Abstract
In the last few decades through an awareness of transfusion transmitted infections (TTI), a majority of countries have mandated serology based blood screening assays for Human immunodeficiency virus (HIV), Hepatitis C virus (HCV), and Hepatitis B virus (HBV). However, despite improved serology assays, the transfusion transmission of HIV, HCV, and HBV continues, primarily due to release of serology negative units that are infectious because of the window period (WP) and occult HBV infections (OBI). Effective mode of nucleic acid technology (NAT) testing of the viruses can be used to minimize the risk of TTIs. This review compiles the examples of NAT testing failures for all three viruses; analyzes the causes for failure, and the suggestions from retrospective studies to minimize such failures. The results suggest the safest path to be individual donation testing (ID) format for highest sensitivity, and detection of multiple regions for rapidly mutating and recombining viruses. The role of blood screening in the context of the donation and transfusion practices in India, the donor population, and the epidemiology is also discussed. World wide, as the public awareness of TTIs increases, as the recipient rights for safe blood are legally upheld, as the possibility to manage diseases such as hepatitis through expensive and prolonged treatment becomes accessible, and the societal responsibility to shoulder the health costs as in the case for HIV becomes routine, there is much to gain by preventing infections than treating diseases.
Keywords: Donor look back, individual donation nucleic acid technology testing, multi pool nucleic acid technology testing, recipient trace back, transfusion transmitted viral infections, window period and occult infections
Introduction
The three major transfusion transmitted infections (TTIs) of viral origin associated with blood transfusion are human immunodeficiency virus (HIV), hepatitis C virus (HCV) and hepatitis B virus (HBV). The mandated blood screening for these in India are serological tests for HBV surface antigen (HBsAg) and antibodies for HCV and HIV-1/2.[1] Although, the mandate for serological screening is important, it does not address the critical window period (WP). The WP is that period of time from infection to the time of detection by a given blood screening assay.[2,3] WP cases for all three viruses are well documented and additionally in the case of HBV, the occult cases as occult HBV infection (OBI) represent the risks and result in TTIs.[3,4,5,6]
The need for safe blood is of utmost importance to the repeat transfusion requiring group of patients with thalassemia, sickle cell anemia and hemophilia. Amongst the chronically transfused patients such as thalassemics, the HBV prevalence as measured by HBsAg presence is 33%,[7] while the prevalence in blood donor population is 1.2%;[8] and the prevalence of HCV in thalassemics as measured by anti-HCV presence is 57%,[9] while the prevalence in blood donor population is 0.4%.[8] These data are amongst the growing evidence that infectious blood screened by serology testing, misses some infectious units during the WP.[3,5] This has necessitated detection of viral nucleic acid (NA) markers, which are propagated prior to serological markers and therefore shortening the infectious WP. As shown in [Table 1], the nucleic acid technology (NAT) enables detection of infectious units at an earlier period of infection that are serology marker negative.[3,10]
Table 1.
Comparison of detection with NAT versus serology testing for HIV-1, HCV and HBV and the window period gain
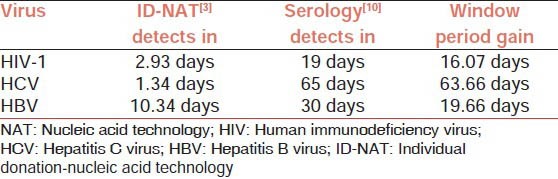
The NAT tests of high sensitivity rely on amplification of intended regions of viral nucleic acids for detection. The objective of NAT testing should be focused on the detection of as few infectious viral copies as possible to provide near zero risk blood for recipients. This is dependent on the analytical sensitivity of the NAT assay for the various genotypes, sub groups, and variant regions. Even more important is the screening sensitivity of the NAT assay which is determined by the mode of practice of the NAT assay. NAT assay can be executed in the format of (a) individual donation (ID), or (b) pools of multiple (MP) donors. ID NAT testing utilizes an aliquot of each sample tested individually, with no dilution due to addition of non-infectious sample aliquots hence providing the highest detection sensitivity equivalent to the analytical sensitivity; whereas for MP NAT testing, pools of various numbers of donations are equally combined prior to testing, with the pool size determining the screening sensitivity. The pool size determines the dilution of the number of virions in a single reactive sample member of a pool, and the decrease in the concentration of the virions in the testing aliquot and the consequent decrease in the probability of detection. Accordingly, in the absence of any intervening strategies such as ultracentrifugation to concentrate the virus, the screening sensitivity of pools of 96 is likely 96 times less than the analytical sensitivity[11] and of pools of 6 has been demonstrated to be six times less than the analytical sensitivity.[12] Screening sensitivity is the pool size dependent detection limit of an assay. In the final analysis, it is the screening sensitivity determined by the pool size that governs the detection status of the assay in the laboratory. For various pool sizes there are examples of reported incidents of TTIs through detection failures, and also theoretical projections that predict the risk days.[3,5,10] The infective potency of HIV, HCV and HBV and that of their sub types also need to be considered in deciding on a test strategy. Typically when components are prepared, the circulating viral copies are the highest in the plasma component, with decreased concentration in the platelet fraction (50 mL residual volume of plasma) and the lowest in packed red blood cells (PRBC) (20 mL residual volume of plasma). In most countries, plasma is optionally quarantined until confirmed to be negative by repeat donation. However, in India, component transfusion is not widely practiced and the largest percentage of transfusion is of whole blood,[13] which carries the same high risk as plasma. The TTI risk in India could be reduced significantly through the appropriate use of platelets and PRBC fractions. Additional ramification of NAT testing extends to TTI recipients who donate organs during WP, which then can affect several more lives.[14]
Transfusion-transmitted infections are usually realized through two mechanisms: (a) repeat donor during the inter-donation interval turning NAT reactive through increased viral load, or sero-reactive through seroconversion, triggering hemovigilance look back investigation and retesting of index and previous donations; (b) recipient acquiring a TTI, and triggering donor trace back hemovigilance and investigation of the index, and pre-post index units. The retesting generally involves (a) NAT testing if only serology testing was performed; (b) testing in ID NAT format if previously tested in MP NAT format; (c) testing by a more sensitive NAT assay; (d) and/or multiple repeats of ID NAT to query larger volume of plasma, which helps in case of low viral load. The causal association of the TTI is typically demonstrated through genomic sequence comparison in the index unit and/or available previous donation aliquots, to that of the recipient.
