Abstract
It has been well documented that mutations in the same retinal disease gene can result in different clinical phenotypes due to difference in the mutant allele and/or genetic background. To evaluate this, a set of consanguineous patient families with Leber congenital amaurosis (LCA) that do not carry mutations in known LCA disease genes was characterized through homozygosity mapping followed by targeted exon/whole-exome sequencing to identify genetic variations. Among these families, a total of five putative disease-causing mutations, including four novel alleles, were found for six families. These five mutations are located in four genes, ALMS1, IQCB1, CNGA3, and MYO7A. Therefore, in our LCA collection from Saudi Arabia, three of the 37 unassigned families carry mutations in retinal disease genes ALMS1, CNGA3, and MYO7A, which have not been previously associated with LCA, and 3 of the 37 carry novel mutations in IQCB1, which has been recently associated with LCA. Together with other reports, our results emphasize that the molecular heterogeneity underlying LCA, and likely other retinal diseases, may be highly complex. Thus, to obtain accurate diagnosis and gain a complete picture of the disease, it is essential to sequence a larger set of retinal disease genes and combine the clinical phenotype with molecular diagnosis.
Keywords: Leber congenital amaurosis, LCA, whole-exome sequencing, SNP, padlock
Introduction
Leber congenital amaurosis (LCA; MIM# 204000) is an early-onset retinal dystrophy that is often diagnosed at birth or within the first year of life. The clinical features of LCA include blindness or severe vision impairment, congenital nystagmus, amaurotic pupils, and reduced or absence of signal in electroretinogram (ERG) [Franceschetti and Dieterle, 1954]. It is estimated to affect one in every 30,000~80,000 individuals and represents 5% of all retinal dystrophies [Dharmaraj et al., 2000; Kaplan et al., 1990; Leber, 1869; Stone, 2007].
LCA is a genetically heterogeneous and predominantly autosomal recessive disease. Up to now, mutations in 16 genes have been associated with LCA: aryl hydrocarbon receptor interacting protein-like 1 (AIPL1), centrosomal protein 290 kDa (CEP290), crumbs homolog 1 (Drosophila) (CRB1), cone-rod homeobox (CRX), guanylate cyclase 2D (GUCY2D), inosine 5′-monophosphate dehydrogenase 1 (IMPDH1), IQ motif containing B1 (IQCB1, also known as NPHP5), Leber congenital amaurosis 5 (LCA5), lecithin retinol acyltransferase (LRAT), orthodenticle homeobox 2 (OTX2), retinal degeneration 3 (RD3), retinol dehydrogenase 12 (RDH12), retinal pigment epithelium-specific protein 65 kDa (RPE65), retinitis pigmentosa GTPase regulator interacting protein 1 (RPGRIP1), spermatogenesis associated 7 (SPATA7), and tubby-like protein 1 (TULP1). [Bowne et al., 2006; den Hollander et al., 2001, 2007a,b, 2006; Dryja et al., 2001; Estrada-Cuzcano et al., 2011; Freund et al., 1998; Friedman et al., 2006; Henderson et al., 2009; Janecke et al., 2004; Marlhens et al., 1997; Mataftsi et al., 2007; Perrault et al., 2004, 1996; Senechal et al., 2006; Sohocki et al., 2000; Stone et al., 2011; Wang et al., 2009]. These genes are involved in various physiological pathways that are important for retinal function, including phototransduction (GUCY2D), maintenance of photoreceptor cell function (AIPL1, TULP1, RD3), visual cycle (LRAT, RPE65, RDH12), centrosomal or ciliary function (CEP290, IQCB1, LCA5, RPGRIP1), retinal development (CRB1, CRX, OTX2), and guanine nucleotide biosynthesis (IMPDH1) [Azadi et al., 2010; den Hollander et al., 2007a, 2006; Dizhoor, 2000; Escobar-Henriques and Daignan-Fornier, 2001; Furukawa et al., 1997; Mehalow et al., 2003; Murga-Zamalloa et al., 2009; Nishida et al., 2003; O’Byrne et al., 2005; Otto et al., 2005; Ramamurthy et al., 2004; Redmond et al., 1998; Thompson et al., 2005; Xi et al., 2005].
Recently, it was reported that certain mutant alleles in syndromic ocular disease genes may cause nonsyndromic LCA. For example, mutations in gene IQCB1 were previously reported to cause Senior-Løken syndrome (SLSN), which is characterized by retinal defects and nephronophthisis [Otto et al., 2005]. However, several nonsense and frameshift mutations in IQCB1 have been found in LCA patients without nephronophthisis or overt renal disease, suggesting that mutations in IQCB1 may cause LCA without having other syndromic phenotypes [Estrada-Cuzcano et al., 2011; Stone et al., 2011]. Considering that many nonsyndromic and syndromic ocular diseases, such as achromatopsia, Alstrom syndrome (ALMS), Batten disease, and SLSN, are associated with “LCA-like ocular phenotypes” [den Hollander et al., 2008], it is conceivable that some percentage of patients diagnosed with LCA are actually caused by either a specific allele or combination of disease gene alleles or mis-assignment due to the absence of syndromic features at the time of diagnosis.
Previously, we collected 37 consanguineous families with recessive LCA from Saudi Arabia. PCR and Sanger sequencing were performed for these families to screen for mutations in all known LCA genes [Li et al., 2009]. Among them, mutations have been identified in nine families. To identify the underlying mutations in the remaining consanguineous LCA disease families, homozygosity mapping using high-density SNP genotyping arrays followed by targeted or whole-exome sequencing was performed. Interestingly, mutations have been identified in four genes, ALMS1, IQCB1 (also known as NPHP5), CNGA3, and MYO7A, in six consanguineous LCA families, accounting for 16% of our collection (six of the 37) (Table 1). Recent studies have linked mutations in IQCB1 with LCA without renal failure [Estrada-Cuzcano et al., 2011; Stone et al., 2011]. Whereas mutations in ALMS1, CNGA3, and MYO7A are known to be associated with syndromic or nonsyndromic eye diseases, they have not been previously linked to LCA. Therefore, our results support an emerging theme that a significant fraction of patients diagnosed with LCA may be accounted for by mutations in syndromic and other retinal disease genes. These results highlight the importance of combining molecular diagnosis with clinical findings for diseases with high genetic heterogeneity in order to obtain accurate diagnosis and devise a proper treatment strategy.
Table 1.
Summary of Mutations Found Segregating with LCA in Six Families
| Mutation # | Protein change | Family | Affected individuals | Gene | OMIM accession number |
|---|---|---|---|---|---|
| c.10945G>T | p.E3649X | KKESH72 | 6 | ALMS1 | 606844 |
| c.1130-1G>C | KKESH24,88 | 2 | IQCB1 | 609237 | |
| c.1479C>A | p.Y493X | KKESH28 | 1 | IQCB1 | 609237 |
| c.1579C>A | p.L527M | KKESH2 | 1 | CNGA3 | 600053 |
| c.578C>T | p.T193I | KKESH34 | 4 | MYO7A | 276903 |
ALMS1 gene sequence [NM_015120.4]. IQCB1 gene sequence [NM_001023570.2]. CNGA3 gene sequence [NM_001298.2]. MYO7A gene sequence [NM_000260.3]. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, according to journal guidelines (www.hgvs.org/mutnomen). The initiation codon is codon 1. None of these mutations are found in normal matching controls or the dbSNP130 or 1,000 Genome databases.
