Abstract
We report a mass spectrometry–based method for the integrated analysis of protein expression, phosphorylation, ubiquitination and acetylation by serial enrichments of different post-translational modifications (SEPTM) from the same biological sample. this technology enabled quantitative analysis of nearly 8,000 proteins and more than 20,000 phosphorylation, 15,000 ubiquitination and 3,000 acetylation sites per experiment, generating a holistic view of cellular signal transduction pathways as exemplified by analysis of bortezomib-treated human leukemia cells.
In recent studies that each focused on one specific type of post-translation modification (PTM), more than 20,000 phosphorylation sites1, 19,000 ubiquitination sites2,3 and 3,600 acetylation sites4 have been reported in human cell lines. PTMs are found in substoichiometric abundance and therefore require enrichment to improve their detection by mass spectrometry. Phosphorylated peptides are usually enriched with immobilized metal affinity chromatography (IMAC)5 or titanium dioxide6; acetylated peptides are enriched with antibodies directed against the acetylated epsilon amino group of lysine residues, K(Ac)7; and ubiquitinated peptides are enriched with antibodies directed against a glycineglycine stub on lysine residues, K(GG), which is the remnant of ubiquitin side chains after a tryptic digest2,8. For relative quantification of PTMs in different biological states, metabolic labeling techniques such as stable-isotope labeling by amino acids in cell culture (SILAC) can be used9. Typically, peptide fractionation strategies such as strong-cation exchange10,11, hydrophilic interaction liquid chromatography12 or isoelectric focusing4 are used to reduce the complexity of PTM samples and increase the depth of coverage.
Here we describe SEPTM, a strategy for serial enrichments of different post-translational modifications. We reasoned that a sequential enrichment strategy in which the flow-through of a first PTM enrichment step is directly used for enrichment of a second PTM, and the flow-through of the second used for a third PTM enrichment step (Fig. 1a), could allow analysis of multiple types of modifications from the same biological sample without decreasing the quality of each individual PTM analysis. We demonstrate SEPTM here for analysis of the phosphoproteome, ubiquitinome and acetylome.
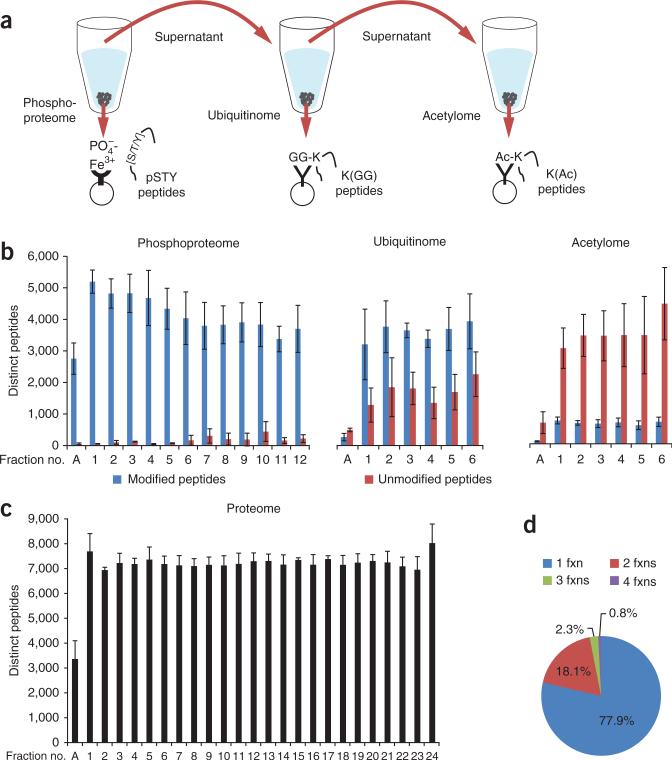
Figure 1.
SEPTM enables deep, quantitative analysis of the PTM-ome of a biological sample. (a) Integrated proteome and PTM-ome profiling by sequential enrichment for phosphorylated peptides with IMAC, for ubiquitinated peptides with K(GG)-specific antibodies and for lysine-acetylated peptides with K(Ac)-specific antibodies. (b) Ideal and even partitioning of phosphorylated, ubiquitinated and acetylated peptides by basic RP fractionation. The specificity of each enrichment step (distinct modified peptides out of total peptides) was >93% for phosphopeptides, >71% for ubiquitinated peptides and >18% for acetylated peptides. (c) We used 5% of the starting samples for whole-proteome analysis; basic RP separation yielded even partitioning of unmodified peptides. Average numbers of PTM sites (b) and proteins (c) per replicate are shown with s.d. error bars. (d) Uniqueness and occurrence of unmodified peptides per fraction (fxn).