As discussed later in this review it is noteworthy that globally an overwhelming majority of TTIs are realized through donors testing positive during repeat donations. This represents the incidence of infection in blood donors; and the recommended time between donations can vary from 6 weeks as in Japan to 3 months in India. However, in India repeat donations is less common and repeat donation at 3-4 months interval is even less common; and donor exclusion through questionnaire and pre and post donation donor counseling is inadequate. Additionally the absence of national donor registry, absence of hemovigilance and recipient follow-up programs, and complications of legal issues render both the donor based and recipient based approaches unenforceable. Hence, at the present time implementing a highly sensitive NAT screening assay for HIV, HCV and HBV is the solution for India to enhance blood safety for these viruses. In view of this, it is imperative to analyze the situations that result in screening detection failures. In this review, we note the NAT detection misses for HIV-1, HCV and HBV, in an effort to understand and suggest a preferred screening strategy.
HIV-1 NAT Blood Screening Detection Failures
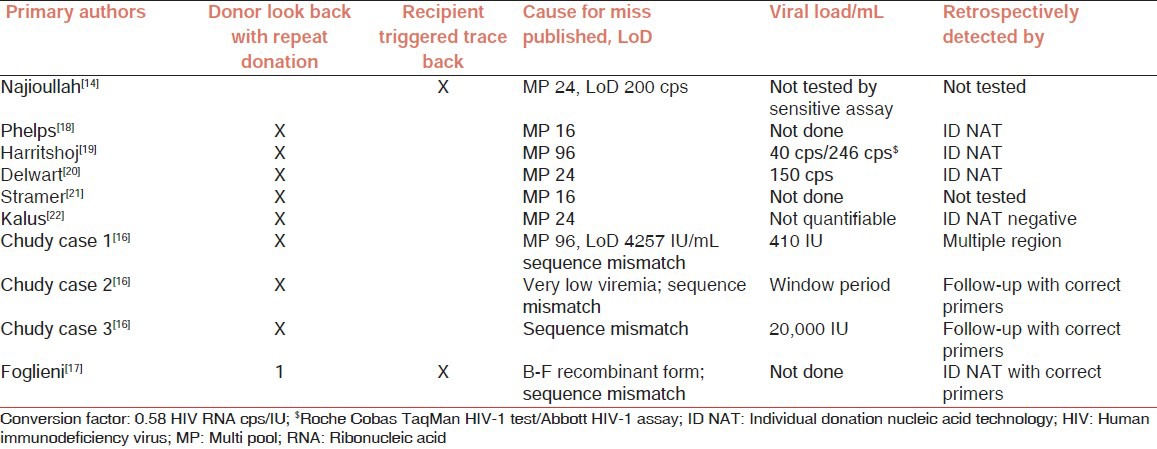
The NAT assays to detect HIV RNA have been successfully practiced for more than a decade, and have been instrumental in curtailing the increase of the disease. Although the NAT HIV screening assays typically have good sensitivity, the occurrence of several TTIs does point to deficiencies. The Paul-Ehrlich-Institut (PEI) has defined minimal NAT HIV sensitivity of 10,000 IU/mL (6,000 cps/mL), which permitted pooled testing and has resulted in several detection failures and release of potentially infectious units.[15] The reasons for HIV NAT detection failures include: (a) decreased sensitivity in pooled format testing,[11,12] (b) the propensity of HIV RNA virus to point mutate[16] and (c) recombination between different subtypes when infected with multiple strains.[17] The mutations and recombinations effect nucleotide changes and if these are in the primer probe binding regions it results in decreased efficiency in isolation, amplification, detection or even complete misses. Table 2 presents HIV-1 RNA cases that were missed by NAT screening,[11,14,16,17,18,19,20,21,22] either because of low viral load, or because of sequence variability and definitely challenge the PEI recommended minimum sensitivity guidelines. Of the ten NAT detection failures seven were tested in pools ranging from 16 to 96. Of these, three were retrospectively proven to be detectable by ID NAT, two were untested by ID NAT, one was ID NAT negative; and four were missed due to sequence mutations resulting in primer probe mismatch. These four were detected by alternate assays, or redesigned assays or by detecting alternate regions. In a study of 18 HIV positive samples spanning a period of 9 years of infection, 12 were either inadequately detected or missed because of recombination between different genotypes.[17] Following these significant number of detection failures due to sequence mismatch, detection of two regions of HIV-1 sequence has been required by PEI (notification to §21a Medicinal Products Law [AMG]). An additional miss in pools of 50 that resulted in TTI reported by Satake et al.[23] is not included in the table, since sufficient details are not available. It is also of importance to note that the realization of all diagnostic misses with a single exception was triggered through seroconversion of the donor detected during repeat donation. In the current Indian blood banking situation which relies on replacement donors, with an insignificant base of repeat voluntary donors and lacks a national donor registry such diagnostic lapses will remain unrealized.