The mutation in the ALMS1 gene is a previously reported allele and all the other mutations are novel. Mutations are validated by Sanger sequencing and segregate with the LCA phenotype within each family. Mutations are not present in normal matching controls. Mutations are within the homozygous regions that we identified for each family by SNP genotyping. All listed genes have been previously associated with syndromic or nonsyndromic eye diseases other than LCA.
Materials and Methods
Sample Collection
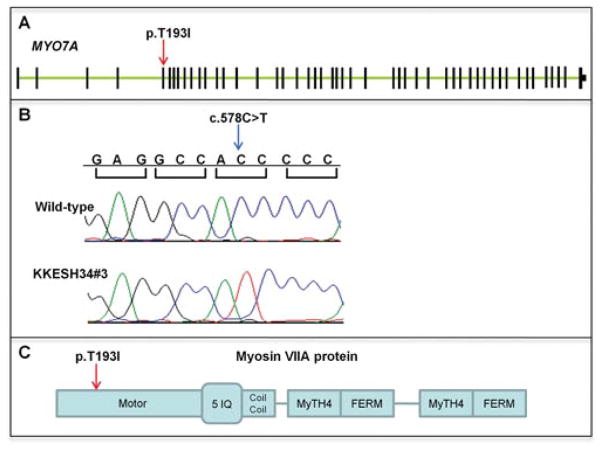
We obtained blood samples and pedigrees after receiving informed consent from all individuals. Approval was obtained from the Institutional Review Boards of the participating centers. LCA families KKESH24, KKESH28, KKESH34, KKESH72, and KKESH88 were obtained by Dr. Lewis through the King Khaled Eye Specialist Hospital (KKESH) in Riyadh, Saudi Arabia. Blood samples were collected from all available family members, and DNA was extracted with the Qiagen blood genomic DNA extraction kit (QIAGEN Inc., Valencia, CA) following the protocol provided by the manufacturer. The pedigrees of these families are shown in Figure 1. The number of affected and unaffected members of each family is listed in Table 2 and can be seen in Figure 1. All of the LCA patients were diagnosed with typical LCA clinical features (Table 2). Patients have experienced vision defects since birth or as early as 2 years old. No significant syndromic phenotypes were observed, except that a patient in KKESH88 showed midfacial hypoplasia and psychomotor delay.
Figure 1.
Pedigrees of six consanguineous LCA KKESH families. Affected: solid symbols; unaffected: open symbols; squares: male; circles: female.
Table 2.
Summary of Six Families with LCA
| Family | Affected individuals | Unaffected individuals | Clinical features |
|---|---|---|---|
| KKESH2 | 1 | 6 | At age 5 months, nystagmus was recorded “shortly after birth,” increasing in amplitude. A nonrecordable ERG was observed at age 10 months old. The patient was noted at 2 years old to have very sluggish pupils. No visual responses were elicited. |
| KKESH72 | 6 | 29 | The patients show irregular horizontal nystagmus, anterior and posterior cuneiform spoke-like cataracts, and mild cortical haze. Patients’ fundus shows vertically oval discs, waxy orange pallor, 3+/4 vascular attenuation, pigmentary maculopathy with central ellipse of gray silvery atrophy, and coarse granular diffuse RPE peripheral atrophy. |
| KKESH24 | 1 | 4 | The patient has nystagmus, hyperopic discs, vascular attenuation, no retinal pigmentation, diffuse RPE atrophy, and nonrecordable ERG. |
| KKESH28 | 1 | 4 | The patient shows nystagmus and eye rubbing. The eyes also have trouble in fixation and following movements. The ERG of the patient is nonrecordable. |
| KKESH88 | 1 | 6 | The patient shows midfacial hypoplasia, enophthalmos, horizontal and rotary nystagmus, variable LET, and LIO overaction. Furthermore, he has hypermetropic discs with hyperemia, moderate vascular attenuation, and sandy RPE throughout. The ERG is nonrecordable. In addition to eye phenotypes, he also shows psychomotor delay. |
| KKESH34 | 4 | 11 | All patients of KKESH34 have poor vision from birth. They all show nystagmus and neuroepithelial atrophy. The ERG of all patients is nonrecordable. |
All affected individuals from these KKESH families present with typical LCA phenotypes.
LET, left esotropia; LIO, left inferior oblique.
Homozygosity Mapping of LCA Patient Families
Homozygosity mapping using high-density SNP arrays was performed on these six consanguineous LCA families (Fig. 1). SNP genotypes were obtained for multiple affected members and unaffected siblings from each pedigree. Homozygous blocks greater than 1 Mb in size were identified from each individual. Based on recessive inheritance models, candidate disease loci are defined by shared homozygous blocks among all affected individuals but those are heterozygous or absent in unaffected siblings. The homozygous regions for six families range from 2000 bp to more than 57 Mb in size (Table 3).
Table 3.
Summary of Homozygous Blocks Identified in Six Families with LCA
| Family | Chr | Start | End | Length |
|---|---|---|---|---|
| KKESH2 | 10 | 4,692,299 | 28,687,145 | 23,994,846 |
| 10 | 95,392,963 | 119,610,549 | 24,217,586 | |
| 2 | 67,335,826 | 87,175,553 | 19,839,727 | |
| 2 | 94,759,484 | 100,814,358 | 6,054,874 | |
| 2 | 237,371,678 | 242,692,820 | 5,321,142 | |
| 13 | 74,774,390 | 97,207,720 | 22,433,330 | |
| 4 | 844,712 | 6,374,355 | 5,529,643 | |
| 4 | 13,417,458 | 48,967,354 | 35,549,896 | |
| 6 | 5,767,191 | 13,849,594 | 8,082,403 | |
| 6 | 22,303,479 | 42,929,919 | 20,626,440 | |
| 16 | 13,703,368 | 21,234,774 | 7,531,406 | |
| 7 | 102,176,469 | 130,176,130 | 27,999,661 | |
| 8 | 103,136,803 | 121,252,831 | 18,116,028 | |
| KKESH72 | 2 | 71,564,649 | 74,339,404 | 2,774,755 |
| KKESH24 | 1 | 27,535,253 | 43,222,769 | 15,687,516 |
| 20 | 53,239,401 | 59,936,745 | 6,697,344 | |
| 3 | 104,855,337 | 104,877,189 | 21,852 | |
| 3 | 122,757,953 | 129,806,851 | 7,048,898 | |
| 7 | 106,234,136 | 126,577,422 | 20,343,286 | |
| KKESH28 | 1 | 185,439,838 | 200,728,605 | 15,288,767 |
| 1 | 200,745,815 | 206,592,720 | 5,846,905 | |
| 11 | 134,433,812 | 134,435,899 | 2,087 | |
| 3 | 118,133,470 | 169,824,936 | 51,691,466 | |
| 12 | 5,363,096 | 19,757,396 | 14,394,300 | |
| 21 | 28,327,299 | 28,339,633 | 12,334 | |
| 21 | 41,134,108 | 46,919,231 | 5,785,123 | |
| 13 | 75,079,282 | 89,305,858 | 14,226,576 | |
| 6 | 23,674,393 | 41,934,989 | 18,260,596 | |
| 16 | 6,564,364 | 8,538,170 | 1,973,806 | |
| 8 | 111,858,228 | 140,521,054 | 28,662,826 | |
| 17 | 46,238,130 | 60,493,874 | 14,255,744 | |
| KKESH88 | 1 | 155,091,425 | 178,231,760 | 23,140,335 |
| 20 | 11,799 | 5,655,548 | 5,643,749 | |
| 21 | 31,374,803 | 40,419,896 | 9,045,093 | |
| 3 | 97,652,153 | 129,099,032 | 31,446,879 | |
| 13 | 46,193,917 | 71,963,849 | 25,769,932 | |
| 5 | 107,956,758 | 112,004,899 | 4,048,141 | |
| 7 | 146,181,172 | 151,027,186 | 4,846,014 | |
| KKESH34 | 11 | 54,631,614 | 91,827,532 | 37,195,918 |
The homozygous regions are shared by affected members and are either heterozygous or are not present in unaffected members. The coordinates are based on Human March 2006 (NCBI36/hg18) Assembly.