To establish the effects of varying the amount of input sample and the extent of first-dimension fractionation on proteome and PTM-ome coverage, we applied SEPTM to SILAC-labeled Jurkat cells treated with the proteasome inhibitor bortezomib (Velcade) (Table 1). SILAC-labeled cell lysates from bortezomib-treated and untreated cells were mixed in equal amounts and digested with trypsin. Using 1 mg of protein lysate per SILAC label and no fractionation before on-line liquid chromatography–tandem mass spectrometry (LC-MS/MS), constituting a protocol we denote as ‘low’ proteome analysis, we identified nearly 2,000 distinct proteins on average per replicate (n = 3) by analysis of a few micrograms of the 1-mg starting sample. Notably, single LC-MS/MS analyses of the serially enriched phosphoproteome, ubiquitinome and acetylome yielded 5,466, 2,078 and 317 fully localized sites, respectively, per replicate.
Table 1.
Relationship among depth of proteome and PTM-ome coverage, input amount of protein lysate, degree of fractionation and mass spectrometry instrument time
| Coverage and input amount | Average number of proteins and site-localized PTMs quantified per replicate; no. of fractions and mass spectrometry runs |
|||
|---|---|---|---|---|
| Proteome | pS, pT, pY | K(GG) | K(Ac) | |
| Low (1 mg per SILAC label) | 1,991 (± 213); 1 | 5,466 (± 615); 1 | 2,078 (± 578); 1 | 317 (± 55); 1 |
| Medium (2.5 mg per SILAC label) | 5,603 (± 129); 8 | 12,913 (± 1,136); 4 | 4,924 (± 1,208); 2 | 799 (± 30); 2 |
| High (7.5 mg per SILAC label) | 7,897 (± 104); 24 | 20,800 (± 1,879); 12 | 15,408 (± 2,000); 6 | 3,190 (± 317); 6 |
Average numbers of quantified proteins and fully localized PTM sites per replicate (n = 3) are shown with s.d. For protein quantification we required at least two unique or razor peptides and ratios per protein. PTM sites were localized in each of the three replicates with a localization probability of >0.75. The sample consisted of SILAC-labeled Jurkat cells stimulated for 4 h with 1 μM bortezomib, a proteasome inhibitor. pS, pT and pY indicate phosphorylation on serine, threonine and tyrosine residues, respectively; K(GG) indicates ubiquitination, and K(Ac) indicates acetylation, on lysine residues. Every mass spectrometry run required 2.5 h of measurement time.
Serial enrichment of larger amounts of input material that we initially fractionated using reversed-phase (RP) chromatography13 at pH 10 (Supplementary Fig. 1a) greatly increased the proteome and PTM-ome depth of coverage (Table 1 and Supplementary Tables 1–3). For ‘high’ proteome coverage analysis with 7.5 mg of protein per SILAC state, we separated samples with basic RP into 72 fractions that we subsequently combined into 24 fractions in a noncontiguous manner13 (Supplementary Fig. 1b and Online Methods). After basic RP separation, we analyzed 5% of each of the 24 fractions by LC-MS/MS. The remaining 95% was subjected to multiple rounds of PTM enrichments. For our high proteome analysis, we combined the original 24 fractions into 12 fractions for phosphoproteome analysis and then into 6 fractions for enrichments of ubiquitinated and acetylated peptides. The combined flow-throughs after IMAC enrichments for phosphopeptides were enriched for ubiquitinated peptides using anti-K(GG) antibody, and the resulting flow-throughs were directly used for anti-K(Ac) enrichments (Fig. 1a).
For ‘medium’ proteome analysis using 2.5 mg of protein per SILAC state, we combined the 72 basic RP fractions into 8 proteome, 4 phosphoproteome and 2 K(GG) and K(Ac) fractions. The total peptide amounts we required per fraction (enrichment) and SILAC state for PTM analysis were >500 μg for IMAC enrichments and >1 mg for K(GG) and K(Ac) enrichments. These minimum starting amounts per fraction were determined from previous studies from our group, which had resulted in numbers of quantified phosphopeptides10 and K(GG) site–containing peptides3 that were equivalent to those from other high-coverage studies at that time1,2.