Table 2.
Summary of NAT testing missed HIV-1 blood screening results of index donations
HCV NAT Blood Screening Detection Failures
NAT testing has offered the highest benefit for HCV detection due to the prolonged interval needed (>60 days) for the development of the serology marker (anti HCV antibody).[3,10] With 0.4% prevalence in the Indian donor population, HCV is the second highest detected TTI.[8] The infectivity of HCV is predicted to be as low as 1 cp/mL[10] and there have been TTI HCV cases with viral loads of below quantifiable levels. The examples include four HCV diagnostic misses all of which resulted in TTIs, two from Germany, one each from Poland and US which were missed by MP NAT screening in pools of 24[24,25,26] and 48.[27] Again, with the exception of Schuttler et al.,[24] all other TTI cases were identified through the donor turning HCV positive during repeat donation. In a look back analysis, TTIs from two instances could have been prevented had the specimens been tested in the single test ID NAT mode to equal the analytical sensitivity.[25,27] The remaining two[24,26] had very low viral load and hence were variably positive even with replicate testing in the ID NAT mode. The estimated total copies transfused, which resulted in infection was as low as 400-500 copies (2.7 cps/IU).[24,26] The minimum detection mandated by PEI for HCV of 5,000 IU/mL (136,500 cps/mL) has made possible pooled testing and will likely result in release of infectious units.[15]
HBV NAT Blood Screening Detection Failures
The benefit of NAT for HBV has been the most challenging to demonstrate for several reasons: (a) HBV has a slow doubling time of 2.7 days resulting in slow increase in viral load, hence the longest WP from infection to detection even for NAT testing;[10] (b) the screening marker of HBsAg being a viral protein is detected early in infection compared to antibody markers, thus showing a less significant WP gain by NAT;[10] (c) the HBsAg protein is produced in abundance as non-deoxyribonucleic acid (DNA) containing defective viral particles possibly at a ratio of 1000 for every DNA containing infectious virion,[28] offering an advantage for the serological detection of abundant HBsAg; and (d) the uncertainty in proving the benefit of HBV NAT in countries that are early implementers, which also have low HBV prevalence.[29,30] Yet, detection failures of HBV with a complex infective status of multiple WPs and occult infections;[6,31,32,33,34,35,36,37] and genetic recombinations[38,39,40] has resulted in highest number of TTIs [Table 3].[31,33,38,40,41,42,43] The delay in adapting HBV ID NAT testing has led to a number of retrospective studies.[6,33,38,40,41,43] Globally, HBV NAT testing has been gaining acceptance for several reasons including: (a) HBsAg, the commonly diagnosed serology marker, unlike antibody markers, is a transiently expressed viral protein detectable late in infection at 30 days and lasting up to 63 days,[10,31] (b) mutations in HBsAg result in serology misses,[33] (c) in certain genotypes such as G, the HBsAg negative WP is substantially prolonged and will be missed by serology tests,[37] (d) in countries with moderate to high HBV prevalence, positivity for antibody markers will be high resulting in high discard rate, which is not viable for blood supply[29,30,44] and finally (e) several studies have demonstrated the benefit of high sensitivity ID NAT testing to detect WP and OBI infections.[30] Factors that necessitate highly sensitive HBV NAT testing in India include (a) relatively high HBsAg positivity of 1.2% in the donor population,[8] (b) a 10-30% anti-Hepatitis B core (HBc) prevalence,[44] (c) the major HBV type being Genotype D, followed by A and C,[45,46] with genotypes D and A documented to remain at low viral load[6,34] and (d) the additional association of HBV genotype D with high occult HBV infections, which is characterized by fluctuating low viral load.[6,35,39,41,42]
Table 3.
