Abstract
Targeted mass spectrometry (MS) is becoming widely used in academia and in pharmaceutical and biotechnology industries for sensitive and quantitative detection of proteins, peptides and post-translational modifications. Here we describe the increasing importance of targeted MS technologies in clinical proteomics and the potential key roles these techniques will have in bridging biomedical discovery and clinical implementation.
Proteomics comprises the large-scale, systematic study of protein structure and function, usually with respect to a defined entity: a pathway, organelle, cell, tissue or organism. Whereas any methods or technologies to systematically interrogate large numbers of proteins can justifiably be considered proteomic approaches, the term is increasingly being used to designate work in which MS is the central technology platform. Clinical proteomics is a loose assemblage of proteomics initiatives unified by their translational nature: that is, their impetus to progress along the path from basic research to medical application. Clinical proteomics experiments typically involve the characterization of proteomes of normal or diseased tissues or biological fluids, thus detailing and quantifying the protein differences that associate with, define or cause the diseased state to illuminate pathobiology, improve disease classification or identify new therapeutic targets. Proteomic biomarker discovery is a familiar instance of clinical proteomics research in which MS-based proteomic approaches are used to identify peptides, proteins or post-translational modifications that support early disease detection, facilitate diagnosis, inform prognosis, guide therapy or monitor disease activity. The ultimate objective of any translational enterprise is clinical implementation, in which knowledge previously gleaned is used to directly drive clinical decision making and intervention. When that implementation involves MS-based measurement of one or more protein-derived analytes, it represents the fullest realization of clinical proteomics.
A defining advantage of MS for discovery or hypothesis generation in clinical proteomics is the capability to confidently identify thousands of proteins in complex biological samples without prespecification of the analytes to be measured. With this broad and unbiased coverage comes the cost of reduced sensitivity and stochastic sampling. As one moves along the translational path, findings must be verified and hypotheses must be tested, requiring that sensitive quantitative protein measurements be made precisely and reliably every time. This crucial phase of clinical proteomics is increasingly achieved by focusing the resources of the mass spectrometer on a defined subset of analytes, an approach called targeted MS.
Targeted MS in the spectrum of MS methods
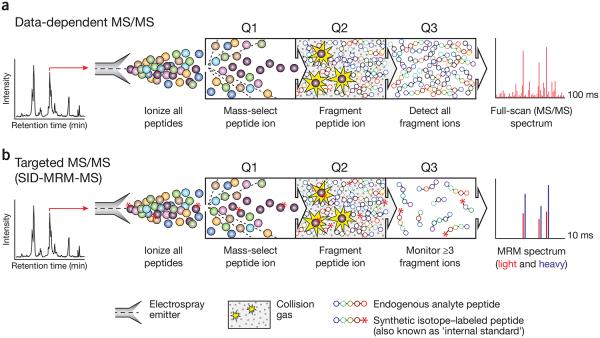
For over four decades, targeted MS approaches have been used to increase the speed, sensitivity and quantitative precision of biomolecule analysis1-3. Targeted MS technologies have been developed, in large part, to overcome the sampling limitations of conventional data-dependent scanning MS analysis used in a discovery-based strategy (Fig. 1). In both approaches, analytes (small molecules, metabolites or peptides) are infused or eluted from a reversed phase column attached to a liquid chromatography instrument and converted to gas phase ions by electrospray ionization. Analyte ions are fragmented in the mass spectrometer (a technique known as tandem MS or MS/MS), and fragment and parent masses are used to establish the identity of the analyte. In data-dependent acquisition, ions are automatically selected for MS/MS based on their signal intensity in the preceding full-scan MS spectrum. Interpretation of the MS/MS spectra provides the amino acid sequences of the selected peptide ions; sequence and parent ion mass–directed database search allows peptide identification. This data collection cycle (typically 2–3 s in duration) is repeated over the entire course of the liquid chromatography (LC)-MS/MS analysis. The principle behind the alternative approach of targeted acquisition is simple: guided by a reference spectrum, an analyte can be identified using only a few selected fragment ions rather than the entire complex content of the MS/MS fragmentation spectrum.
Figure 1.
