Abstract
Introduction:
For nucleic acid testing (NAT) of blood donations, either the blood samples can be pooled together in a batch of six or eight prior to testing (mini-pool-NAT [MP-NAT]), or the tests can be run on every individual sample (individual donor-NAT [ID-NAT]). It has been debated in various studies whether pooling of samples results in decreased sensitivity of detection as the volume of individual samples gets lesser in a pool. The objective of this study was to investigate the effect of dilution on the sensitivity of tests.
Materials and Methods:
The study was performesd on nine plasma samples which were hepatitis B reactive exclusively by Procleix Ultrio Plus and not by Procleix Ultrio or serology. These nine exclusive UltrioPlus ID-NAT yield samples were diluted in 1:2, 1:4. 1:6 and 1:8 dilutions using previously tested negative plasma and each dilution of every sample along with archived undiluted sample were retested in three replicates with Procleix Ultrio Plus Assay.
Results:
Among NAT yield samples, 88.88% of the samples were detected when retested in ID-NAT in undiluted form. Samples with higher viral load (sample 5 and 6) were detected by all dilutions. When samples with viral load below 20 IU/mL were tested in dilutions of 1:6 or 1:8, only 9 out of 27 replicates (33.33%) were detected. This means that more than 67% of low viral load samples were missed by MP-NAT of 1:6 or 1:8 dilution out of total NAT yield samples.
Conclusion:
Individual Donor NAT is ideal methodology for NAT as dilution due to pooling may miss samples with low viral load as evident in this study.
Keywords: Hepatitis B surface antigen, hepatitis B virus, nucleic acid testing, pooling of samples
Introduction
Hepatitis B virus (HBV) is a major human pathogen that may cause acute and chronic hepatitis, cirrhosis and hepatocellular carcinoma. HBV can be transmitted by blood from asymptomatic donors with acute HBV infections who have not yet developed hepatitis B surface antigen (HBsAg) or anti-HBc (i.e., donors in the seronegative window period), when HBV deoxyribonucleic acid (DNA) can be detected in the donor's blood.[1,2] It is estimated that approximately 1 million people die each year from HBV infection and related complications and approximately 350 million people are chronic carriers of HBV.[3,4] Confidence in results of transfusion transmitted infections in a blood bank is of critical importance. Although enzyme-linked immunosorbent assay (ELISA) blood screening technology relies on the detection of serological markers, these markers may not appear in the blood until up to 3 months after an infection, leaving a “window period” in which a risk of transfusion-transmitted infection is increased. Nucleic acid testing (NAT) assay reduces this window period by detecting the presence of the viral ribonucleic acid (RNA) or DNA directly. Depending on the sensitivity of the test, implementation of HBV NAT has the potential to reduce the risk of infection to levels similar to those for human immunodeficiency virus (HIV) and hepatitis C virus (HCV).
For NAT of blood donations, either the blood samples can be pooled together in a batch of 6 or 8 prior to testing to screen a large number of donations with few tests (mini-pool nucleic acid testing [MP-NAT]) or the tests can be run on every individual sample (individual donor nucleic acid testing [ID-NAT]). It has been debated in various studies whether pooling of samples results in decreased sensitivity of detection as the volume of individual sample gets lesser in a pool.[5,6] Therefore, greater the number of samples in a pool, lesser is the sensitivity of detection of the test. Furthermore, the replication rate of HBV is very low, with a mean doubling time of 2.6 days[7] and the viral load is also very low during the window phase. Therefore, more sensitive tests are required for HBV detection than HIV or HCV. The objective of this study was to investigate the effect of dilution on the sensitivity of tests with low viral load, serology negative, ID-NAT HBV reactive samples.
Materials and Methods
The study was performed on nine plasma samples, which were exclusive Procleix Ultrio Plus (UP) HBV yields i.e., NAT Ultrio Plus Assay reactive but serology and Procleix Ultrio non-reactive during a Procleix Ultrio Plus pilot study[8] conducted on blood donors for a period of 1 year, from September 2011 to September 2012, at All India Institute of Medical Sciences (AIIMS), New Delhi, India. In that study, 6021 samples were screened by Procleix Ultrio Plus and Ultrio Assays and routine serology tests. Out of 6021 samples, 12 blood donor samples were only reactive with Procleix Ultrio Plus, but negative with Procleix Ultrio and serology. Out of these 12 Ultrio Plus yields, three samples were not available for further analysis.
The samples were ID NAT tested by transcription mediated amplification technology (TMA) using Procleix Ultrio Plus Assay (Novartis, Emeryville, CA) for simultaneous detection of HIV-1, HCV-RNA and HBV-DNA. All the nine samples were discriminated using Procleix Ultrio Plus Discriminatory assay at the AIIMS, New Delhi.
Serological tests for HIV-1, HCV and HBV were done using Bio-Rad Genscreen plus HIV Ag-Ab, Bio Rad Laboratories, Steenvoorde, France; BioMerieux Hepanostika HBsAg Ultra and BioMerieux Hepanostika HCV Ultra, M/s BioMerieux, B.V. Boseind, The Netherlands. Real time polymerase chain reaction (PCR) test for HBV viral load using COBAS Ampliprep and COBAS Taqman was performed on the nine samples at Dr. Lal's Path Lab, Rohini, Delhi (College of American Pathologists and National Accreditation Board for Testing and Calibration Laboratories accredited) to quantify the virus in each of the sample.