We found that basic-pH RP separation provided nearly ideal partitioning of post-translationally modified (Fig. 1b) and unmodified (Fig. 1c) peptides, with each fraction contributing similar numbers of identified peptides. Nearly 80% of all peptides were unique in each fraction for the proteome (Fig. 1d) and phosphoproteome, and 90% were unique in each fraction for the ubiquitinome and acetylome (Supplementary Fig. 2). The numbers of identified and localized K(GG) and K(Ac) sites increased more than linearly with higher input amounts and numbers of basic-pH RP fractions as well as instrument time (Table 1). In contrast, the numbers of identified peptides/proteins and phosphosites increased less than linearly with analysis time. This effect is likely due to the far greater complexity of the proteome and enriched phosphopeptide fractions, which are undersampled by the mass spectrometry instrument to a greater extent than are K(GG) and K(Ac) peptide fractions, and such undersampling can lead to missing data.
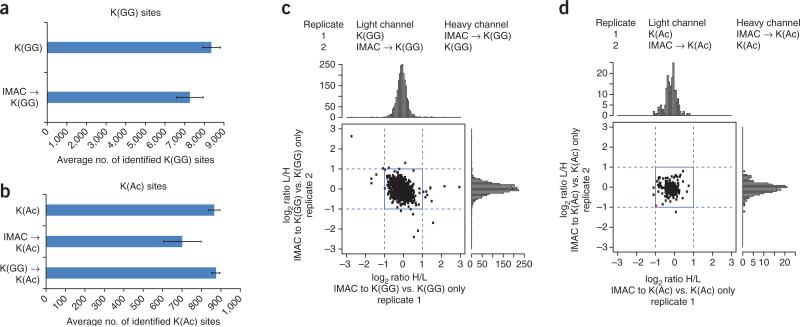
To evaluate potential losses of PTMs in the SEPTM strategy relative to conventional enrichment from separate samples, we compared the number of identified PTM sites after PTM enrichment with and without a preceding enrichment step for a different PTM in a label-free experiment. We chose to enrich for phospho- before ubiquitin- or acetyl-site peptides because the solvent used for IMAC enrichment is volatile (unlike those used for immunoaffinity capture) and can be directly evaporated. Initial enrichment of phosphopeptides by IMAC followed by enrichment of ubiquitinated and acetylated peptides using antibodies decreases identified K(GG)- and K(Ac)-sites by 13% and 19%, respectively (Fig. 2a,b; Supplementary Table 4). K(GG) enrichment before K(Ac) enrichment had no noticeable effect on the number of identified K(Ac)-sites. Using SILAC quantification, we further tested if the relatively small losses observed after IMAC enrichment could be ascribed to mechanical sample handling or a specific depletion of a subset of K(GG)- or K(Ac)-modified peptides. No specific decrease of K(GG)- or K(Ac)-site SILAC ratios was observed when sequential enrichment in one SILAC state was compared to single enrichment in the other SILAC state (Fig. 2c,d; Supplementary Table 4). Incomplete solubility of peptides in the highly organic IMAC binding buffer may explain losses after the primary IMAC enrichment.
Figure 2.
Comparison of nonserial to serial enrichments for K(GG) and K(Ac) enrichments. (a,b) Comparison of the number of identified K(GG) (a) and K(Ac) (b) sites. Label-free, tryptic HeLa peptides (2.5 mg) were either singly or doubly enriched for the indicated PTM enrichments. Each identified modification-site value is that of the last PTM enriched in the sample. s.d. error bars were calculated for three process replicates. (c,d) SILAC ratio comparisons of doubly versus singly K(GG)-enriched (c) or K(Ac)-enriched (d) samples. SILAC heavy (H)- and light (L)-labeled HeLa peptides (2.5 mg) were subjected to either one or two rounds of differential PTM enrichment (as indicated). log2 SILAC ratios of replicates 1 and 2 are visualized as scatter plots and associated histograms. We used a moderated t-test (P < 0.05) to detect any statistically significant up- or downregulated sites among 2,131 K(GG) sites (c) and 193 K(Ac)-sites (d). Only one data point had a P <0.05; this is colored red in the scatter plot. The second replicates were performed with an inverted SILAC labeling strategy.
We evaluated the capability of the SEPTM approach to study PTM co-regulation in Velcade-treated Jurkat cells, as it is known that proteasome inhibition leads to alterations in protein abundance and ubiquitination2,14, and may also indirectly affect protein phosphorylation and acetylation. In our “high” coverage experiment, we quantified an average of 7,897 proteins, 20,800 phosphorylation-sites, 15,408 K(GG)-sites and 3,190 K(Ac)-sites per replicate (Table 1) with an overlap across all three replicates of 94% on protein, 66% on phosphorylation-site, 44% on K(GG)-site and 55% on K(Ac)-site level (Supplementary Fig. 3). Interestingly, when we performed MS/MS searches for all three PTMs at the same time for each data set, we observed that only ca. 0.3% of all post-translationally modified tryptic peptides contain multiple different types of modifications (such as phospho- plus ubiquitin- or acetyl-sites; Supplementary Table 5), indicating that few peptides with multiple distinct modifications are being lost by serial enrichment, further supporting the efficacy of the SEPTM approach.