Comparison of conventional data-dependent analysis to targeted MRM-MS on a triple quadrupole mass spectrometer. (a) In a data-dependent MS experiment, digested proteins are loaded on a reversed-phase column attached to a liquid chromatography setup and eluted via electrospray to yield gas-phase ions. At any given point in the chromatographic separation many tens to hundreds of peptides are eluting nearly simultaneously. A full-scan MS spectrum is acquired and informs collection of subsequent MS/MS scans in which 4–10 ions observed in the MS spectrum are automatically selected on the basis of their signal intensity (Q1) for fragmentation by collision with inert gas (Q2). The complete array of fragment ions is detected (Q3), which constitutes the full-scan MS/MS spectrum (far right). (b) In an SID-MRM-MS analysis, proteotypic peptides uniquely representing proteins of interest are predefined together with their most informative fragment ions. Peptides are selected for fragmentation (Q1 and Q2), and fragment ions are selected for detection (Q3) based on a user-specified list of targeted precursor-fragment pairs (‘transitions’). Synthetic peptides containing stable-isotope labels can be spiked in as standards (asterisks). Comparing labeled to unlabeled peak area (far right) provides precise relative quantification of the endogenous analyte.
In the earliest implementation of targeted MS, multiple ion monitoring, signals for a few selected ions were extracted from previously collected full-scan MS data and used to identify and quantify analytes1. With the development of the triple quadrupole mass analyzer4, it became both possible and practical to accomplish this during data collection, by rapidly mass selecting and fragmenting specific precursor ions representing analytes of interest and monitoring signals for only a few predefined fragment ions for each analyte (Fig. 1b). In a contemporary multiple reaction monitoring (MRM) experiment (also commonly referred to as selected reaction monitoring), each fragment ion from an analyte needs to be sampled for only a few milliseconds to obtain interpretable spectra. More than 100 precursor-product ion pairs (referred to as ‘transitions’) can thus be recorded per second in MRM, enabling targeted MS analysis of many tens of analytes in a time frame much shorter than the peak width of their chromatographic elution. Whereas in early MRM implementations the instrument cycled continuously through the entire set of transitions, recent software and hardware improvements now support scheduled analysis of subsets of transitions based on established retention times of the analytes in the chromatographic system used5, allowing hundreds of different analytes to be targeted and analyzed in a single MRM analysis. In a properly designed and implemented MRM ‘assay’, analytes are consistently measured by their specified transitions; absence of detection means that the analyte is below detection limits. In contrast, absence of detection in a data-dependent MS analysis (Fig. 1a) can mean either that the analyte is below detection limits or that it was not sampled.
Analysis of proteins and their post-translational modifications by MRM is based on detection of peptides derived by digestion of the protein (Fig. 2). The highest detection confidence and measurement precision for peptides in complex samples is obtained by combining stable isotope dilution (SID)—an analytical chemistry technique in which a known concentration of an isotope-labeled compound is added to the sample before analysis—with MRM to yield SIDMRM5-9. In this approach, synthetic versions of each analyte peptide containing an amino acid labeled with a stable isotope (for example, 15N or 13C) are used as internal standards. The labeled internal standards separate by chromatography and fragment identically to their native counterparts but are distinguished in the MS and MS/MS spectra by the increased masses of the peptide and of fragment ions containing the labeled amino acid. Confidence that the correct analyte is measured using just a few fragment ions from the peptide is increased by the requirements that the labeled internal standard and endogenous analyte elute together in chromatography and that their monitored fragment ions have the same relative abundance. The sensitivity of MRM-MS is greater than that of discovery proteomics methods because the signal from the selected ions accumulates for longer periods of time in the mass spectrometer.
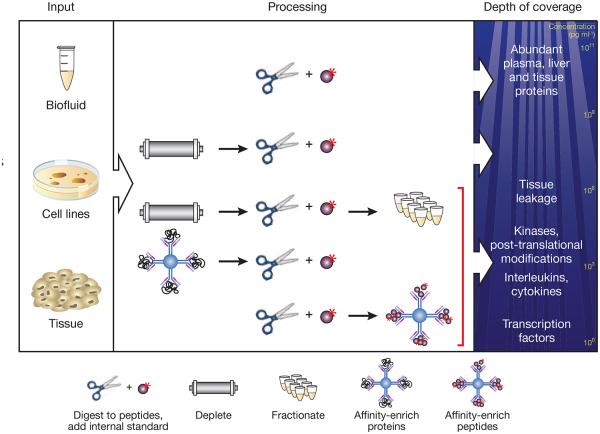
Figure 2.
