146, c = 49.9 |
| Resolution (Å) | 25.87–2.82 (2.95–2.82) |
| Total No. of observations | 85516 (8354) |
| No. of unique reflections | 17306 (1782) |
| Multiplicity | 4.9 (4.7) |
| 〈I/σ(I)〉 | 18.6 (5.2) |
| Completeness (%) | 92 (66) |
| R merge † | 0.05 (0.26) |
| Mosaicity (°) | 0.6 |
R
merge = 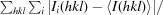
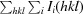 , where Ii(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the average intensity of the i observations.
, where Ii(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the average intensity of the i observations.
2.5. Structure solution and refinement
Molecular-replacement calculations were carried out using Phaser (McCoy et al., 2007 ▶) as implemented in the CCP4 package (Winn et al., 2011 ▶) and all data in the resolution range 25.87–2.82 Å were used in the calculations. The structure of another FFRP from M. tuberculosis, Rv3291c (PDB entry 2ivm; Shrivastava & Ramachandran, 2007 ▶), was used as the search model following a sequence-homology search. Phaser gave an unambiguous solution with four monomers in the crystallographic asymmetric unit. The initial model was refined using the maximum-likelihood method implemented in REFMAC5 (Murshudov et al., 2011 ▶).
3. Results and discussion
The FFRP family is implicated in the control of multiple cellular functions, with small molecules such as amino acids functioning as effectors. The exact physiological roles and the identity of many of the target genes of these regulators remain unidentified. As a first step to gain insights into its molecular mechanisms, Rv2779c was successfully cloned into pET-21d and overexpressed in E. coli C41 (DE3) cells. Purified protein was obtained by a two-step protocol consisting of affinity and size-exclusion chromatography. The molecular weight of 21.4 kDa for a subunit of His6-tagged Rv2779c was confirmed by 12% SDS–PAGE. Size-exclusion chromatography experiments are in agreement with an octameric association in solution for the protein. Crystals suitable for X-ray analysis were obtained by the hanging-drop vapour-diffusion method in 0.2 M trisodium citrate dihydrate, 20% PEG 3350. The crystals diffracted to 2.8 Å resolution and belonged to space group P21212, with unit-cell parameters a = 99.6, b = 146, c = 49.9 Å. The crystal mosaicity was around 0.6°, with an overall data completeness of 92%. Assuming that the asymmetric unit contains a tetramer, the calculated Matthews coefficient is 2.29 Å3 Da−1 (Matthews, 1968 ▶), corresponding to 46% solvent content. A sequence-based homology search of Rv2779c against the Protein Data Bank (http://www.rcsb.org) using BLAST (http://blast.ncbi.nlm.nih.gov) showed that Rv2779c has 25% sequence identity to Rv3291c (PDB entry 2ivm; Shrivastava & Ramachandran, 2007 ▶), which was thus used as a search model for molecular replacement. A total of 5% of the reflections were used for the calculation of R free (Brünger, 1992 ▶) in the refinement. Initial rigid-body refinement with REFMAC5 (Murshudov et al., 2011 ▶) using the model ouput by Phaser gave an R work of 36% and an R free of 42%. Examination of the crystal symmetry reveals that the four subunits in the asymmetric unit associate to form an octamer broadly similar to that reported for FFRPs such as Rv3291c. Further structural refinement and model building are currently under way.
Acknowledgments
AD is the recipient of junior and senior research fellowships from the Indian Council of Medical Research, New Delhi. Funding from the Council of Scientific and Industrial Research, India (network project SPLenDID, BSC0104) and the Department of Biotechnology, India (National Bioscience Award 2010 grant to RR; No. GAP0083) are acknowledged. This article bears CSIR–CDRI communication No. 8579.
References
- Beloin, C., Jeusset, J., Révet, B., Mirambeau, G., Le Hégarat, F. & Le Cam, E. (2003). J. Biol. Chem. 278, 5333–5342. [DOI] [PubMed]
- Betts, J. C., Lukey, P. T., Robb, L. C., McAdam, R. A. & Duncan, K. (2002). Mol. Microbiol. 43, 717–731. [DOI] [PubMed]
- Bradford, U. K. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Brinkman, A. B., Ettema, T. J. G., de Vos, W. M. & van der Oost, J. (2003). Mol. Microbiol. 48, 287–294. [DOI] [PubMed]
- Brünger, A. T. (1992). Nature (London), 355, 472–475. [DOI] [PubMed]
- Calvo, J. M. & Matthews, R. G. (1994). Microbiol. Rev. 58, 466–490. [DOI] [PMC free article] [PubMed]
- Cole, S. T. et al. (1998). Nature (London), 393, 537–544.
- Ettema, T. J. G., Brinkman, A. B., Tani, T. H., Rafferty, J. B. & Van Der Oost, J. (2002). J. Biol. Chem. 277, 37464–37468. [DOI] [PubMed]
- Flynn, J. L. & Chan, J. (2003). Curr. Opin. Immunol. 15, 450–455. [DOI] [PubMed]
- Hasan, S., Daugelat, S., Srinivasa Rao, P. S. & Schreiber, M. (2006). PLoS Comput. Biol. 2, e61. [DOI] [PMC free article] [PubMed]
- Jeong, J.-A., Baek, E.-Y., Kim, S. W., Choi, J.-S. & Oh, J. -I. (2013). J. Bacteriol. 195, 3610–3620. [DOI] [PMC free article] [PubMed]
- Koike, H., Ishijima, S. A., Clowney, L. & Suzuki, M. (2004). Proc. Natl Acad. Sci. USA, 101, 2840–2845. [DOI] [PMC free article] [PubMed]
- Leonard, P. M., Smits, S. H., Sedelnikova, S. E., Brinkman, A. B., de Vos, W. M., van der Oost, J., Rice, D. W. & Rafferty, J. B. (2001). EMBO J. 20, 990–997. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nakano, N., Kumarevel, T., Matsunaga, E., Shinkai, A., Kuramitsu, S. & Yokoyama, S. (2007). Acta Cryst. F63, 964–966. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ren, J., Sainsbury, S., Combs, S. E., Capper, R, G., Jordan, P, W., Berrow, N. S., Stammers, D. K., Saunders, N, J. & Owens, R. J. (2007). J. Biol. Chem. 282, 14655–14664. [DOI] [PubMed]
- Shrivastava, T., Dey, A. & Ramachandran, R. (2009). J. Mol. Biol. 392, 1007–1019. [DOI] [PubMed]
- Shrivastava, T. & Ramachandran, R. (2007). Nucleic Acids Res. 35, 7324–7335. [DOI] [PMC free article] [PubMed]
- Thaw, P., Sedelnikova, S. E., Muranova, T., Wiese, S., Ayora, S., Alonso, J. C., Brinkman, A. B., Akerboom, J., van der Oost, J. & Rafferty, J. B. (2006). Nucleic Acids Res. 34, 1439–1449. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.