Our high-coverage proteome analysis revealed only minor changes in protein expression upon proteasome inhibition, which is in agreement with previous reports2,14. Only 0.8% of all proteins were found to be reproducibly regulated in all three replicates (moderated t-test false discovery rate of P < 0.02; Supplementary Fig. 4a). Nearly all 59 proteins were upregulated; among these, 15 are annotated as cell-cycle regulators in Gene Ontology (GO term: 0007049), including proteins that control cell-cycle entry (Mapk6, β-catenin, Myc and cyclin-D3), replication (Cdc6 and Msh6), spindle assembly (Zwint and Nusap1) or the anaphase-promoting complex (Fbxo5, Rassf1 and Bub1).
For PTM analysis, we normalized all SILAC ratios of PTM sites to their corresponding protein expression ratios and considered only high-quality PTM sites that were fully localized and detected in all three replicates. In agreement with the results of previous studies2,14, extensive changes in protein ubiquitination were detected, with 3,316 out of 5,678 (59%) K(GG) sites observed to be altered (Supplementary Fig. 4a). Ubiquitination changes affected virtually all major cellular pathways and machineries (Supplementary Table 6). We also detected strong induction of phosphorylation upon bortezomib treatment. We found 376 out of 10,493 (3.6%) phosphorylated serine, threonine and tyrosine sites to be regulated, and gene enrichment analysis showed an enrichment of proteins involved in transcription, cytoskeleton organization, nuclear transport and cell-cycle regulation (Supplementary Table 6). The acetylome analysis revealed no significant regulation among 1,554 K(Ac) sites. However, we found that 414 of these K(Ac) sites occurred on lysine residues that were also found to be ubiquitinated.
To examine potential PTM cross-regulation, we analyzed linear signature motifs in a window of ±15 amino acids around all identified PTM sites of proteins that were upregulated after bortezomib treatment on either the protein or phosphosite level using MotifX15, but we did not observe any significantly occurring motifs. A direct search for known phosphodegron motifs16 on proteins that were upregulated after bortezomib treatment revealed two transcription factors, Myc and Klf2, with the F-box protein Fbw7 (Cdc4) recognition sequences LPpTPPLpSPSRR (Myc Thr58 and Ser62) and LLpTPXXpS (Klf2 Thr244 and Ser248). In addition, Myc was found to be ubiquitinated at Lys67 and Lys163 and acetylated at Lys163.
To illustrate co-regulation of cellular processes by changes in protein abundance and PTMs, we analyzed all proteins that (i) were upregulated on either the protein or PTM-site level and (ii) had experimental data in curated protein-protein interaction databases supporting that they interact with each other (Supplementary Fig. 4b). The interaction network revealed six major, partially interconnected protein networks that regulate cell cycle, replication, transcription, translation and the proteasome. Ubiquitination and phosphorylation affect all of these processes, whereas protein abundance changes occur mainly for cell-cycle regulators. The key transcriptional regulators that are upregulated on single or multiple phosphorylation sites include components of the RNA polymerase II transcription machinery, such as Polr2c (pSer124), Gtf2e2 (pSer18) and Taf2g (pSer85). Among the most highly upregulated kinase phosphosites (Supplementary Table 7) was Ser614 on Cdk12, a cyclin-dependent kinase that is known to regulate gene expression through phosphorylation of RNA polymerase II (ref. 17).
The SEPTM technology we present integrates whole-proteome and deep, quantitative analysis of multiple, biologically important PTMs in a unified pipeline. As such, it provides a level of coverage similar to that of individual PTM analyses. SEPTM will be useful for obtaining a holistic view of signal transduction processes across time and perturbational conditions and will be generically applicable to any biological model system and PTM. In principle, the SEPTM technology can be extended to include additional PTMs, such as phosphotyrosine or arginine/lysine methylation, as long as sufficient starting material and good enrichment methods are available.