Protein and peptide enrichment strategies to increase sensitivity and specificity of analyte detection in SID-MRM-MS. After extraction from tissues, cell lines or biofluids (left), proteins are digested into peptides. To achieve the highest detection confidence and measurement precision for peptides in these complex samples, synthetic versions of each analyte peptide containing an amino acid labeled with a stable isotope (for example, 15N or 13C) are added as internal standards (top line, center). Additional strategies may be required to achieve desired detection sensitivity in complex matrices. In plasma, depletion of the 12–70 most abundant plasma proteins using immunoaffinity depletion columns can result in limits of detection of ~100 ng ml−1 (second line, center), representing ‘middling’ depths in the plasma peptide ocean (right). Biomarkers in diseases such as cancer and cardiovascular disease are commonly present in the low nanogram per milliliter range and can be robustly detected by coupling depletion to limited fractionation (third line, center), albeit at the expense of reducing throughput and introducing variability and analytical complexity. A different strategy to enhance sensitivity relies on immunoaffinity reagents to enrich target analytes. In one implementation the intact protein is captured using an anti-protein antibody before digestion (fourth line, center). Alternatively, a proteotypic peptide derived from the protein can be captured using an anti-peptide antibody to the target peptide of interest (bottom line, center).
Peptide and protein concentrations are derived from ratios of the peak areas measured for each internal standard and endogenous peptide (Fig. 1b). These measurements typically have high precision, but they may have low accuracy as the release efficiency of analyte peptides from a protein during proteolysis depends on sequence context and is highly variable and because labeled peptides are not subjected to all the same sample preparation steps as the endogenous sample. In most cases today MRM is used to precisely and consistently measure relative changes in the levels of targeted analytes across samples rather than to accurately establish the amount or concentration in a sample. In cases where the accurate (or ‘absolute’) amount of a peptide and the corresponding protein are needed, such as in clinical measurements, stable isotope–labeled proteins or protein surrogates in which the tryptic peptide analytes are surrounded by their native sequences can be used as reference standards for accurate quantification when added at the start of sample processing10.
Targeted MS in biomedicine and its adoption in proteomics
The recent explosion of interest in clinical and translational applications of targeted MS for peptide and protein measurements has its roots in a technology revolution a generation ago. In the 1960s, immunoassays were used routinely to measure small molecules such as metabolites, hormones and drugs in blood, urine and other body fluids3. Because the emerging targeted MS methods offered decisive advantages, they progressively replaced immunoassays in clinical research laboratories even though these changes required deployment of costly instrumentation and knowledgeable personnel to run the instruments and analyze the data. In contrast to immunoassay-based methods, developing MS-based assays for new analytes was simple and fast, and, rather than measuring only one analyte at a time using different tests, a single LC-MS/MS analysis could be used to measure many tens of small molecules. Today, millions of LC-MS/MS assays are run annually in clinical laboratories worldwide, the vast majority based on MRM1,3.
The adoption of targeted MS for analysis of peptides and proteins has been driven by many of the same positive attributes that were recognized for small molecules. Desiderio and colleagues first demonstrated the use of targeted MS for the analysis of bioactive peptides2 in the 1980s. Application of these methods to the analysis of proteins and post-translational modifications began to accelerate in the late 1990s and has rapidly expanded over the past decade, enabled by improvements in the capabilities of LC-MS systems and in methods to prepare tissue and biofluid samples for MS analysis.
Experimental challenges
The most substantial challenge in using MRM-MS for targeted peptide analysis in clinical proteomics applications is the prevalence of interferences from other peptides and small molecules in the sample matrix. This problem, although well studied for small-molecule analysis, is both less well recognized and far more severe for peptide analysis11, chiefly because peptide MRM analyses are typically carried out in an ocean of many hundreds of thousands to millions of peptides produced by digestion of the 10,000 or more proteins (not counting modified forms) found in blood and other tissues. Interference manifests itself in two ways: ion suppression and transition interference. Suppression decreases the ion current response of an analyte in an unpredictable and nonreproducible way. Suppression effects increase with the complexity of the biological matrix. Transition interference is caused by peptides or other sample constituents that have both a precursor mass and one or more fragment ion masses that are identical or nearly identical to those being monitored for an analyte of interest. Software has been introduced that facilitates detection of interference and identification of unreliable transitions11,12. These programs use additional information inherent to the analysis to build confidence in assignment, including co-elution of analyte and standard based on simultaneous appearance of their respective transitions; the relative ratio of the selected fragment ions to one another compared to those observed for the internal standard; statistical measures of assay precision; and other independent scoring methods.