Whole-Exome Sequencing
Whole-exome capture sequencing provides an alternative way to sequence large numbers of coding regions at a lower price than traditional PCR and Sanger sequencing. One affected member from each family was chosen for NimbleGen whole-exome capture followed by Illumina HiSeq paired-end sequencing. For each patient, a total of about 4.4 to 100 million reads were uniquely mapped to the targeted gene-coding regions with an average coverage of 9× to 467×. SNPs were identified based on filtering criteria (posterior probability cutoff = 0.9, minimum coverage = 3). A total of 52,188–370,000 SNPs were identified for further analysis (Supp. Tables S1–S6).
PCR and Direct Sequencing
Direct PCR and sequencing were used to validate the mutations found in human ALMS1 (RefSeq: NM_015120.4; MIM# 606844), IQCB1 (RefSeq: NM_001023570.2; MIM# 609237), CNGA3 (RefSeq: NM_001298.2; MIM# 600053), and MYO7A (Ref-Seq: NM_000260.3; MIM# 276903). Primers were designed using primer design tool Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) [Rozen and Skaletsky, 2000]. Each amplicon was 300–500 base pairs in length and was sequenced directly with an ABI3730 machine in both forward and reverse directions. Each read was aligned to the reference sequence and base changes were identified with the Sequencher program.
For mutations, nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, according to journal guidelines (www.hgvs.org/mutnomen). The initiation codon is codon 1.
All mutations identified in this study have been submitted into The WayStation (http://www.centralmutations.org/) database or LOVD v 2.0 database [Fokkema et al., 2011].
Results
Candidate disease loci in consanguineous families were determined based on the property of identity-by-descent (IBD). To identify underlying mutations in the LCA family collection from Saudi Arabia, homozygosity mapping and targeted exon/whole-exome sequencing were performed. The pedigrees of the six families, KKESH2, KKESH24, KKESH28, KKESH34, KKESH72, and KKESH88, reported here are shown in Figure 1. The clinical phenotypes of affected members of these families are listed in Table 2.
A Nonsense Mutation in ALMS1 in the KKESH72 Family
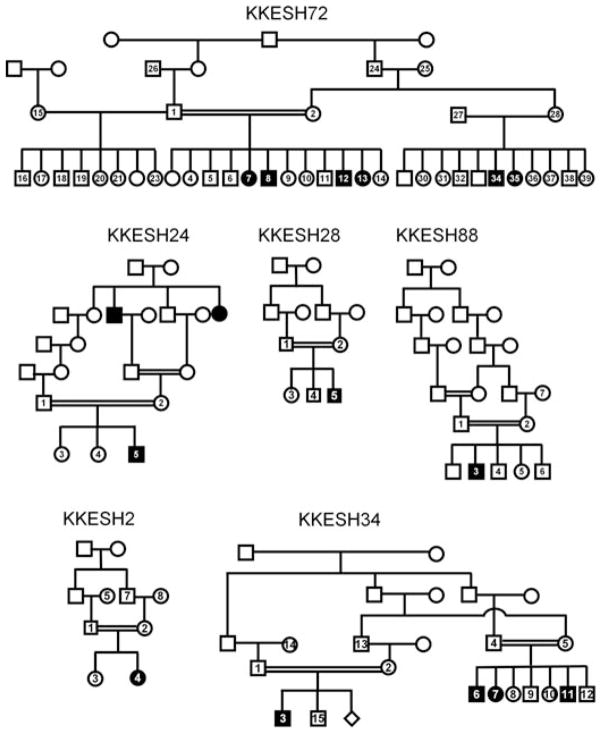
As shown in Figure 1, one of the biggest families in our collection is KKESH72. It is a four generation family containing six affected and 29 unaffected members. Four of the affected and one of the unaffected members were genotyped using SNP arrays. Although multiple homozygous blocks are present in each individual, only one homozygous region located on chromosome 2 from 71.5 to 74.3 Mb is shared among all four affected members and is either heterozygous or absent in the unaffected individual (Table 3). Consistent with previous data, no known LCA gene is located within this region, indicating that this region must contain a novel LCA disease gene. A custom-designed padlock probe set covering all exons in this region was used for capture sequencing as described in the methods section. A total of 4.45 million reads were generated, with an average sequencing coverage of 467×. For individual KKESH72#13, a total of 15 homozygous rare variants were detected that cause amino acid (aa) changes and reside within the homozygous block. Among them, only one nonsense mutation was identified. This mutation is located in the sixteenth exon of ALMS1 (c.10945G>T, p. E3649X) (Fig. 2A and B and Supp. Table S1). Interestingly, this nonsense mutation has been previously reported to be associated with ALMS [Marshall et al., 2007]. This mutation was confirmed by direct Sanger sequencing (Fig. 2B) and segregates with the disease in this family where affected individuals are homozygous for this mutation and other unaffected individuals are either heterozygous or do not carry the mutation. ALMS1 encodes a protein of 4,169 aa that localizes to centrosomes and ciliary basal bodies, and is important for normal cilium function [Li et al., 2007]. This nonsense mutation in exon 16 will either give rise to a protein with a truncated C-terminus missing 520 aa that is likely to have partial function or cause complete loss of function due to nonsense-mediated mRNA decay [Li et al., 2007]. To further check the potential effect of this mutation in splicing, ESEfinder 3.0 [Cartegni et al., 2003; Smith et al., 2006] was used to identify potential exonic splicing enhancers (ESE) around this variant. Interestingly, three potential ESEs are identified that might be affected by the mutation (Supp. Table S7). In the case that the 16th exon is skipped due to the mutation, it will lead to shift of the open reading frame and truncation of the protein. Multiple mutations in human ALMS1 have been reported to cause the ALMS, which is characterized by cone–rod retinal dystrophy, cardiomyopathy, type 2 diabetes mellitus, obesity, and hearing loss [Bond et al., 2005; Collin et al., 2002; Hearn et al., 2002; Marshall et al., 2007]. Patients in the KKESH72 family are reported to have nystagmus, cataracts, fundus defects, and other retinal defects (Table 2). In contrast to the phenotype in ALMS, no other syndromic phenotypes such as hearing loss or obesity were observed.
Figure 2.