ONLINE METHODS
Preparation of SILAC-labeled Jurkat and HeLa S3 cells
SILAC-labeled cells were cultured at a concentration of 106 cells/mL in RPMI-1640 medium (Caisson labs), supplemented with 15 mg/L methionine, 200 mg/L proline, 10% (Jurkat) or 5% (HeLa S3) dialyzed FBS (Sigma F-0392), 1× penicillin/streptomycin (Gibco, 10378-016), and either normal l-lysine (K0) and l-arginine (R0) or heavy-labeled 13C6-15N2 lysine (K8) and 13C6-15N4 arginine (R10). Lysine and arginine were supplemented at concentrations of 40 mg/L and 120 mg/L, respectively. Labeled Jurkat cells were harvested after 8 cell doublings and were stimulated for 4 h with either 1 μM proteasome inhibitor bortezomib (Velcade; Takeda-Millennium Pharmaceuticals) or the same volume of dimethylsulfoxide. We analyzed three replicates, two of which were full process replicates on the same biological sample (replicates 1 and 3), with the third being a label swap in which Jurkat cells were Velcade-treated in the light rather than the heavy SILAC labeling state (replicate 2).
Cell lysis and peptide digestion
After drug treatment, cells were washed once with PBS and lysed for 30 min in ice-cold lysis urea buffer (8 M urea; 75 mM NaCl, 50 mM Tris HCl pH 8.0, 1 mM EDTA, 2 μg/mL aprotinin (Sigma, A6103), 10 μg/mL leupeptin (Roche, #11017101001), 1 mM PMSF (Sigma, 78830), 10 mM NaF, 5 mM sodium butyrate, 5 mM iodoacetamide (Sigma, A3221), Phosphatase Inhibitor Cocktail 2 (1:100, Sigma, P5726), Phosphatase Inhibitor Cocktail 3 (1:100, Sigma, P0044). Lysates were centrifuged at 20,000g for 10 min, and protein concentrations of the clarified lysates were measured via BCA assay (Pierce, 23227). From this procedure, Jurkat cell lysates produced ~8 mg of protein per 10 million cells. Light- and heavy-labeled lysates were then combined in a 1:1 protein ratio.
Protein disulfide bonds of the combined lysates were reduced for 45 min with 5 mM dithiothreitol (Thermo Scientific, 20291) and alkylated for 45 min with 10 mM iodoacetamide. Samples were then diluted 1:4 with 50 mM Tris HCl, pH 8.0, to reduce the urea concentration to 2 M. Lysates were digested overnight at room temperature with trypsin in a 1:50 enzyme-to-substrate ratio (Promega, V511X) on a shaker. Peptide mixtures were acidified to a final volumetric concentration of 1% formic acid (Fluka, 56302) and centrifuged at 2,000g for 5 min to pellet urea that had precipitated out of solution. Peptide mixtures were desalted on tC18 SepPak columns (Waters, 500 mg WAT036790). Columns were conditioned with 1 × 5 ml 100% acetonitrile and 1 × 5 ml 50% acetonitrile/0.1% formic acid washes, and equilibrated with 4 × 5 ml 0.1% trifluoroacetic acid (Fluka, TX1276-6). After loading the sample onto the column, samples were desalted with 3× 5 ml 0.1% trifluoroacetic acid washes and 1 × 5 ml 1% formic acid wash. Peptides were eluted from the column with 2 × 3 ml 50% acetonitrile/0.1% formic acid. 15 mg, 5 mg, and 2 mg peptide aliquots were then generated for high, medium, and low-scale coverage analyses. Eluted peptide samples were placed in a vacuum concentrator to evaporate the elution solvent and produce purified peptide samples.
Basic reversed-phase separation of peptides and generation of proteome samples for analysis
To reduce peptide complexity, samples were separated by basic reversed-phase chromatography18. We found that, unlike any other pre-fractionation technique, basic RP separation facilitates generation of equal and unique fractions of modified and unmodified peptide mixtures at the same time. Post-translational modifications such as phosphorylation, ubiquitination and acetylation bias the length and charge state of peptides. In our experience, separation techniques that rely on the charge state or pKa value of peptides, such as SCX or IEF, do not provide the level of uniqueness/fraction nor the equivalency of modified peptide numbers/fraction obtained using basic RP separation with non-contiguous fraction combining. The degree of fractionation reported in Table 1 is meant to provide useful starting points for investigators; further fractionation may be of some value in increasing the numbers of identified PTMs, at the cost of using more instrument time. Our decision to use the highest level of fractionation for proteome followed by phosphoproteome analysis and the lowest for K(GG) and K(Ac) analysis was guided by the numbers of identified MS/MS scans in each LC-MS/MS run as a measure for sample complexity. The average numbers of identified MS/MS scans pre–LC-MS/MS run for unfractionated samples decreased in the following order: proteome (29,563) > phosphoproteome (15,252) > K(GG) (7,787) > K(Ac) (4,603).