Although assays can be configured without the use of labeled internal standard peptides5, in this case measurement precision and confidence in identification both suffer, especially in complex plasma and tissue matrices. Expense is frequently raised as an objection to the use of isotope-labeled peptides that currently cost $250–500 for 1–2 milligrams of purified and quantified material. However this amount of peptide is enough to run 10,000 or more assays, potentially bringing the cost of peptide on a per-analyte, per-assay basis down to a few pennies. Additional substantial savings can be achieved by configuring initial assays with unpurified labeled peptides now offered by many companies. In our view, when the cost of biological and especially clinical follow-up on an incorrect identification is considered, the cost of labeled peptides needed for internal standards becomes unsubstantial and well-justified. Furthermore, transferability of assays across laboratories to measure proteins in plasma is presently only possible through the use of labeled internal reference standards9. Certainly it is likely that clinically deployed MRM assays will require the use of labeled internal standard peptides and/or labeled proteins to establish that the test is measuring the analyte of interest and is free of interference. We anticipate that use of isotope-labeled peptides as internal standards will continue to increase, leading to additional reductions in the cost of synthetic peptides.
Probing the depths of the plasma and tissue proteomes
Once an MRM assay been developed it has a high probability of being applicable in any context in which measurement of the target protein is desirable. This has motivated the development of public repositories containing configured MRM assays13,14. However, although MRM assays can be configured to measure peptides and modified peptides from nearly any protein, developing assays that achieve the desired sensitivity in clinical proteomics applications is not guaranteed. The problem is not primarily one of insufficient instrument sensitivity. Rather, it is signal-to-biological-noise ratio caused by sample complexity and the wide dynamic range of protein abundance in sample matrices such as plasma and tissue. Multiple strategies have therefore been developed to improve sensitivity and specificity of peptide detection and quantification in complex matrices (Fig. 2).
In the case of plasma and solid tissue (which is often admixed with or contaminated by plasma), depletion of the 12–70 most abundant plasma proteins using immunoaffinity depletion columns has become standard, despite inherent risks that the depleted abundant proteins are informative or that informative lower-abundance proteins might be bound to the targeted proteins and incidentally lost. Depth of detection can be additionally increased using two or more dimensions of chromatography-based separation8,15. By combining one or more of these sample-processing steps with MRM, it is possible to quantitatively measure proteins in blood that are present in the high picogram to low nanogram per milliliter concentration. Even greater sensitivities have been achieved for small numbers of analytes using highly targeted and focused chromatographic isolation methods16,17, but the generality of these approaches has yet to be demonstrated.
Immunoaffinity enrichment of target analytes short-circuits the need for abundant protein depletion and fractionation before SID-MRM-MS. In such an approach (Fig. 2) either the intact protein is captured using an anti-protein antibody18 or a peptide derived from the protein is captured using an anti-peptide antibody that has been raised to the target peptide of interest19-21. In either case only a single capture antibody is required as the mass spectrometer substitutes for a secondary detection antibody, providing absolute sequence specificity. The protein capture method is limited by the availability of antibodies capable of selective and sensitive immunoprecipitation of the protein from tissues and plasma. Immunoaffinity enrichment of peptides usually requires generation of custom antibodies for each target. Although this is a lengthy and expensive process, the combination of peptide immunoaffinity enrichment with MRM is very versatile. Both the labeled reference and endogenous forms of the analyte peptide are simultaneously enriched with the anti-peptide antibody in a method called stable isotope standards and capture by anti-peptide antibodies (SISCAPA); after elution their relative amounts can be measured by SID-MRM. The affinity of the anti-peptide capture antibody needs to be high, but its selectivity need not be high because the mass spectrometer can readily distinguish and quantify the analyte peptide of interest despite the binding of other peptides in the digested sample. SISCAPA assays can be highly multiplexed22 and throughput can be improved by coupling SISCAPA to magnetic bead–handling robotics to automate peptide capture, wash and elution steps20. These assays have proven to be robust and reproducible across laboratories, with detection limits of about one nanogram of protein per milliliter of plasma and assay coefficient of variation of 15% or less23. Of note, comparable advantages accrue to the immuno-MALDI–time of flight MS method, which couples affinity enrichment to the alternate matrix-assisted laser desorption and ionization (MALDI) MS interface24.