Summary of NAT testing missed HBV blood screening and repository sample results of index donations
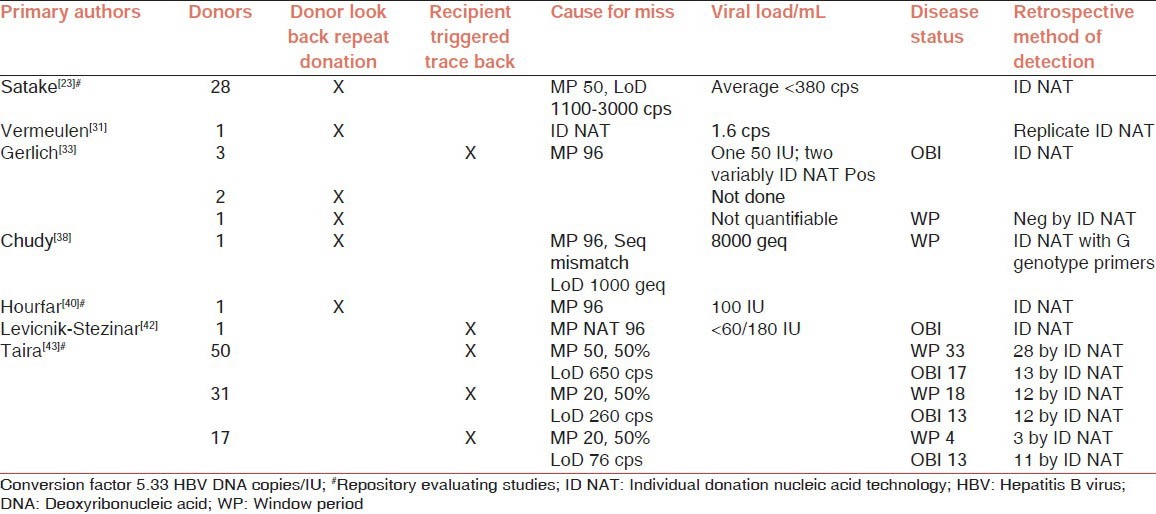
Typically in countries such as Japan with medium HBV endemicity where pooled NAT testing is practiced, the HBV TTIs are kept at low numbers through additional interventions.[41] These include: (a) several types of mandatory HBV serological assays (b) with cut off definitions for anti-HBc to remove possibly infectious units and (c) preferred concentration of anti-HBs to neutralize low infectious DNA, (c) practicing only 6 week interval for repeat donation, enabling identification of WP infections and variably HBV infective occult donors within short intervals.[41,47] Despite these interventions as shown in Table 3, the retrospective study by Taira et al.[43] indicated that 86% of WP and 88% of OBI TTIs resulting from screening as MP20 could have been averted with ID NAT. Several countries also recommend HBV vaccination for repeat volunteer donors, however reports of vaccine breakthrough cases indicate that this may not be a sufficiently safe path.[48] Since all these checks and balances and preventive measures are missing in India, the only solution is to have a HBV NAT assay practiced in a format to have the highest sensitivity.[4]
Comparative Evaluation of ID versus MP NAT Performance with Clinical Specimens
In addition to screening misses, several comparative studies evaluating ID NAT versus pool testing, using the same reagent, thereby excluding the variability of reagent and platforms have been executed. Shan et al.[49] comparing ID NAT with pools of 16 using Procleix Duplex reagent observed that the WP extended by 2.1 days for HIV and 2.4 days for HCV for MP16. González et al.[35] observed a 5.1 fold greater HBV yield by ID NAT versus MP8 by the Procleix Ultrio assay. Yang et al.[50] identified 3-fold higher HBV yields by ID NAT versus MP4 also using the Procleix Ultrio assay. Over 4 years of blood donor screening at South African Blood Services, 336 ID-NAT yields that were negative for HBsAg had been identified and testing these in MP16 established that 237 (71%) of the yields would have been missed.[5] Using the Procleix Ultrio Plus assay Vermeulen et al. observed a decrease in detection of HIV NAT yields as follows: 10% in MP4, 20% in MP8, 30% in MP16.[51] Grabarczyk et al.[52] compared HBV NAT yields for MP6 with Cobas TaqScreen MPX versus Cobas Ampliscreen in MP24 and observed 12.6 fold increase for HBV WP and 6.6 fold increase for OBI NAT yields. Roth[4] observed that despite enriching interventions such as centrifugation, MP96 missed 22% of the ID NAT detectable samples.