For basic RP separation, desalted peptides were reconstituted in 20 mM ammonium formate, pH 10, (high scale, 1.8 mL; medium scale, 900 μL; low scale samples did not require peptide partitioning) and centrifuged at 10,000g to clarify the mixture before it was transferred into autosampler tubes. Basic reversed-phase chromatography was conducted on Zorbax 300 Å Extend-C18 columns, using an Agilent 1100 Series HPLC instrument. High-scale separations were performed on a 9.4 mm × 250 mm column (Agilent, 5 μm bead size), whereas medium-scale separations were performed on a 4.6 mm × 250 mm column (Agilent, 3.5 μm bead size). It had been previously determined that 15 mg and 5 mg were the maximum peptide amounts that could be loaded onto each respective column without observing significant peptide loss. Prior to each separation, columns were monitored for efficient separation with standard mixtures containing 6 peptides. Solvent A (2% acetonitrile, 5 mM ammonium formate, pH 10), and a nonlinear increasing concentration of solvent B (90% acetonitrile, 5 mM ammonium formate, pH 10) were used to separate peptides by their hydrophobicity at a high pH. For separation of 5 mg total peptides on a 4.6 × 250 mm Zorbax Extend column we used a flow rate of 1 ml/min and increased the percentage of solvent B in a nonlinear gradient with 4 different slopes (0% for 9 min; 0% to 6% in 4 min; 6% to 28.5% in 50 min; 28.5% to 34% in 5.5 min; 34% to 60% in 13 min; 60% for 8.5 min). For separation of 15 mg total peptides on a 9.4 × 250 mm Zorbax Extend column we used a flow rate of 3 ml/min and increased the percentage of solvent B in a nonlinear gradient with 4 different slopes (0% for 2 min; 0% to 10% in 5 min; 10% to 27% in 34 min; 27% to 31% 4 min; 31% to 39% in 4 min; 39% to 60% in 7 min; 60% for 8 min). Eluted peptides were collected in 96 × 2 mL deepwell plates (Whatman, #7701-5200) with 1 min (= 1 ml) fractions for the 4.6 mm column and 40 s (= 2 ml) fractions for the 9.4 mm column. Early eluting peptides were collected in fraction “A”, which is a combined sample of all fractions collected before any major UV-214 signals were detected.
High- and medium-scale peptide samples were combined into 24 and 8 subfractions, respectively, to be used for proteome analysis. Subfractions were achieved in a serpentine, concatenated pattern, combining eluted fractions from the beginning, middle, and end of the run to generate subfractions of similar complexities that contain hydrophilic as well as hydrophobic peptides. For high-scale proteome analysis every 24th fraction was combined (1,25,49; 2,26,50; ...), and for medium-scale proteome analysis every 8th fraction was combined (1,9,17,25,33,41,49,57,65; 2,10,18,26,34,42,50,58,66; ...). Subfractions were acidified to a final concentration of 1% formic acid, and 5% of the volumetric samples were reserved for proteome analysis.
The remaining 95% of each of the original 72 subfractions from bRP (above) were further combined before enrichment for PTM analyses as follows. For high-scale (deep) analysis of the phosphoproteome, every 12th fraction was combined (1,13; 2,14; ...) to generate 12 fractions. For medium-scale (moderate analysis depth) analysis every 4th fraction was combined (1,5; 2,6, ...) to generate 4 fractions. Peptide fractions were subsequently dried by vacuum sublimation in a lyophilizer. Owing to the low starting concentration of ammonium formate, no additional desalting step before phospho-enrichment was necessary. In the small-scale (low coverage) study, a total of 2 mg of protein digest was used. 10 μg of reconstituted sample was used for proteome analysis with the remainder going through serial enrichment as described, below. We have also analyzed an early eluting hydrophilic fraction, (fraction A in Fig. 1 and Supplementary Fig. 1a) and found that although this fraction does not contain significant numbers of unmodified, ubiquitinated or acetylated peptides, it does contain a large number of multiply phosphorylated peptides and is therefore important to analyze for the phosphoproteome analysis.