Targeted MS in verification of candidate biomarkers
The portfolio of targeted MS in translational science is expanding dramatically, for instance, with its increasing use to monitor nodes in signaling cascades, canonical cancer pathways or other biologically important networks13. With a trio of reagents for each node (a conventional proteotypic peptide to control for global expression, and phosphorylated and nonphosphorylated forms of a phosphopeptide to assess phosphorylation stoichiometry), pathway activity can be dissected and precisely quantified. Despite such important new roles, biomarker candidate verification remains among the signal applications of targeted MS in clinical proteomics.
Over the past decade proposed biomarkers have been derived largely from genomics and proteomics experiments in which very large numbers of transcript or protein measurements are made on comparatively small numbers of samples, creating conditions that favor detection of spurious differences unrelated to the disease-specific variables of interest. Cancer, the focus of many biomarker discovery efforts, poses a special undersampling problem because the often extreme molecular heterogeneity of disease means that even informative markers may be expressed to a different extent in only a subset of cases. These challenges are compounded in MS-based proteomics discovery experiments because stochastic sampling of complex proteomes means that proteins are inconsistently observed across samples and when observed are likely to have their abundance estimated by different component peptides, with attendant quantitative imprecision. Discovery based on transcript expression amounts avoids this complication but identifies differences that are upstream of protein expression and so may be (literally) lost in translation. It has therefore become increasingly well recognized that the legions of differentially expressed analytes emerging from ‘omics’ discovery experiments are candidates, not biomarkers, and require additional credentialing using precise, quantitative methods in larger sample numbers, a process we have called “verification”25.
The low probability that any particular biomarker candidate will be a functional biomarker suggests that a successful biomarker development enterprise will require a large proportion (ideally, all) of the candidates to be evaluated in verification studies. This highlights a serious mismatch between the verification capacity requirements of biomarker development and the still conventional approach of antibody-based verification. Though antibody-based methods, particularly well-characterized enzyme-linked immunosorbent assays, are suitable tools for verification, qualified antibodies are generally unavailable especially for new biomarker candidates and are prohibitively slow and costly to develop26, leaving the vast majority of potential markers without a straightforward means for verification. Reversed-phase protein arrays27 are potentially powerful verification tools but are limited to the few hundred antibodies of sufficient quality that are currently available. Technology solutions ranging from microbead suspensions to planar arrays coupled with microscale or nanoscale fluidics can be used to improve antibody multiplexing and sample economy28,29 but do not address the fundamental problem of a dearth of highly specific affinity reagents and are probably better positioned for clinical deployment of limited multianalyte assays or real-time point of care tests than as verification solutions. Aptamer-based approaches have theoretical advantages of specific, stable binding without substantial crossreactivity and so hold promise for a range of proteomics applications30,31, but their utility for biomarker candidate verification has not yet been convincingly demonstrated.
In contrast, targeted MS methods using MRM-MS approaches are in many respects ideally suited to candidate verification25,32,33. Analytically validated MRM assays that use internal standards can be configured for up to tens of proteins per month (depending on the extent of enrichment required for detection23), much faster than immunoassay development. Multiplexing is straightforward, with the ability to quantify hundreds of analytes in a single LC-MRM-MS run5 and up to 50 different analytes by peptide immuno-MRM20,22. Multiplexing capacity may be increased and overall platform robustness may be enhanced through the use of higher flow rates (such as 0.5 milliliters per minute) with ultra-high-performance liquid chromatography rather than the nanoflow chromatography (≤300 nanoliters per minute) that is typically used in this application. The consequent reduction in overall sensitivity of quantification can be ameliorated by loading more sample onto the larger columns used with ultra-high-performance liquid chromatography. This substitution could facilitate verification as well as the later stages of clinical validation and implementation34.