Nonsense mutations and splicing changes are identified in ALSM1 and IQCB1. A: Exon–intron structure of ALSM1. Exons are shown as black boxes. The nonsense mutation p.E3649X is located in the sixteenth exon (red arrow). B: Sequence traces of control and affected members. A homozygous nonsense mutation from G to T in c.10945 is identified in affected member KKESH72#13 (blue arrow). C: Exon–intron structure of IQCB1. Exons are shown as black boxes. The splice acceptor site change is located at the 5′ boundary of the 12th exon and the nonsense mutation p.Y493X is located in the fourteenth exon (red arrows). D: Sequence traces of control and affected members. A homozygous splicing change in c.1130-1 is identified in affected members KKESH24#5 and KKESH88#3, and a homozygous nonsense mutation in c.1479 is identified in affected member KKESH28#5 (blue arrows).
A Nonsense Mutation and a Splicing Change in IQCB1 in Three KKESH Families
It has been recently reported that some of the mutant alleles in IQCB1 cause nonsyndromic LCA [Estrada-Cuzcano et al., 2011; Stone et al., 2011]. KKESH24, KKESH88, and KKESH28 are three consanguineous LCA families with one affected member in each family and four, six, and four unaffected members, respectively (Fig. 1). Based on homozygosity mapping, multiple homozygous blocks were identified for each family (Table 3). Whole-exome sequencing was conducted for one affected member from each family: KKESH24#5, KKESH88#5, and KKESH28#5. For the three KKESH-affected individuals, a total of 107, 117, and 81 million reads were generated, resulting average sequence coverage of 40×, 21×, and 30×, respectively. A total of 6, 4, and 27 homozygous rare variants that cause aa changes reside in each family’s homozygous block (Supp. Tables S2–S4). Among these candidate SNPs, only one obvious loss-of-function mutation is found for each family, all of which locate in IQCB1. One mutation is a novel splice acceptor site change (c.1130-1G>C) in KKESH24#5 and KKESH88#5, and the other is a novel nonsense mutation (c.1479C>A, p.Y493X) in KKESH28#5 (Fig. 2C and D). Both mutations are confirmed by direct Sanger sequencing (Fig. 2D) and are likely to be pathogenic. First, both mutations segregate with the disease in each family, with affected members being homozygous for the mutations and other unaffected individuals as either heterozygous or not carrying the mutation. Second, these mutations are rare as they are not found in 200 matching controls, the dbSNP130 database, or the 1,000 Genome database. Third, IQCB1 encodes an IQ-domain protein, which is important for cilium function and colocalizes with RPGR at the connecting cilia of photoreceptors [Otto et al., 2005]. Human mutations in IQCB1 are associated with LCA [Estrada-Cuzcano et al., 2011; Stone et al., 2011] and SLSN [Otto et al., 2005], which is characterized by nephronophthisis and retinal degeneration. The patients in these three KKESH families show typical LCA phenotypes such as nystagmus, nonrecordable ERGs, and other visual defects. However, no kidney defects were observed at the time of diagnosis.
A Missense Mutation in CNGA3 in the KKESH2 Family
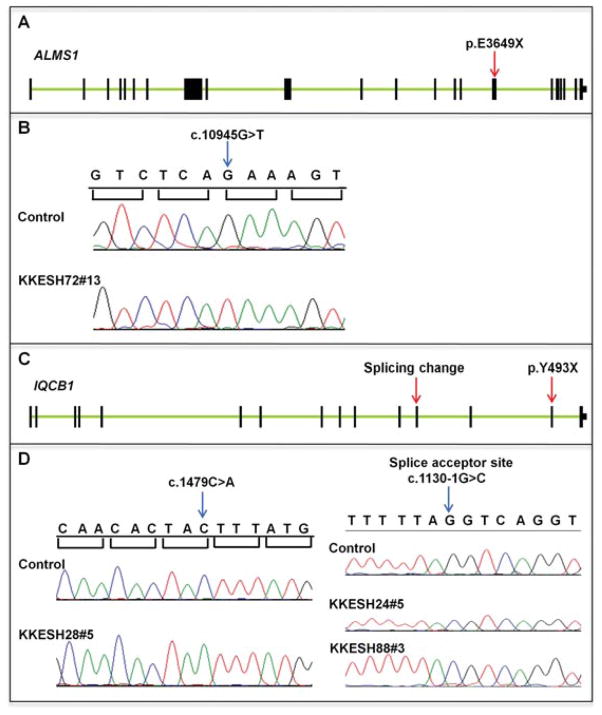
KKESH2 is a four generation family with one affected and six unaffected members (Fig. 1). One unaffected and one affected member were genotyped by SNP arrays. Ten homozygous blocks ranging from 5.3 to 35.5 Mb were identified in the affected member and are not present in the unaffected member (Table 3). Whole-exome sequencing was conducted for one affected individual, KKESH2#4. A total of 100 million reads were generated with 36× coverage. SNP calling reveals 11 homozygous rare variants that lead to aa changes and reside within the homozygous region (Supp. Table S5). Based on gene annotation, we focus on a missense change (c.1579C>A, p.L527M) in the CNGA3 gene. This variant was confirmed by direct Sanger sequencing (Fig. 3A and B) and is likely to be pathogenic. First, it is located within the homozygous region chr2: 94.7–100.8 Mb and segregates with the disease in this family, given the affected member is homozygous for this mutation and other unaffected individuals are either heterozygous or do not carry the mutation. Second, this variant is rare as it is absent in all 200 normal matching controls. In addition, it is not recorded in the dbSNP130 database and the 1,000 Genome database. Third, CNGA3 encodes a member of the family of cyclic nucleotide-gated channel alpha 3 proteins, which are important for normal vision and olfactory signaling transduction. The p.L527M mutation resides in the cGMP-binding domain of CNGA3, which is important for the cyclic nucleotide gating mechanism (Fig. 3C). This mutated leucine is conserved across vertebrate species, from human to stickleback, further suggesting functional importance of this residue (Fig. 3D). Human mutations in this gene are reported in hereditary cone photoreceptor disorder, which is characterized by cone photoreceptor dysfunction, and achromatopsia, which leads to early vision loss, nystagmus, photophobia, color blindness, but has normal scotopic responses [Johnson et al., 2004; Wissinger et al., 2001]. Affected members of the KKESH2 family show early-onset nystagmus (at 5 months old), sluggish pupils, no visual response, and nonrecordable ERG at 10 months of age (Table 2). Taken together, these data suggest that the p.L527M mutation in CNGA3 is associated with the LCA phenotype in the KKESH2 family.
Figure 3.
A missense mutation is identified in CNGA3. A: Exon–intron structure of CNGA3. Exons are shown as black boxes. The nonsense mutation p.L527M is located in the eighth exon (red arrow). B: Sequence traces of control and affected members. A homozygous missense mutation in c.1579 is identified in affected member KKESH2#4 (blue arrow). C: Schematic view of the CNGA3 protein structure. The p.L527M mutation is located within the intracellular cGMP-binding site. D: Amino acid alignment among 11 vertebrate species. The mutated leucine is located within this region and is conserved across all species.