In order to understand the performance of the commercial reagents in the commonly practiced format, the Procleix Ultrio Plus assay in the ID NAT format has been compared with the TaqScreen assay in the MP6 format. For HIV-1 NAT yields, the decrease in detection from ID NAT with Procleix Ultrio Plus assay was 17% in pools of 6 by Cobas TaqScreen assay.[51] For HBV the overall decrease was 30%, with more pronounced decrease during 2nd WP[31] characterized by low viral load (49% decrease) and OBI (33% decrease) HBV infection status.[53]
Globally, public awareness of TTIs following MP NAT testing has resulted in increased adoption of ID NAT testing.[10] Nearly 22 countries, constituting 60% of NAT testing countries around the world practice ID NAT. The remaining countries practice various pooled configurations.[54] With automation and increased through-put in the past 4 years, the trend has been towards changing from MP NAT to ID NAT. The ability to successfully implement ID NAT in India and its impact on interdicting release of infectious units has been demonstrated in a multicenter study with centralized testing[55] and on-site screening studies.[56] Although the argument of affordability of ID NAT has been the premise for promotion of pool testing, the residual risk of the resulting TTIs places a serious burden on the health-care, finances, society and human life. The cost of ART treatment substantially borne by the government could be decreased by improved blood screening to prevent potential HIV TTIs. The situation for Hepatitis is even grimmer, since the manifestation of disease takes a long time and there is no national policy for patient counseling or treatment. The treatment costs for hepatocellular carcinoma and decompensated cirrhosis are the highest in the health-care management.[57] Through health economics it appears to be more cost beneficial to screen blood with ID NAT to interdict HBV and HCV.
Conclusion
This paper has reviewed the current global use of NAT to screen the blood supply. Clearly many nations have embraced NAT and after realizing the utility of ID NAT have reduced their pool sizes.[54] Although it is important to consider the affordability of NAT, global experience strongly suggests that well-designed health economic studies, when performed in India, will-definitely point to the use of ID NAT. In the interest of public health and transfusion safety, immediate urgent attention should be paid to ID NAT.
Acknowledgement
The author thanks Dr. Jerry A. Holmberg for valuable suggestions.
Footnotes
Source of Support: Venkatakrishna Shyamala of Research Diagnostics is an independent consultant and part-time consultant with Novartis Vaccines and Diagnostics
Conflicting Interest: None declared.
References
- 1.Regulatory requirements of blood and /or its components including blood products. [last accessed on 2013 Oct 10]. Available from: http://cdsco.nic.in/html/guideline.htm .
- 2.Weusten JJ, van Drimmelen HA, Lelie PN. Mathematic modeling of the risk of HBV, HCV, and HIV transmission by window-phase donations not detected by NAT. Transfusion. 2002;42:537–48. doi: 10.1046/j.1537-2995.2002.00099.x. [DOI] [PubMed] [Google Scholar]
- 3.Weusten J, Vermeulen M, van Drimmelen H, Lelie N. Refinement of a viral transmission risk model for blood donations in seroconversion window phase screened by nucleic acid testing in different pool sizes and repeat test algorithms. Transfusion. 2011;51:203–15. doi: 10.1111/j.1537-2995.2010.02804.x. [DOI] [PubMed] [Google Scholar]
- 4.Roth WK. Hepatitis B and blood transfusion. ISBT Sci Ser. 2007;2:178–83. [Google Scholar]
- 5.Vermeulen M, Lelie N, Reddy R. Recent insights in testing for transfusion transmissible viral infections. ISBT Sci Ser. 2011;6:229–33. [Google Scholar]
- 6.Allain JP, Mihaljevic I, Gonzalez-Fraile MI, Gubbe K, Holm-Harritshøj L, Garcia JM, et al. Infectivity of blood products from donors with occult hepatitis B virus infection. Transfusion. 2013;53:1405–15. doi: 10.1111/trf.12096. [DOI] [PubMed] [Google Scholar]
- 7.Singh H, Pradhan M, Singh RL, Phadke S, Naik SR, Aggarwal R, et al. High frequency of hepatitis B virus infection in patients with beta-thalassemia receiving multiple transfusions. Vox Sang. 2003;84:292–9. doi: 10.1046/j.1423-0410.2003.00300.x. [DOI] [PubMed] [Google Scholar]
- 8.2010. [Last accessed on 2013 Oct 10]. p. 74. http://aidsdatahub.org/en/india-reference-library/item/12112-annual-cmis-bulletin-2008-09-national-aids-control-organisationindia .