Several studies have now demonstrated the central importance and effective deployment of MRM-MS as a verification tool for candidates in the context of a comprehensive discovery-to-verification biomarker pipeline. Working in a mouse model of breast cancer, Whiteaker et al.35 developed conventional MRM assays to 56 protein targets including 49 high-confidence biomarker candidates. The median analytical coefficient of variation for all assays was 5.7% at limits of quantification (LOQ), representative of the capabilities of this method. Using a single multiplexed assay, 46 of the targeted peptide analytes were above LOQ in >50% of plasma samples from mice bearing clinically apparent tumors; 30 proteins were significantly elevated in breast cancer and so were preliminarily verified in the context of the model. Addona et al. used MRM for preclinical verification of four new and several established markers of cardiovascular disease derived from short-term longitudinal coronary sinus and peripheral blood sampling of patients undergoing planned therapeutic myocardial injury15. Only three of the 52 candidates initially selected for quantitative assay development by MRM-MS had available reagents suitable for construction of enzyme-linked immunosorbent assays. The MRM-MS assays were used to measure the target analytes present at low nanogram per milliliter amounts in plasma.
Other groups have demonstrated the use of MRM for verification of previously described but insufficiently verified biomarker candidates in cardiovascular disease36 and cancer37. Wang38 used enrichment with variant-agnostic anti-protein antibodies coupled with MRM-MS to measure cancer-specific mutant proteins that were difficult to assess by other means. In the largest-scale demonstration of the potential of targeted MS for candidate verification yet undertaken, Hüttenhain14 developed MRM assays for 1,157 proteins associated with cancer and measured 73 of the target proteins in a crude plasma digest, 182 in depleted plasma and 408 in urine. Using a subset of MRM assays that were directed to four of the five proteins that comprise the US Food and Drug Administration–approved, antibody-based OVA1 biomarker test for ovarian cancer risk, they showed significant (P < 0.01) differential expression in the expected direction in a set of 83 plasma samples from 67 individuals with ovarian cancer and 16 with benign ovarian tumors. In the same analysis they monitored 30 additional proteins for which there was evidence of association with ovarian cancer, showing significant (P < 0.01) cancer versus control differences for 19 of them and demonstrating the use of MRM both to confirm and to extend established antibody-based assays.
Clinical implementation of targeted MS
To many in the field of proteomics, the pivotal question has been not whether but when targeted MS will be broadly adopted as a tool for clinical measurement of protein analytes, supplementing not supplanting the current use of immunoassays. This is less hubris or provincialism than a practical assessment of the advantages of the method, similar to what motivated its adoption for small-molecule measurements a generation ago. There are no fundamental technical obstacles to its adoption in clinical laboratories. Data demonstrating key analytical requirements such as low assay coefficients of variation, interlaboratory reproducibility and means to accurately measure amounts of targeted analytes have continued to accrue9,15,16,20-22. Clinical translation is therefore largely a problem of engineering rather than radical invention.
The first major inroads have now begun in the predicted fashion—namely, in the context of a clinically important analyte that has been recalcitrant to conventional immunoassays and with assay deployment initially centralized to reference laboratories. The seminal instance involved development of a peptide immunoaffinity MRM-MS assay for thyroglobulin39. Thyroglobulin levels are used to monitor disease activity in some subtypes of thyroid cancer. However, 20% of the population has circulating anti-thyroglobulin antibodies that interfere with the immunoassay and result in erroneous test results. The MS-based method involves digestion of all proteins to peptides before capture of a unique peptide from thyroglobulin using an anti-peptide antibody, thereby removing interferences from anti-thyroglobulin antibodies. Immuno-MRM assays for thyroglobulin are being developed by ARUP Laboratories and Quest Diagnostics, both of which are involved in the large-scale development and application of clinical tests. Lab-based immuno-MRM assays have been developed for similar reasons to measure parathyroid hormone in blood3 and total pepsin or pepsinogen in saliva21.
Future developments will likely include the development of diagnostics and clinical predictors based on targeted MS-based detection of multiplexed panels of proteins or modified peptides that are either difficult or costly to translate into conventional immunoassays because suitable antibodies prove too difficult to produce or because the multiplex level required is greater (for example, >5) than immunoassays can readily support. Such tests will face the same requirements for clinical adoption as any other assay (reagents for calibration and standardization, demonstration of accuracy and robustness, and so on), with potential new elements such as standards to assess the digestion efficiency of the clinical sample. More widespread and general deployment, for instance, to hospital laboratories and for a wider range of protein measurements, seems a more distant eventuality. In addition to a greater number of validated targets and high-performance assays based on qualified reagents, the development of robust, turnkey instruments is needed as well as more intelligent software for robust data analysis without expert oversight. This will require cooperation from the MS vendors who will need to clearly see the opportunity before being willing to make the investments. Participation and cooperation from regulatory agencies will also be essential in promoting development of new tests using new devices.