A Missense Mutation in MYO7A in the KKESH34 Family
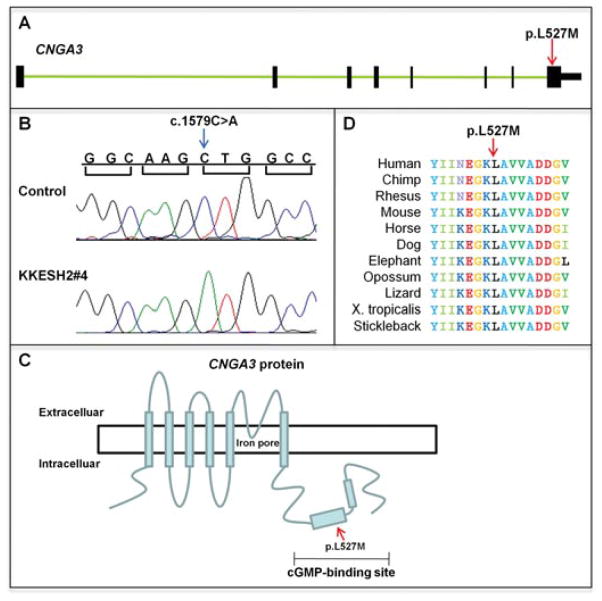
KKESH34 is a large consanguineous family containing four affected and 11 unaffected individuals (Fig. 1). All the affected and three unaffected members were genotyped by SNP arrays. The only homozygous region that was shared by affected members and not present in unaffected members resides on chr11: 5463161491827532, with a size of 37.2 Mb (Table 3). Since the size of the candidate region is large, whole-exome sequencing was performed for the patient KKESH34#6. A total of 7.7 million reads were generated, with coverage of 27×. A total of six homozygous rare variants that lead to aa changes were identified within the candidate region (Supp. Table S6). Based on conservation score (data not shown) and gene annotations, we focus on a novel missense variant (c.578C>T, p.T193I) in the MYO7A gene (Fig. 4A and B). This variant is rare, as it is absent from 200 matching controls, the dbSNP database, and 1,000 Genome database. It segregates with the disease in this family, given that all the affected members are homozygous for this mutation and other unaffected individuals are either heterozygous or do not carry the mutation. MYO7A encodes an unconventional myosin, myosin VIIA, which is involved in the transportation of opsin in photoreceptor cilia [Liu et al., 1999]. Also, it was recently demonstrated that myosin VIIA protein may affect the localization and function of the visual retinoid cycle enzyme, RPE65, which has been associated with LCA [Lopes et al., 2011]. The mutation identified (p.T193I) is a novel allele residing in the motor domain, which is responsible for binding filamentous actin and hydrolyzing ATP (Fig. 4C). Human mutations in MYO7A are associated with Usher syndrome, characterized by deafness and progressive vision loss. All patients of KKESH34 have had poor vision since birth. They all also have nystagmus, neuroepithelial atrophy, and nonrecordable ERGs (Table 2). However, patients in the KKESH34 family do not exhibit hearing loss. Therefore, it is likely that this mutation in MYO7A will only partially affect protein function, resulting in retinal specific clinical phenotype.
Figure 4.
A missense mutation is identified in MYO7A. A: Exon–intron structure of MYO7A. Exons are shown as black boxes. The missense mutation p.T193I is located in the fifth exon (red arrow). B: Sequence traces of control and affected members. A homozygous missense mutation in c.578 is identified in affected member KKESH34#3 (blue arrow). C: Schematic view of the MYO7A protein structure. The p.T193I mutation is located within the motor domain.
Discussion
We performed homozygosity mapping and whole-exome sequencing to identify mutations affecting a collection of consanguineous LCA families. Our experiments show that homozygosity mapping coupled with whole-exome sequencing is a very powerful tool for identifying mutations, even for families with a small number of affected members. For example, in the KKESH28 family, there was only one affected member and four unaffected members (Fig. 1 and Table 2). Homozygosity mapping alone discovered 12 homozygous blocks whose widths range from 2087 bp to 51.7 Mb, making the approach of PCR and Sanger sequencing impractical (Table 3). By using whole-exome sequencing, we were able to discover 27 rare SNPs that lead to aa changes and reside within the homozygous regions (Supp. Table S4). Among these candidate SNPs, a nonsense mutation (c.1479C>A, p.Y493X) was identified, suggesting this approach can be effective in identifying mutations in families with a small number of affected members.
Interestingly, we have identified families carrying mutations in ALMS1, IQCB1, CNGA3, and MYO7A genes that have been previously associated with other syndromic or nonsyndromic retinal diseases whose phenotypes overlap with that of LCA. The mutations include a previously reported nonsense mutation (c.10945G>T, p. E3649X) in ALMS1 in family KKESH72, a novel splicing change (c.1130-1G>C) in IQCB1 in two families, KKESH24 and KKESH88, a novel nonsense mutation (c.1479C>A, p.Y493X) of IQCB1 in family KKESH28, a novel missense mutation (c.1579C>A, p.L527M) in CNGA3 in family KKESH2, and a novel missense mutation (c.578C>T, p.T193I) in MYO7A in family KKESH34 (Table 1). Based on the phenotype of the patients, it is likely that in each case these missense mutations result in partial loss-of-function of the gene. These alleles are not only useful for molecular diagnoses, but further study of the functional consequence of these mutations will likely provide additional insight into the molecular mechanisms by which these proteins act.
Our findings support the emerging theme that a significant fraction of patients diagnosed with LCA may carry mutations in other syndromic or nonsyndromic retinal disease genes. This is likely due to several reasons. First, it is possible that patients diagnosed as having LCA are too young at the time of diagnosis to present with other syndromic phenotypes. For example, patients who carry mutations in IQCB1 may not show kidney defects until they are in their teens. Second, it is possible that different alleles in the same gene can result in different clinical presentations. For example, patients in family KKESH34 carry a missense mutation (c.578C>T, p.T193I) in MYO7A and show typical LCA phenotypes without hearing problems. It is possible that this mutation results in a partial loss-of-function allele that is only critical for function in the eye. Indeed, similar phenomena have been observed for many retinal disease genes. For example, partial loss-of-function mutations in CEP290 lead to LCA [Coppieters et al., 2010; den Hollander et al., 2006], while null alleles are also associated with recessive Meckel syndrome [Baala et al., 2007; Frank et al., 2008], Bardet-Biedl syndrome [Leitch et al., 2008], and Joubert syndrome [Sayer et al., 2006; Valente et al., 2006]. This phenomenon is also observed in other LCA causal genes, such as CRX [Freund et al., 1997; Swain et al., 1997] and RDH12 [Fingert et al., 2008; Janecke et al., 2004]. Third, it is possible that the same mutation can lead to a different clinical presentation, presumably due to modifications from the genetic background and/or the environment. In our report, the nonsense mutation (c.10945G>T, p. E3649X) in the ALMS1 gene identified in family KKESH72 has been previously reported to cause ALMS [Marshall et al., 2007]. However, none of the patients in this family show other syndromic phenotypes, such as hearing loss or obesity, which are typical for ALMS. It is possible that the different genetic background in the LCA patients from family KKESH72 lead to the different clinical presentations. Indeed, it has been reported recently that a common allele in RPGIP1L can modify the retinal degeneration phenotype in ciliopathies [Katsanis et al., 2001; Khanna et al., 2009]. In addition, in this report, the same mutation in IQCB1 has been identified in both KKESH24#5 and KKESH88#3 patients. It is interesting to note that while the eye phenotype in both patients is quite similar, KKESH88#3 also shows additional neural defects such as midfacial hypoplasia and psychomotor delay. With the whole-exome sequencing data, it is possible to check if KKESH88#3 carries potential genetic modifiers that can potentially explain the clinic phenotype. Indeed, in KKESH88#3, rare variants have been found in gene ACAT1 and FGFR2, which is associated with psychomotor delay and midfacial hypoplasia, respectively (Supp. Table S8) [Korman, 2006; Tartaglia et al., 1997].