- 9.Mathur M, Wanjari K, Turbadkar D. Seroprevalence of HIV, hepatitis C and hepatitis B in multitransfused thalassemics. Indian J Med Microbiol. 2008;26:205–6. doi: 10.4103/0255-0857.40552. [DOI] [PubMed] [Google Scholar]
- 10.Kleinman SH, Lelie N, Busch MP. Infectivity of human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus and risk of transmission by transfusion. Transfusion. 2009;49:2454–89. doi: 10.1111/j.1537-2995.2009.02322.x. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt M, Korn K, Nübling CM, Chudy M, Kress J, Horst HA, et al. First transmission of human immunodeficiency virus Type 1 by a cellular blood product after mandatory nucleic acid screening in Germany. Transfusion. 2009;49:1836–44. doi: 10.1111/j.1537-2995.2009.02203.x. [DOI] [PubMed] [Google Scholar]
- 12.Assal A, Barlet V, Deschaseaux M, Dupont I, Gallian P, Guitton C, et al. Comparison of the analytical and operational performance of two viral nucleic acid test blood screening systems: Procleix Tigris and cobas s 201. Transfusion. 2009;49:289–300. doi: 10.1111/j.1537-2995.2008.01965.x. [DOI] [PubMed] [Google Scholar]
- 13.Marwaha N. Whole blood and component use in resource poor settings. Biologicals. 2010;38:68–71. doi: 10.1016/j.biologicals.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Najioullah F, Barlet V, Renaudier P, Guitton C, Crova P, Guérin JC, et al. Failure and success of HIV tests for the prevention of HIV-1 transmission by blood and tissue donations. J Med Virol. 2004;73:347–9. doi: 10.1002/jmv.20097. [DOI] [PubMed] [Google Scholar]
- 15.Nübling CM, Heiden M, Chudy M, Kress J, Seitz R, Keller-Stanislawski B, et al. Experience of mandatory nucleic acid test (NAT) screening across all blood organizations in Germany: NAT yield versus breakthrough transmissions. Transfusion. 2009;49:1850–8. doi: 10.1111/j.1537-2995.2009.02212.x. [DOI] [PubMed] [Google Scholar]
- 16.Chudy M, Weber-Schehl M, Pichl L, Jork C, Kress J, Heiden M, et al. Blood screening nucleic acid amplification tests for human immunodeficiency virus Type 1 may require two different amplification targets. Transfusion. 2012;52:431–9. doi: 10.1111/j.1537-2995.2011.03281.x. [DOI] [PubMed] [Google Scholar]
- 17.Foglieni B, Candotti D, Guarnori I, Raffaele L, Berzuini A, Spreafico M, et al. A cluster of human immunodeficiency virus Type 1 recombinant form escaping detection by commercial genomic amplification assays. Transfusion. 2011;51:719–30. doi: 10.1111/j.1537-2995.2010.02942.x. [DOI] [PubMed] [Google Scholar]
- 18.Phelps R, Robbins K, Liberti T, Machuca A, Leparc G, Chamberland M, et al. Window-period human immunodeficiency virus transmission to two recipients by an adolescent blood donor. Transfusion. 2004;44:929–33. doi: 10.1111/j.1537-2995.2004.03364.x. [DOI] [PubMed] [Google Scholar]
- 19.Harritshoj LH, Dickmeiss E, Hansen MB, Ullum H, Jorgensen LB, Gerstoft J. Transfusion-transmitted human immunodeficiency virus infection by a Danish blood donor with a very low viral load in the preseroconversion window phase. Transfusion. 2008;48:2026–8. doi: 10.1111/j.1537-2995.2008.01847.x. [DOI] [PubMed] [Google Scholar]
- 20.Delwart EL, Kalmin ND, Jones TS, Ladd DJ, Foley B, Tobler LH, et al. First report of human immunodeficiency virus transmission via an RNA-screened blood donation. Vox Sang. 2004;86:171–7. doi: 10.1111/j.0042-9007.2004.00416.x. [DOI] [PubMed] [Google Scholar]
- 21.Stramer SL. Current risks of transfusion-transmitted agents: A review. Arch Pathol Lab Med. 2007;131:702–7. doi: 10.5858/2007-131-702-CROTAA. [DOI] [PubMed] [Google Scholar]
- 22.Kalus U, Edelmann A, Pruss A, Hofmann J, Kiesewetter H, Krüger DH, et al. Noninfectious transfusion of platelets donated before detection of human immunodeficiency virus RNA in plasma. Transfusion. 2009;49:435–9. doi: 10.1111/j.1537-2995.2008.02012.x. [DOI] [PubMed] [Google Scholar]
- 23.Satake M. Infectious risks associated with the transfusion of blood components and pathogen inactivation in Japan. Int J Hematol. 2004;80:306–10. doi: 10.1532/ijh97.04118. [DOI] [PubMed] [Google Scholar]
- 24.Schüttler CG, Caspari G, Jursch CA, Willems WR, Gerlich WH, Schaefer S. Hepatitis C virus transmission by a blood donation negative in nucleic acid amplification tests for viral RNA. Lancet. 2000;355:41–2. doi: 10.1016/S0140-6736(99)04719-4. [DOI] [PubMed] [Google Scholar]
- 25.Taylor C, Price TH, Strong DM. Possible HCV transmission from blood screened by pooled nucleic acid testing. Transfusion. 2002;42(Suppl):9S S30–030E. [Google Scholar]