These nine exclusive Ultrio Plus ID-NAT yield samples were diluted in 1:2, 1:4. 1:6 and 1:8 dilutions using previously tested negative plasma (by ELISA and NAT) and each dilution of every sample along with undiluted sample were retested in three replicates with Procleix Ultrio Plus Assay.
Results
Out of 9 samples, 8 showed the presence of HBV DNA by real time PCR while one sample was not detected. This may be due to the low sensitivity of PCR kits as compared to TMA based Procleix Ultrio Plus kits as suggested by few studies for HCV.[9] Among the eight samples, which were detected by PCR, six had less than 20 IU/mL viral load, one sample had 810 IU/mL and another sample had 50 IU/mL viral load.
All the nine samples were further retested in undiluted and dilutions in triplicate and results observed mentioned in Table 1 below. The percentage of detection of samples by each dilution is also being shown in Figure 1.
Table 1.
Profile of 9 HBV NAT yield samples
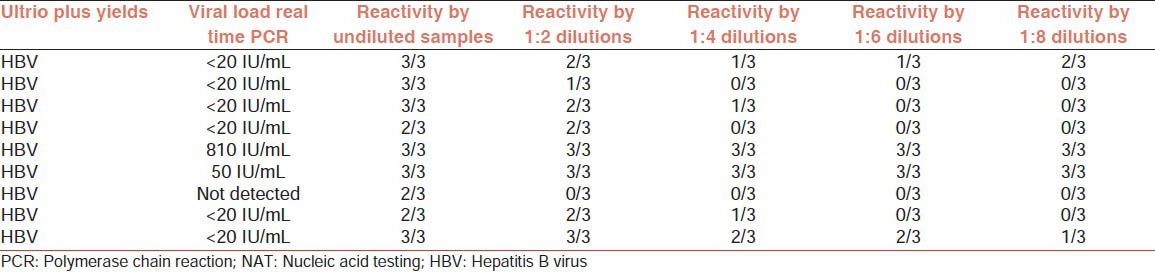
Figure 1.
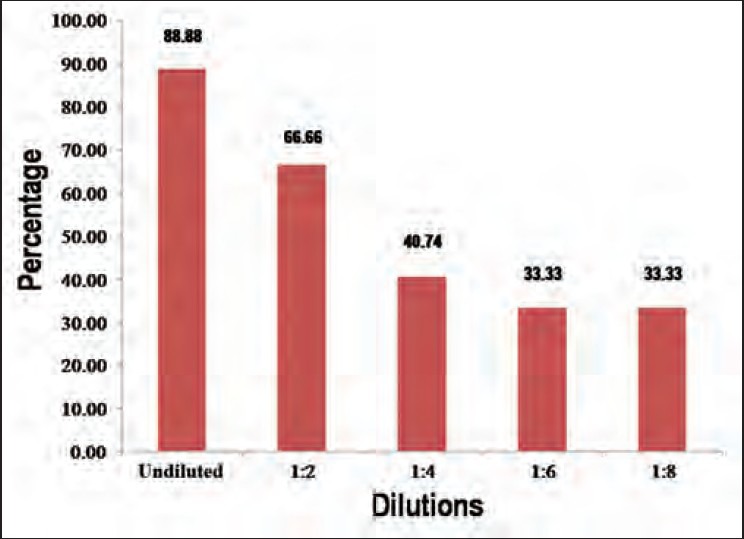
Percentage of detection by various dilutions
Discussion
Scientific models estimate that NAT reduces the infectious window period by 35-91% for HIV-1, HCV and HBV with individual donation testing, while only 17-87% with mini-pool (pools of 16) nucleic acid testing.[10,11] This study reaffirms the statement that dilution of samples results in lesser sensitivity of test and lesser chances of detection of viral nucleic acid.
Among NAT yield samples, 88.88% of the samples were detected when retested in ID-NAT or undiluted format. The lower detection can be explained by low viral load and Poisson distribution. The same explanation also goes for one case where 1:8 dilution has shown better detection than 1:4 and 1:6 dilutions. Samples with higher viral load (sample five and six) were detected by all dilutions. When samples with a viral load below 20 IU/mL were tested in dilutions of 1:6 or 1:8, only 6 out of 36 replicates (16.66%) were detected. This means that more than 83% low viral load samples were missed by MP-NAT of 1:6 or 1:8 dilution out of total NAT yield samples. Dilution effects lower viral load samples and could lead to them being undetected when diluted or pooled. A recent similar dilution study from South African National Board Services, South Africa shows that MP 6 NAT may miss 53% of low viral load HBV NAT yield cases.[12]
Various studies from Australia and the other parts of the world have shown that mini-pool NAT screening would probably not detect HBV DNA in low level chronic carriers.[13] However, single donor NAT may offer some sensitivity gain for WP detection.[14] In India, a high number of NAT yields are associated with low viral load especially with HBV and is apparent from the fact that out of 20 Ultrio Plus and Ultrio Yields for our initial Ultrio Plus pilot study, at least 7 or 35% were low viral loads and were a mix of the window period and occult HBV cases. Individual Donor NAT is ideal methodology for NAT as dilution due to pooling may miss samples with low viral load as evident in this study.