Proteomics continues to rapidly evolve through invention of improved sample-handling methods, use of higher-efficiency chromatography and the introduction of faster, more sensitive and precise MS technologies. These advances have greatly increased sample-analysis throughput while reducing undersampling and providing more consistent and reproducible peptide measurements. As the capabilities of MS-based proteomics technologies continue to improve, the line between what constitutes ‘discovery’ and ‘verification’ is blurring, leading to what has been recently described as ‘platform convergence’33. High-resolution, high-mass-accuracy MS systems once confined to the data-dependent discovery realm are increasingly being used for targeted MS analysis with the benefits of higher specificity and lower potential for false positive identifications40-42. These attributes will be of particular importance to clinical adoption of targeted MS approaches. A happy consequence is the anticipated elimination of boundaries between discovery and verification, replaced by a continuum enabled by a single integrated LC-MS/MS platform. This may define the point at which these maturing technologies reach adulthood.
ACKNOWLEDGMENTS
We thank L. Gaffney for her help with graphics. This work was supported in part by the Broad Institute of MIT and Harvard, and by grants from the US National Cancer Institute (U24CA160034, part of the Clinical Proteomics Tumor Analysis Consortium initiative, to S.A.C.) and National Heart, Lung, and Blood Institute (HHSN268201000033C and R01HL096738 to S.A.C.). S.A.C. and M.A.G. acknowledge the financial support of the Women’s Cancer Research Fund of the Entertainment Industry Foundation and the Komen Foundation for funding portions of this work.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Lawson AM. The scope of mass spectrometry in clinical chemistry. Clin. Chem. 1975;21:803–824. [PubMed] [Google Scholar]
- 2.Zhu X, Desiderio DM. Peptide quantification by tandem mass spectrometry. Mass Spectrom. Rev. 1996;15:213–240. doi: 10.1002/(SICI)1098-2787(1996)15:4<213::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 3.Grebe SKG, Singh RJJ. LC-MS/MS in the clinical laboratory—where to from here? Clin. Biochem. Rev. 2011;32:5–31. [PMC free article] [PubMed] [Google Scholar]
- 4.Yost RA, Enke CG. Triple quadrupole mass spectrometry for direct mixture analysis and structure elucidation. Anal. Chem. 1979;51:1251–1264. doi: 10.1021/ac50048a002. [DOI] [PubMed] [Google Scholar]
- 5.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 6.Barr JR, et al. Isotope dilution–mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin. Chem. 1996;42:1676–1682. [PubMed] [Google Scholar]
- 7.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics. 2007;6:2212. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Addona TA, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brun V, Masselon C, Garin J, Dupuis A. Isotope dilution strategies for absolute quantitative proteomics. J. Proteomics. 2009;72:740–749. doi: 10.1016/j.jprot.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Abbatiello SE, Mani DR, Keshishian H, Carr SA. Automated detection of inaccurate and imprecise transitions in quantitative assays of peptides by multiple monitoring mass spectrometry. Clin. Chem. 2010;56:291–305. doi: 10.1373/clinchem.2009.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reiter L, et al. mProphet: automated data processing and statistical validation of large-scale SRM experiments. Nat. Methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 13.Remily-Wood ER, et al. A database of reaction monitoring mass spectrometry assays elucidating therapeutic response in cancer. Proteomics Clin. Appl. 2011;5:383–396. doi: 10.1002/prca.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hüttenhain R, et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci. Transl. Med. 2012;4:142ra94. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Addona TA, et al. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nat. Biotechnol. 2011;29:635–643. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortin T, et al. Clinical quantitation of prostate-specific antigen biomarker in the low nanogram/milliliter range by conventional bore liquid chromatography-tandem mass spectrometry multiple reaction monitoring coupling and correlation with ELISA tests. Mol. Cell. Proteomics. 2009;8:1006–1015. doi: 10.1074/mcp.M800238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi T, et al. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc. Natl. Acad. Sci. USA. 2012;109:15395–15400. doi: 10.1073/pnas.1204366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berna MJ, et al. Quantification of NTproBNP in rat serum using immunoprecipitation and LC/MS/MS: a biomarker of drug-induced cardiac hypertrophy. Anal. Chem. 2008;80:561–566. doi: 10.1021/ac702311m. [DOI] [PubMed] [Google Scholar]