In summary, it is evident that in addition to the 16 known LCA genes, mutations in several other syndromic or nonsyndromic eye disease genes may also lead to the LCA phenotype. Due to the high genetic heterogeneity of LCA, it is likely to be informative to sequence a larger set of retinal genes along with the known LCA genes. Combining accurate molecular diagnoses with the clinical phenotypes of LCA patients will be an essential step to proper treatment of this disease in the future.
Supplementary Material
Acknowledgments
Contract grant sponsor: Retina Research Foundation and the National Eye Institute (R01EY018571 to R.C.).
We are indebted to John Cavender, M.D., the Research Director of the King Khalid Eye Specialist Hospital at the time of this study, to the Research Council of KKESH for financial support, and to the staff of the KKESH Research Department for their diligent commitment to this program. In addition, we thank the family reported here for their willing cooperation in this study. We would like to thank Dr. Molly Bray for the SNP genotyping. Dr. Lewis is a Senior Scientific Investigator of Research to Prevent Blindness, New York. HW was supported by postdoctoral fellowship F32EY19430.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Azadi S, Molday LL, Molday RS. RD3, the protein associated with Leber congenital amaurosis type 12, is required for guanylate cyclase trafficking in photoreceptor cells. Proc Natl Acad Sci USA. 2010;107:21158–21163. doi: 10.1073/pnas.1010460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baala L, Audollent S, Martinovic J, Ozilou C, Babron MC, Sivanandamoorthy S, Saunier S, Salomon R, Gonzales M, Rattenberry E, Esculpavit C, Toutain A, Moraine C, Parent P, Marcorelles P, Dauge MC, Roume J, Le Merrer M, Meiner V, Meir K, Menez F, Beaufrere AM, Francannet C, Tantau J, Sinico M, Dumez Y, MacDonald F, Munnich A, Lyonnet S, Gubler MC, Genin E, Johnson CA, Vekemans M, Encha-Razavi F, Attie-Bitach T. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am J Hum Genet. 2007;81:170–179. doi: 10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Flintoff K, Higgins J, Scott S, Bennet C, Parsons J, Mannon J, Jafri H, Rashid Y, Barrow M, Trembath R, Woodruff G, Rossa E, Lynch S, Sheilds J, Newbury-Ecob R, Falconer A, Holland P, Cockburn D, Karbani G, Malik S, Ahmed M, Roberts E, Taylor G, Woods CG. The importance of seeking ALMS1 mutations in infants with dilated cardiomyopathy. J Med Genet. 2005;42:e10. doi: 10.1136/jmg.2004.026617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne SJ, Sullivan LS, Mortimer SE, Hedstrom L, Zhu J, Spellicy CJ, Gire AI, Hughbanks-Wheaton D, Birch DG, Lewis RA, Heckenlively JR, Daiger SP. Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2006;47:34–42. doi: 10.1167/iovs.05-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin GB, Marshall JD, Ikeda A, So WV, Russell-Eggitt I, Maffei P, Beck S, Boerkoel CF, Sicolo N, Martin M, Nishina PM, Naggert JK. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alstrom syndrome. Nat Genet. 2002;31:74–78. doi: 10.1038/ng867. [DOI] [PubMed] [Google Scholar]
- Coppieters F, Casteels I, Meire F, De Jaegere S, Hooghe S, van Regemorter N, Van Esch H, Matuleviciene A, Nunes L, Meersschaut V, Walraedt S, Standaert L, Coucke P, Hoeben H, Kroes HY, Vande Walle J, de Ravel T, Leroy BP, De Baere E. Genetic screening of LCA in Belgium: predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum Mutat. 2010;31:E1709–E1766. doi: 10.1002/humu.21336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, Heckenlively JR, van den Born LI, de Kok YJ, van der Velde-Visser SD, Kellner U, Jurklies B, van Schooneveld MJ, Blankenagel A, Rohrschneider K, Wissinger B, Cruysberg JR, Deutman AF, Brunner HG, Apfelstedt-Sylla E, Hoyng CB, Cremers FP. Leber congenital amaurosis and retinitis pigmentosa with Coats-like exudative vasculopathy are associated with mutations in the crumbs homologue 1 (CRB1) gene. Am J Hum Genet. 2001;69:198–203. doi: 10.1086/321263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, Koenekoop RK, Mohamed MD, Arts HH, Boldt K, Towns KV, Sedmak T, Beer M, Nagel-Wolfrum K, McKibbin M, Dharmaraj S, Lopez I, Ivings L, Williams GA, Springell K, Woods CG, Jafri H, Rashid Y, Strom TM, van der Zwaag B, Gosens I, Kersten FF, van Wijk E, Veltman JA, Zonneveld MN, van Beersum SE, Maumenee IH, Wolfrum U, Cheetham ME, Ueffing M, Cremers FP, Inglehearn CF, Roepman R. Mutations in LCA5, encoding the ciliary protein lebercilin, cause Leber congenital amaurosis. Nat Genet. 2007a;39:889–895. doi: 10.1038/ng2066. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, Zonneveld MN, Strom TM, Meitinger T, Brunner HG, Hoyng CB, van den Born LI, Rohrschneider K, Cremers FP. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, Lopez I, Yzer S, Zonneveld MN, Janssen IM, Strom TM, Hehir-Kwa JY, Veltman JA, Arends ML, Meitinger T, Musarella MA, van den Born LI, Fishman GA, Maumenee IH, Rohrschneider K, Cremers FP, Koenekoop RK. Identification of novel mutations in patients with Leber congenital amaurosis and juvenile RP by genome-wide homozygosity mapping with SNP microarrays. Invest Ophthalmol Vis Sci. 2007b;48:5690–5698. doi: 10.1167/iovs.07-0610. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Dharmaraj SR, Silva ER, Pina AL, Li YY, Yang JM, Carter CR, Loyer MK, El-Hilali HK, Traboulsi EK, Sundin OK, Zhu DK, Koenekoop RK, Maumenee IH. Mutational analysis and clinical correlation in Leber congenital amaurosis. Ophthalmic Genet. 2000;21:135–150. [PubMed] [Google Scholar]
- Dizhoor AM. Regulation of cGMP synthesis in photoreceptors: role in signal transduction and congenital diseases of the retina. Cell Signal. 2000;12:711–719. doi: 10.1016/s0898-6568(00)00134-0. [DOI] [PubMed] [Google Scholar]
- Dryja TP, Adams SM, Grimsby JL, McGee TL, Hong DH, Li T, Andreasson S, Berson EL. Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am J Hum Genet. 2001;68:1295–1298. doi: 10.1086/320113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M, Daignan-Fornier B. Transcriptional regulation of the yeast gmp synthesis pathway by its end products. J Biol Chem. 2001;276:1523–1530. doi: 10.1074/jbc.M007926200. [DOI] [PubMed] [Google Scholar]