- 26.Kretzschmar E, Chudy M, Nübling CM, Ross RS, Kruse F, Trobisch H. First case of hepatitis C virus transmission by a red blood cell concentrate after introduction of nucleic acid amplification technique screening in Germany: A comparative study with various assays. Vox Sang. 2007;92:297–301. doi: 10.1111/j.1423-0410.2007.00903.x. [DOI] [PubMed] [Google Scholar]
- 27.Grabarczyk P, Gronowska A, Brojer E, Letowska M, Radziwon P. Sequence analysis confirmation of transfusion-transmitted hepatitis C by red blood cells that tested negative by minipool hepatitis C virus nucleic acid testing. Transfusion. 2007;47:1102–4. doi: 10.1111/j.1537-2995.2007.01263.x. [DOI] [PubMed] [Google Scholar]
- 28.Gerlich WH, Glebe D, Schüttler CG. Deficiencies in the standardization and sensitivity of diagnostic tests for hepatitis B virus. J Viral Hepat. 2007;14(Suppl 1):16–21. doi: 10.1111/j.1365-2893.2007.00912.x. [DOI] [PubMed] [Google Scholar]
- 29.Comanor L, Holland P. Hepatitis B virus blood screening: Unfinished agendas. Vox Sang. 2006;91:1–12. doi: 10.1111/j.1423-0410.2006.00773.x. [DOI] [PubMed] [Google Scholar]
- 30.Dwyre DM, Fernando LP, Holland PV. Hepatitis B, hepatitis C and HIV transfusion-transmitted infections in the 21st century. Vox Sang. 2011;100:92–8. doi: 10.1111/j.1423-0410.2010.01426.x. [DOI] [PubMed] [Google Scholar]
- 31.Vermeulen M, Dickens C, Lelie N, Walker E, Coleman C, Keyter M, et al. Hepatitis B virus transmission by blood transfusion during 4 years of individual-donation nucleic acid testing in South Africa: Estimated and observed window period risk. Transfusion. 2012;52:880–92. doi: 10.1111/j.1537-2995.2011.03355.x. [DOI] [PubMed] [Google Scholar]
- 32.Gerlich WH, Bremer C, Saniewski M, Schüttler CG, Wend UC, Willems WR, et al. Occult hepatitis B virus infection: Detection and significance. Dig Dis. 2010;28:116–25. doi: 10.1159/000282074. [DOI] [PubMed] [Google Scholar]
- 33.Gerlich WH, Wagner FF, Chudy M, Harritshoj LH, Lattermann A, Wienzek S, et al. HBsAg non-reactive HBV infection in blood donors: Transmission and pathogenicity. J Med Virol. 2007;79:S32–6. [Google Scholar]
- 34.Garmiri P, Rezvan H, Abolghasemi H, Allain JP. Full genome characterization of hepatitis B virus strains from blood donors in Iran. J Med Virol. 2011;83:948–52. doi: 10.1002/jmv.21772. [DOI] [PubMed] [Google Scholar]
- 35.González R, Torres P, Castro E, Barbolla L, Candotti D, Koppelman M, et al. Efficacy of hepatitis B virus (HBV) DNA screening and characterization of acute and occult HBV infections among blood donors from Madrid, Spain. Transfusion. 2010;50:221–30. doi: 10.1111/j.1537-2995.2009.02343.x. [DOI] [PubMed] [Google Scholar]
- 36.Louisirirotchanakul S, Oota S, Khuponsarb K, Chalermchan W, Phikulsod S, Chongkolwatana V, et al. Occult hepatitis B virus infection in Thai blood donors. Transfusion. 2011;51:1532–40. doi: 10.1111/j.1537-2995.2010.03023.x. [DOI] [PubMed] [Google Scholar]
- 37.Candotti D, Grabarczyk P, Ghiazza P, Roig R, Casamitjana N, Iudicone P, et al. Characterization of occult hepatitis B virus from blood donors carrying genotype A2 or genotype D strains. J Hepatol. 2008;49:537–47. doi: 10.1016/j.jhep.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 38.Chudy M, Schmidt M, Czudai V, Scheiblauer H, Nick S, Mosebach M, et al. Hepatitis B virus genotype G monoinfection and its transmission by blood components. Hepatology. 2006;44:99–107. doi: 10.1002/hep.21220. [DOI] [PubMed] [Google Scholar]
- 39.Mahgoub S, Candotti D, El Ekiaby M, Allain JP. Hepatitis B virus (HBV) infection and recombination between HBV genotypes D and E in asymptomatic blood donors from Khartoum, Sudan. J Clin Microbiol. 2011;49:298–306. doi: 10.1128/JCM.00867-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hourfar MK, Jork C, Schottstedt V, Weber-Schehl M, Brixner V, Busch MP, et al. Experience of German Red Cross blood donor services with nucleic acid testing: Results of screening more than 30 million blood donations for human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus. Transfusion. 2008;48:1558–66. doi: 10.1111/j.1537-2995.2008.01718.x. [DOI] [PubMed] [Google Scholar]
- 41.Satake M, Taira R, Yugi H, Hino S, Kanemitsu K, Ikeda H, et al. Infectivity of blood components with low hepatitis B virus DNA levels identified in a lookback program. Transfusion. 2007;47:1197–205. doi: 10.1111/j.1537-2995.2007.01276.x. [DOI] [PubMed] [Google Scholar]