Footnotes
Source of Support: Hemogenomics Pvt Ltd has helped financially to get the sample tested from a CAP accredited Lab
Conflicting Interest: Mr Sourit Chakroborty is an employee of Hemogenomics Pvt Ltd.
References
- 1.Roth WK, Weber M, Petersen D, Drosten C, Buhr S, Sireis W, et al. NAT for HBV and anti-HBc testing increase blood safety. Transfusion. 2002;42:869–75. doi: 10.1046/j.1537-2995.2002.00128.x. [DOI] [PubMed] [Google Scholar]
- 2.Kleinman SH, Lelie N, Busch MP. Infectivity of human immunodeficiency virus-1, hepatitis C virus, and hepatitis B virus and risk of transmission by transfusion. Transfusion. 2009;49:2454–89. doi: 10.1111/j.1537-2995.2009.02322.x. [DOI] [PubMed] [Google Scholar]
- 3.Mahoney FJ. Update on diagnosis, management, and prevention of hepatitis B virus infection. Clin Microbiol Rev. 1999;12:351–66. doi: 10.1128/cmr.12.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schüttler CG, Caspari G, Jursch CA, Willems WR, Gerlich WH, Schaefer S. Hepatitis C virus transmission by a blood donation negative in nucleic acid amplification tests for viral RNA. Lancet. 2000;355:41–2. doi: 10.1016/S0140-6736(99)04719-4. [DOI] [PubMed] [Google Scholar]
- 5.Najioullah F, Barlet V, Renaudier P, Guitton C, Crova P, Guérin JC, et al. Failure and success of HIV tests for the prevention of HIV-1 transmission by blood and tissue donations. J Med Virol. 2004;73:347–9. doi: 10.1002/jmv.20097. [DOI] [PubMed] [Google Scholar]
- 6.Yang MH, Li L, Hung YS, Hung CS, Allain JP, Lin KS, et al. The efficacy of individual-donation and minipool testing to detect low-level hepatitis B virus DNA in Taiwan. Transfusion. 2010;50:65–74. doi: 10.1111/j.1537-2995.2009.02357.x. [DOI] [PubMed] [Google Scholar]
- 7.Yoshikawa A, Gotanda Y, Itabashi M, Minegishi K, Kanemitsu K, Nishioka K, et al. HBV NAT positive [corrected] blood donors in the early and late stages of HBV infection: Analyses of the window period and kinetics of HBV DNA. Vox Sang. 2005;88:77–86. doi: 10.1111/j.1423-0410.2005.00602.x. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee K. Procleix ultrio plus assay evaluation experience for Indian Donor population. Transfusion. 2012;52(Suppl s3):218 A. [Google Scholar]
- 9.Comanor L, Anderson F, Ghany M, Perrillo R, Heathcote EJ, Sherlock C, et al. Transcription-mediated amplification is more sensitive than conventional PCR-based assays for detecting residual serum HCV RNA at end of treatment. Am J Gastroenterol. 2001;96:2968–72. doi: 10.1111/j.1572-0241.2001.04669.x. [DOI] [PubMed] [Google Scholar]
- 10.Busch MP. Evolving approaches to estimate risks of transfusiontransmitted viral infections: Incidence-window period model after ten years. In: Dax EM, Farrugia A, Vyas GN, editors. Advances in Transfusion Safety. 127. IV. Basel: Developments in Biologicals, Karger; 2007. pp. 87–112. [PubMed] [Google Scholar]
- 11.Kleinman SH, Busch MP. Assessing the impact of HBV NAT on window period reduction and residual risk. J Clin Virol. 2006;36(Suppl 1):S23–9. doi: 10.1016/s1386-6532(06)80005-3. [DOI] [PubMed] [Google Scholar]
- 12.Vermeulen M, Coleman C, Mitchel J, Reddy R, van Drimmelen H, Ficket T, et al. Sensitivity of individual donation- and minipool-nucleic acid amplification test options in detecting window period and occult hepatitis B virus infections. Transfusion. 2013 Oct;53(Suppl 3):2459–66. doi: 10.1111/trf.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinman SH, Kuhns MC, Todd DS, Glynn SA, McNamara A, DiMarco A, et al. Frequency of HBV DNA detection in US blood donors testing positive for the presence of anti-HBc: Implications for transfusion transmission and donor screening. Transfusion. 2003;43:696–704. doi: 10.1046/j.1537-2995.2003.00391.x. [DOI] [PubMed] [Google Scholar]
- 14.Sato S, Ohhashi W, Ihara H, Sakaya S, Kato T, Ikeda H. Comparison of the sensitivity of NAT using pooled donor samples for HBV and that of a serologic HBsAg assay. Transfusion. 2001;41:1107–13. doi: 10.1046/j.1537-2995.2001.41091107.x. [DOI] [PubMed] [Google Scholar]