- Estrada-Cuzcano A, Koenekoop RK, Coppieters F, Kohl S, Lopez I, Collin RW, De Baere EB, Roeleveld D, Marek J, Bernd A, Rohrschneider K, van den Born LI, Meire F, Maumenee IH, Jacobson SG, Hoyng CB, Zrenner E, Cremers FP, den Hollander AI. IQCB1 mutations in patients with Leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2011;52:834–839. doi: 10.1167/iovs.10-5221. [DOI] [PubMed] [Google Scholar]
- Fingert JH, Oh K, Chung M, Scheetz TE, Andorf JL, Johnson RM, Sheffield VC, Stone EM. Association of a novel mutation in the retinol dehydrogenase 12 (RDH12) gene with autosomal dominant retinitis pigmentosa. Arch Ophthalmol. 2008;126:1301–1307. doi: 10.1001/archopht.126.9.1301. [DOI] [PubMed] [Google Scholar]
- Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- Franceschetti A, Dieterle P. Diagnostic and prognostic importance of the electroretinogram in tapetoretinal degeneration with reduction of the visual field and hemeralopia. Confin Neurol. 1954;14:184–186. [PubMed] [Google Scholar]
- Frank V, den Hollander AI, Bruchle NO, Zonneveld MN, Nurnberg G, Becker C, Du Bois G, Kendziorra H, Roosing S, Senderek J, Nurnberg P, Cremers FP, Zerres K, Bergmann C. Mutations of the CEP290 gene encoding a centrosomal protein cause Meckel-Gruber syndrome. Hum Mutat. 2008;29:45–52. doi: 10.1002/humu.20614. [DOI] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Freund CL, Wang QL, Chen S, Muskat BL, Wiles CD, Sheffield VC, Jacobson SG, McInnes RR, Zack DJ, Stone EM. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- Friedman JS, Chang B, Kannabiran C, Chakarova C, Singh HP, Jalali S, Hawes NL, Branham K, Othman M, Filippova E, Thompson DA, Webster AR, Andreasson S, Jacobson SG, Bhattacharya SS, Heckenlively JR, Swaroop A. Premature truncation of a novel protein, RD3, exhibiting subnuclear localization is associated with retinal degeneration. Am J Hum Genet. 2006;79:1059–1070. doi: 10.1086/510021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Hearn T, Renforth GL, Spalluto C, Hanley NA, Piper K, Brickwood S, White C, Connolly V, Taylor JF, Russell-Eggitt I, Bonneau D, Walker M, Wilson DI. Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alstrom syndrome. Nat Genet. 2002;31:79–83. doi: 10.1038/ng874. [DOI] [PubMed] [Google Scholar]
- Henderson RH, Williamson KA, Kennedy JS, Webster AR, Holder GE, Robson AG, FitzPatrick DR, van Heyningen V, Moore AT. A rare de novo nonsense mutation in OTX2 causes early onset retinal dystrophy and pituitary dysfunction. Mol Vis. 2009;15:2442–2447. [PMC free article] [PubMed] [Google Scholar]
- Janecke AR, Thompson DA, Utermann G, Becker C, Hubner CA, Schmid E, McHenry CL, Nair AR, Ruschendorf F, Heckenlively J, Wissinger B, Nurnberg P, Gal A. Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat Genet. 2004;36:850–854. doi: 10.1038/ng1394. [DOI] [PubMed] [Google Scholar]
- Johnson S, Michaelides M, Aligianis IA, Ainsworth JR, Mollon JD, Maher ER, Moore AT, Hunt DM. Achromatopsia caused by novel mutations in both CNGA3 and CNGB3. J Med Genet. 2004;41:e20. doi: 10.1136/jmg.2003.011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J, Bonneau D, Frezal J, Munnich A, Dufier JL. Clinical and genetic heterogeneity in retinitis pigmentosa. Hum Genet. 1990;85:635–642. doi: 10.1007/BF00193589. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- Khanna H, Davis EE, Murga-Zamalloa CA, Estrada-Cuzcano A, Lopez I, den Hollander AI, Zonneveld MN, Othman MI, Waseem N, Chakarova CF, Maubaret C, Diaz-Font A, MacDonald I, Muzny DM, Wheeler DA, Morgan M, Lewis LR, Logan CV, Tan PL, Beer MA, Inglehearn CF, Lewis RA, Jacobson SG, Bergmann C, Beales PL, Attie-Bitach T, Johnson CA, Otto EA, Bhattacharya SS, Hildebrandt F, Gibbs RA, Koenekoop RK, Swaroop A, Katsanis N. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korman SH. Inborn errors of isoleucine degradation: a review. Mol Genet Metab. 2006;89:289–299. doi: 10.1016/j.ymgme.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Leber T. Uber retinitis pigmentosa und angeborene amaurose. Albrecht Von Graefes Arch Ophthalmol. 1869;15:1–25. [Google Scholar]
- Leitch CC, Zaghloul NA, Davis EE, Stoetzel C, Diaz-Font A, Rix S, Alfadhel M, Lewis RA, Eyaid W, Banin E, Dollfus H, Beales PL, Badano JL, Katsanis N. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- Li G, Vega R, Nelms K, Gekakis N, Goodnow C, McNamara P, Wu H, Hong NA, Glynne R. A role for Alstrom syndrome protein, alms1, in kidney ciliogenesis and cellular quiescence. PLoS Genet. 2007;3:e8. doi: 10.1371/journal.pgen.0030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang H, Peng J, Gibbs RA, Lewis RA, Lupski JR, Mardon G, Chen R. Mutation survey of known LCA genes and loci in the Saudi Arabian population. Invest Ophthalmol Vis Sci. 2009;50:1336–1343. doi: 10.1167/iovs.08-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Udovichenko IP, Brown SD, Steel KP, Williams DS. Myosin VIIa participates in opsin transport through the photoreceptor cilium. J Neurosci. 1999;19:6267–6274. doi: 10.1523/JNEUROSCI.19-15-06267.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes VS, Gibbs D, Libby RT, Aleman TS, Welch DL, Lillo C, Jacobson SG, Radu RA, Steel KP, Williams DS. The Usher 1B protein, MYO7A, is required for normal localization and function of the visual retinoid cycle enzyme, RPE65. Hum Mol Genet. 2011;20:2560–2570. doi: 10.1093/hmg/ddr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlhens F, Bareil C, Griffoin JM, Zrenner E, Amalric P, Eliaou C, Liu SY, Harris E, Redmond TM, Arnaud B, Claustres M, Hamel CP. Mutations in RPE65 cause Leber’s congenital amaurosis. Nat Genet. 1997;17:139–141. doi: 10.1038/ng1097-139. [DOI] [PubMed] [Google Scholar]
- Marshall JD, Hinman EG, Collin GB, Beck S, Cerqueira R, Maffei P, Milan G, Zhang W, Wilson DI, Hearn T, Tavares P, Vettor R, Veronese C, Martin M, So WV, Nishina PM, Naggert JK. Spectrum of ALMS1 variants and evaluation of genotype-phenotype correlations in Alstrom syndrome. Hum Mutat. 2007;28:1114–1123. doi: 10.1002/humu.20577. [DOI] [PubMed] [Google Scholar]
- Mataftsi A, Schorderet DF, Chachoua L, Boussalah M, Nouri MT, Barthelmes D, Borruat FX, Munier FL. Novel TULP1 mutation causing leber congenital amaurosis or early onset retinal degeneration. Invest Ophthalmol Vis Sci. 2007;48:5160–5167. doi: 10.1167/iovs.06-1013. [DOI] [PubMed] [Google Scholar]