IMAC phospho-enrichment of peptides
In a method adapted from Ficarro et al.5 iron-chelated IMAC beads were prepared from Ni-NTA superflow agarose beads (Qiagen, #1018611) that were stripped of nickel with 100 mM EDTA and incubated in an aqueous solution of 10 mM FeCl3 (Sigma, 451649). Dried phosphopeptide fractions (high scale: 12; medium scale: 4) were reconstituted in 50% acetonitrile/0.1% trifluoroacetic acid and then diluted 1:1 with 100% acetonitrile/0.1% trifluoroacetic acid to obtain a final 80% acetonitrile/0.1% TFA peptide solution at a concentration of 0.5 μg/μl. Peptide mixtures were enriched for phosphorylated peptides with 10 μL IMAC beads for each sample for 30 min. Enriched IMAC beads were loaded on Empore C18 silica-packed stage tips (3M, 2315) as has been described previously11. Stage tips were equilibrated with 2 × 100 μL washes of methanol, 2 × 50 μL washes of 50% acetonitrile/0.1% formic acid, and 2 × 100 μL washes of 1% formic acid. Samples were then loaded onto stage tips and washed twice with 50 μL of 80% acetonitrile/0.1% trifluoroacetic acid and 100 μL of 1% formic acid. Phosphorylated peptides were eluted from IMAC beads with 3 × 70 μL washes of 500 mM dibasic sodium phosphate, pH 7.0, (Sigma, S9763) and washed twice with 100 μL of 1% formic acid before being eluted from stage tips with 60 μL 50% acetonitrile/0.1% formic acid. All washes were performed on a tabletop centrifuge at a maximum speed of 3,500g.
For subsequent serial enrichment for ubiquitinated (K(GG)) and lysine-acetylated (K(Ac)) peptides, flow-throughs after IMAC enrichment were retained and combined in a stepwise fashion to generate 6 large-scale and 2 medium-scale fractions: For large-scale analysis every 6th IMAC supernatant fraction was combined (1,7; 2,8; ...), and for medium scale analysis every 2nd IMAC supernatant fraction was combined (1,3; 2,4).
K(GG) and K(Ac) enrichment of peptides
K(GG) antibody beads (Cell Signaling Technology, 5662S) were cross-linked with dimethyl pimelimidate using a method adapted from Udeshi et al.3. Peptide fractions were reconstituted in 1.4 mL of IAP buffer (50 mM MOPS, 50 mM NaCl, 10 mM Na2HPO4, pH 7) on ice, and samples were clarified by centrifugation at 10,000g. K(GG) beads were washed four times with IAP buffer on ice. Fractions were enriched for K(GG) peptides with 62.5 μg of K(GG) antibody per fraction for 1 h by end-over-end turning at 4 °C. K(GG) beads were pelleted at 1,000g and peptide mixtures were saved for K(Ac) enrichment. Beads were washed 3 times with ice-cold IAP buffer and 2 times with ice-cold PBS before enriched peptides were eluted from antibody beads with two 5-min incubation steps with 50 μL 0.15% trifluoroacetic acid. Elution mixtures were then desalted on Empore C18 silica bead Stagetips19. Stage tips were equilibrated with 2 × 100 μL washes of methanol, 2 × 50 μL washes of 50% acetonitrile/0.1% formic acid, and 2 × 100 μL washes of 1% formic acid. Samples were then loaded onto stage tips and washed twice with 100 μL of 1% formic acid before being eluted with 60 μL 50% acetonitrile/0.1% formic acid.
K(Ac) antibody beads (ImmuneChem, ICP0338) were washed four times with IAP buffer on ice. Peptide fractions taken from the K(GG) enrichment procedure were enriched with 100 μg of K(Ac) antibody per fraction and incubated for 1 h by end-over-end turning at 4 °C. K(Ac) enriched peptides were washed, eluted and desalted as described above for K(GG)-peptides.
LC-MS/MS analysis
All peptide samples were separated on an online nanoflow EASY-nLC 1000 UHPLC system (Thermo Fisher Scientific) and analyzed on a benchtop Orbitrap Q Exactive mass spectrometer (Thermo Fisher Scientific). Ten percent of each proteome (containing ~1 μg) and fifty percent of each phosphopeptide, K(GG) peptide, and K(Ac) peptide sample were injected onto a capillary column (Picofrit with 10 μm tip opening / 75 μm diameter, New Objective, PF360-75-10-N-5) packed in-house with 20 cm C18 silica material (1.9 μm ReproSil-Pur C18-AQ medium, Dr. Maisch GmbH, r119.aq). The UHPLC setup was connected with a custom-fit microadapting tee (360 μm, IDEX Health & Science, UH-753), and capillary columns were heated to 50 °C in column heater sleeves (Phoenix-ST) to reduce backpressure during UHPLC separation. Injected peptides were separated at a flow rate of 200 nL/min with a linear 80 min gradient from 100% solvent A (3% acetonitrile, 0.1% formic acid) to 30% solvent B (90% acetonitrile, 0.1% formic acid), followed by a linear 6 min gradient from 30% solvent B to 90% solvent B. Each sample was run for 150 min, including sample loading and column equilibration times. Data-dependent acquisition was obtained using Xcalibur 2.2 software in positive ion mode at a spray voltage of 2.00 kV. MS1 Spectra were measured with a resolution of 70,000, an AGC target of 3e6 and a mass range from 300 to 1800 m/z. Up to 12 MS2 spectra per duty cycle were triggered at a resolution of 17,500, an AGC target of 5e4, an isolation window of 2.5 m/z and a normalized collision energy of 25. Peptides that triggered MS2 scans were dynamically excluded from further MS2 scans for 20 s.