- 42.Levicnik-Stezinar S, Rahne-Potokar U, Candotti D, Lelie N, Allain JP. Anti-HBs positive occult hepatitis B virus carrier blood infectious in two transfusion recipients. J Hepatol. 2008;48:1022–5. doi: 10.1016/j.jhep.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 43.Taira R, Satake M, Momose S, Hino S, Suzuki Y, Murokawa H, et al. Residual risk of transfusion-transmitted hepatitis B virus (HBV) infection caused by blood components derived from donors with occult HBV infection in Japan. Transfusion. 2013;53:1393–404. doi: 10.1111/j.1537-2995.2012.03909.x. [DOI] [PubMed] [Google Scholar]
- 44.Makroo RN, Chowdhry M, Bhatia A, Arora B, Rosamma NL. Hepatitis B core antibody testing in Indian blood donors: A double-edged sword! Asian J Transfus Sci. 2012;6:10–3. doi: 10.4103/0973-6247.95043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vivekanandan P, Abraham P, Sridharan G, Chandy G, Daniel D, Raghuraman S, et al. Distribution of hepatitis B virus genotypes in blood donors and chronically infected patients in a tertiary care hospital in southern India. Clin Infect Dis. 2004;38:e81–6. doi: 10.1086/383144. [DOI] [PubMed] [Google Scholar]
- 46.Kumar A, Dwivedi M, Misra SP, Misra V, Narang S, Pandey R, et al. Distribution of hepatitis B virus genotypes and its association with severity of liver disease in patients with chronic hepatitis B in Uttar Pradesh, India. Indian J Virol. 2011;22:24–8. doi: 10.1007/s13337-011-0034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto C, Tadokoro K, Fujimura K, Hirakawa S, Mitsunaga S, Juji T. Analysis of HBV infection after blood transfusion in Japan through investigation of a comprehensive donor specimen repository. Transfusion. 2001;41:878–84. doi: 10.1046/j.1537-2995.2001.41070878.x. [DOI] [PubMed] [Google Scholar]
- 48.Stramer SL, Wend U, Candotti D, Foster GA, Hollinger FB, Dodd RY, et al. Nucleic acid testing to detect HBV infection in blood donors. N Engl J Med. 2011;364:236–47. doi: 10.1056/NEJMoa1007644. [DOI] [PubMed] [Google Scholar]
- 49.Shan H, Ren FR, Zhao HY, Zhang YZ, Wen GX, Yao FZ, et al. A multi-Chinese blood center study testing serologic-negative donor samples for hepatitis C virus and human immunodeficiency virus with nucleic acid testing. Transfusion. 2007;47:2011–6. doi: 10.1111/j.1537-2995.2007.01424.x. [DOI] [PubMed] [Google Scholar]
- 50.Yang MH, Li L, Hung YS, Hung CS, Allain JP, Lin KS, et al. The efficacy of individual-donation and minipool testing to detect low-level hepatitis B virus DNA in Taiwan. Transfusion. 2010;50:65–74. doi: 10.1111/j.1537-2995.2009.02357.x. [DOI] [PubMed] [Google Scholar]
- 51.Vermeulen M, Coleman C, Mitchel J, Reddy R, van Drimmelen H, Fickett T, et al. Comparison of human immunodeficiency virus assays in window phase and elite controller samples: Viral load distribution and implications for transmission risk. Transfusion. 2013;53:2384–98. doi: 10.1111/trf.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grabarczyk P, Letowska M, Kopacz A, Liszewski G, Sulkowska E, Brojer E. Effectiveness of various minipool nucleic acid test systems for hepatitis B virus detection in blood donors. Transfusion. 2009;49:1494–5. doi: 10.1111/j.1537-2995.2009.02225.x. [DOI] [PubMed] [Google Scholar]
- 53.Vermeulen M, Coleman C, Mitchel J, Reddy R, van Drimmelen H, Ficket T, et al. Sensitivity of individual-donation and minipool nucleic acid amplification test options in detecting window period and occult hepatitis B virus infections. Transfusion. 2013;53:2459–66. doi: 10.1111/trf.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth WK, Busch MP, Schuller A, Ismay S, Cheng A, Seed CR, et al. International survey on NAT testing of blood donations: Expanding implementation and yield from 1999 to 2009. Vox Sang. 2012;102:82–90. doi: 10.1111/j.1423-0410.2011.01506.x. [DOI] [PubMed] [Google Scholar]
- 55.Makroo RN, Choudhury N, Jagannathan L, Parihar-Malhotra M, Raina V, Chaudhary RK, et al. Multicenter evaluation of individual donor nucleic acid testing (NAT) for simultaneous detection of human immunodeficiency virus -1 & hepatitis B & C viruses in Indian blood donors. Indian J Med Res. 2008;127:140–7. [PubMed] [Google Scholar]
- 56.Agarwal N, Chatterjee K, Coshic P, Borgohain M. Nucleic acid testing for blood banks: An experience from a tertiary care centre in New Delhi, India. Transfus Apher Sci. 2013;49:482–4. doi: 10.1016/j.transci.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 57.van Hulst M, Hubben GA, Sagoe KW, Promwong C, Permpikul P, Fongsatitkul L, et al. Web interface-supported transmission risk assessment and cost-effectiveness analysis of postdonation screening: A global model applied to Ghana, Thailand, and the Netherlands. Transfusion. 2009;49:2729–42. doi: 10.1111/j.1537-2995.2009.02351.x. [DOI] [PubMed] [Google Scholar]