- Mehalow AK, Kameya S, Smith RS, Hawes NL, Denegre JM, Young JA, Bechtold L, Haider NB, Tepass U, Heckenlively JR, Chang B, Naggert JK, Nishina PM. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum Mol Genet. 2003;12:2179–2189. doi: 10.1093/hmg/ddg232. [DOI] [PubMed] [Google Scholar]
- Murga-Zamalloa CA, Swaroop A, Khanna H. RPGR-containing protein complexes in syndromic and non-syndromic retinal degeneration due to ciliary dysfunction. J Genet. 2009;88:399–407. doi: 10.1007/s12041-009-0061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- O’Byrne SM, Wongsiriroj N, Libien J, Vogel S, Goldberg IJ, Baehr W, Palczewski K, Blaner WS. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT) J Biol Chem. 2005;280:35647–35657. doi: 10.1074/jbc.M507924200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O’Toole JF, Helou J, Attanasio M, Utsch B, Sayer JA, Lillo C, Jimeno D, Coucke P, De Paepe A, Reinhardt R, Klages S, Tsuda M, Kawakami I, Kusakabe T, Omran H, Imm A, Tippens M, Raymond PA, Hill J, Beales P, He S, Kispert A, Margolis B, Williams DS, Swaroop A, Hildebrandt F. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- Perrault I, Hanein S, Gerber S, Barbet F, Ducroq D, Dollfus H, Hamel C, Dufier JL, Munnich A, Kaplan J, Rozet JM. Retinal dehydrogenase 12 (RDH12) mutations in leber congenital amaurosis. Am J Hum Genet. 2004;75:639–646. doi: 10.1086/424889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault I, Rozet JM, Calvas P, Gerber S, Camuzat A, Dollfus H, Chatelin S, Souied E, Ghazi I, Leowski C, Bonnemaison M, Le Paslier D, Frezal J, Dufier JL, Pittler S, Munnich A, Kaplan J. Retinal-specific guanylate cyclase gene mutations in Leber’s congenital amaurosis. Nat Genet. 1996;14:461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- Ramamurthy V, Niemi GA, Reh TA, Hurley JB. Leber congenital amaurosis linked to AIPL1: a mouse model reveals destabilization of cGMP phosphodiesterase. Proc Natl Acad Sci USA. 2004;101:13897–13902. doi: 10.1073/pnas.0404197101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma JX, Crouch RK, Pfeifer K. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat Genet. 1998;20:344–351. doi: 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sayer JA, Otto EA, O’Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, Utsch B, Khanna H, Liu Y, Drummond I, Kawakami I, Kusakabe T, Tsuda M, Ma L, Lee H, Larson RG, Allen SJ, Wilkinson CJ, Nigg EA, Shou C, Lillo C, Williams DS, Hoppe B, Kemper MJ, Neuhaus T, Parisi MA, Glass IA, Petry M, Kispert A, Gloy J, Ganner A, Walz G, Zhu X, Goldman D, Nurnberg P, Swaroop A, Leroux MR, Hildebrandt F. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- Senechal A, Humbert G, Surget MO, Bazalgette C, Arnaud B, Arndt C, Laurent E, Brabet P, Hamel CP. Screening genes of the retinoid metabolism: novel LRAT mutation in leber congenital amaurosis. Am J Ophthalmol. 2006;142:702–704. doi: 10.1016/j.ajo.2006.04.057. [DOI] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Sohocki MM, Bowne SJ, Sullivan LS, Blackshaw S, Cepko CL, Payne AM, Bhattacharya SS, Khaliq S, Qasim Mehdi S, Birch DG, Harrison WR, Elder FF, Heckenlively JR, Daiger SP. Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat Genet. 2000;24:79–83. doi: 10.1038/71732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM. Leber congenital amaurosis - a model for efficient genetic testing of heterogeneous disorders: LXIV Edward Jackson memorial lecture. Am J Ophthalmol. 2007;144:791–811. doi: 10.1016/j.ajo.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Stone EM, Cideciyan AV, Aleman TS, Scheetz TE, Sumaroka A, Ehlinger MA, Schwartz SB, Fishman GA, Traboulsi EI, Lam BL, Fulton AB, Mullins RF, Sheffield VC, Jacobson SG. Variations in NPHP5 in patients with nonsyndromic leber congenital amaurosis and Senior-Loken syndrome. Arch Ophthalmol. 2011;129:81–87. doi: 10.1001/archophthalmol.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, Sieving PA, Zack DJ. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Di Rocco C, Lajeunie E, Valeri S, Velardi F, Battaglia PA. Jackson-Weiss syndrome: identification of two novel FGFR2 missense mutations shared with Crouzon and Pfeiffer craniosynostotic disorders. Hum Genet. 1997;101:47–50. doi: 10.1007/s004390050584. [DOI] [PubMed] [Google Scholar]
- Thompson DA, Janecke AR, Lange J, Feathers KL, Hubner CA, McHenry CL, Stockton DW, Rammesmayer G, Lupski JR, Antinolo G, Ayuso C, Baiget M, Gouras P, Heckenlively JR, den Hollander A, Jacobson SG, Lewis RA, Sieving PA, Wissinger B, Yzer S, Zrenner E, Utermann G, Gal A. Retinal degeneration associated with RDH12 mutations results from decreased 11-cis retinal synthesis due to disruption of the visual cycle. Hum Mol Genet. 2005;14:3865–3875. doi: 10.1093/hmg/ddi411. [DOI] [PubMed] [Google Scholar]
- Valente EM, Silhavy JL, Brancati F, Barrano G, Krishnaswami SR, Castori M, Lancaster MA, Boltshauser E, Boccone L, Al-Gazali L, Fazzi E, Signorini S, Louie CM, Bellacchio E, Bertini E, Dallapiccola B, Gleeson JG. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- Wang H, den Hollander AI, Moayedi Y, Abulimiti A, Li Y, Collin RW, Hoyng CB, Lopez I, Abboud EB, Al-Rajhi AA, Bray M, Lewis RA, Lupski JR, Mardon G, Koenekoop RK, Chen R. Mutations in SPATA7 cause Leber congenital amaurosis and juvenile retinitis pigmentosa. Am J Hum Genet. 2009;84:380–387. doi: 10.1016/j.ajhg.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissinger B, Gamer D, Jagle H, Giorda R, Marx T, Mayer S, Tippmann S, Broghammer M, Jurklies B, Rosenberg T, Jacobson SG, Sener EC, Tatlipinar S, Hoyng CB, Castellan C, Bitoun P, Andreasson S, Rudolph G, Kellner U, Lorenz B, Wolff G, Verellen-Dumoulin C, Schwartz M, Cremers FP, Apfelstedt-Sylla E, Zrenner E, Salati R, Sharpe LT, Kohl S. CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet. 2001;69:722–737. doi: 10.1086/323613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Pauer GJ, Marmorstein AD, Crabb JW, Hagstrom SA. Tubby-like protein 1 (TULP1) interacts with F-actin in photoreceptor cells. Invest Ophthalmol Vis Sci. 2005;46:4754–4761. doi: 10.1167/iovs.05-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.