- 19.Anderson NL, et al. Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by anti-peptide antibodies (SISCAPA) J. Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 20.Whiteaker JR, et al. Evaluation of large scale quantitative proteomic assay development using peptide affinity-based mass spectrometry. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.005645. M110.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neubert H, Gale J, Muirhead D. Online high-flow peptide immunoaffinity enrichment and nanoflow LC-MS/MS: assay development for total salivary pepsin/pepsinogen. Clin. Chem. 2010;56:1413–1423. doi: 10.1373/clinchem.2010.144576. [DOI] [PubMed] [Google Scholar]
- 22.Whiteaker JR, et al. Sequential multiplexed analyte quantification using peptide immunoaffinity enrichment coupled to mass spectrometry. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.015347. M111.015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn E, et al. Inter-laboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.013854. M111.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sparbier K, et al. Immuno-MALDI-TOF MS: new perspectives for clinical applications of mass spectrometry. Proteomics. 2009;9:1442–1450. doi: 10.1002/pmic.200800616. [DOI] [PubMed] [Google Scholar]
- 25.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 26.Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin. Chem. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Angulo AM, et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clin. Proteomics. 2011;8:11. doi: 10.1186/1559-0275-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng AHC, Uddayasankar U, Wheeler AR. Immunoassays in microfluidic systems. Anal. Bioanal. Chem. 2010;397:991–1007. doi: 10.1007/s00216-010-3678-8. [DOI] [PubMed] [Google Scholar]
- 29.Ellington AA, Kullo IJ, Bailey KR, Klee GG. Antibody-based protein multiplex platforms: technical and operational challenges. Clin. Chem. 2010;56:186–193. doi: 10.1373/clinchem.2009.127514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brody EN, Gold L, Lawn RM, Walker JJ, Zichi D. High-content affinity-based proteomics: unlocking protein biomarker discovery. Expert Rev. Mol. Diagn. 2010;10:1013–1022. doi: 10.1586/erm.10.89. [DOI] [PubMed] [Google Scholar]
- 31.Thiviyanathan V, Gorenstein DG. Aptamers and the next generation of diagnostic reagents. Proteomics Clin. Appl. advance online publication 23 October 2012 (doi:10.1002/prca.201200042) [DOI] [PMC free article] [PubMed]
- 32.Makawita S, Diamandis EP. The bottleneck in the cancer biomarker pipeline and protein quantification through mass spectrometry–based approaches: current strategies for candidate verification. Clin. Chem. 2010;56:212–222. doi: 10.1373/clinchem.2009.127019. [DOI] [PubMed] [Google Scholar]
- 33.Smith RD. Mass spectrometry in biomarker applications: from untargeted discovery to targeted verification, and implications for platform convergence and clinical application. Clin. Chem. 2012;58:528–530. doi: 10.1373/clinchem.2011.180596. [DOI] [PubMed] [Google Scholar]
- 34.Percy AJ, Chambers AG, Yang J, Domanski D, Borchers CH. Comparison of standard-flow and nano-flow liquid chromatography systems for MRM-based quantitation of putative plasma biomarker proteins. Anal. Bioanal. Chem. 2012;404:1089–1101. doi: 10.1007/s00216-012-6010-y. [DOI] [PubMed] [Google Scholar]
- 35.Whiteaker JR, et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat. Biotechnol. 2011;29:625–634. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domanski D. et al. MRM-based multiplexed quantitation of 67 putative cardiovascular disease biomarkers in human plasma. Proteomics. 2012;12:1222–1243. doi: 10.1002/pmic.201100568. [DOI] [PubMed] [Google Scholar]
- 37.Pan S, et al. Multiplex targeted proteomic assay for biomarker detection in plasma: a pancreatic cancer biomarker case study. J. Proteome Res. 2012;11:1937–1948. doi: 10.1021/pr201117w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, et al. Mutant proteins as cancer-specific biomarkers. Proc. Natl. Acad. Sci. USA. 2011;108:2444–2449. doi: 10.1073/pnas.1019203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin. Chem. 2008;54:1796–1804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaffe JD, et al. Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol. Cell. Proteomics. 2008;7:1952–1962. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;11:1–17. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallien S, et al. Targeted proteomic quantification on quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics. advance online publication 7 September 2012 (doi:10.1074/mcp.O112.019802) [DOI] [PMC free article] [PubMed]