Identification and quantification of proteins
All mass spectra were analyzed with MaxQuant software version 1.3.0 (ref. 20). using a human UniProt database. MS/MS searches for the proteome data sets were performed with the following parameters: Oxidation of methionine and protein N-terminal acetylation as variable modifications; carbamidomethylation as fixed modification. For PTM data sets additional variable modifications were searched: Phosphorylation of serine, threonine and tyrosine residues for IMAC enriched samples; diglycine modification of lysine residues for K(GG) enriched samples; and epsilon-acetylated lysine for K(Ac) enriched samples. To study co-occurrence of different PTMs on the same peptides, phosphorylation, di-glycine modification and acetylation were searched simultaneously in a separate MS/MS search. Trypsin/P was selected as the digestion enzyme, and a maximum of 3 labeled amino acids and 2 missed cleavages per peptide were allowed. The mass tolerance for precursor ions was set to 20 p.p.m. for the first search (used for nonlinear mass re-calibration) and 6 p.p.m. for the main search. Fragment ion mass tolerance was set to 20 p.p.m. For identification we applied a maximum FDR of 1% separately on protein, peptide and PTM-site level. We required 2 or more unique/razor peptides for protein identification and a ratio count of 2 or more for protein quantification per replicate measurement. PTM-sites were considered to be fully localized when they were measured with a localization probability >0.75 in each of the three replicates. To assign regulated proteins and PTM-sites we used the Limma package in the R environment to calculate moderated t-test P values corrected by the Benjamini Hochberg method, as described previously14.
LC-MS/MS raw files
All raw data are available at ftp://ftp.broadinstitute.org/distribution/proteomics/public_datasets/.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the Broad Institute of MIT and Harvard and by the following grants from the US National Institutes of Health: grant U24CA160034 from the National Cancer Institute Clinical Proteomics Tumor Analysis Consortium Initiative (to S.A.C.) and grants HHSN268201000033C and R01HL096738 from the National Heart, Lung, and Blood Institute (to S.A.C.).
Footnotes
AUTHOR CONTRIBUTIONS
P.M. and S.A.C. developed the SEPTM strategy and conceived of the study; P.M., J.W.Q. and J.P. performed experiments; P.M., J.W.Q., J.P., N.D.U., K.R.C., D.R.M., M.W.B., M.A.G., J.D.J. and S.A.C. contributed to experimental design and data analysis; and P.M. and S.A.C. wrote the manuscript with input from all authors.
Supplementary information is available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Olsen JV, et al. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 2.Kim W, et al. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Udeshi ND, et al. Mol. Cell Proteomics. 2013;12:825–831. doi: 10.1074/mcp.O112.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhary C, et al. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 5.Ficarro SB, et al. Anal. Chem. 2009;81:4566–4575. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinkse MW, Lemeer S, Heck AJ. Methods Mol. Biol. 2011;753:215–228. doi: 10.1007/978-1-61779-148-2_14. [DOI] [PubMed] [Google Scholar]
- 7.Kim SC, et al. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 8.Xu G, Paige JS, Jaffrey SR. Nat. Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong SE, et al. Mol. Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 10.Mertins P, et al. Mol. Cell Proteomics. 2012;11:M111, 014423. doi: 10.1074/mcp.M111.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villén J, Gygi SP. Nat. Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNulty DE, Annan RS. Mol. Cell Proteomics. 2008;7:971–980. doi: 10.1074/mcp.M700543-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, et al. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Udeshi ND, et al. Mol. Cell Proteomics. 2012;11:148–159. doi: 10.1074/mcp.M111.016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz D, Gygi SP. Nat. Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 16.Hunter T. Mol. Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 17.Cheng SW, et al. Mol. Cell Biol. 2012;32:4691–4704. doi: 10.1128/MCB.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang F, Shen Y, Camp DG, II, Smith RD. Expert Rev. Proteomics. 2012;9:129–134. doi: 10.1586/epr.12.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rappsilber J, Mann M, Ishihama Y. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 20.Cox J, et al. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.