Abstract
The rapid expansion of genomics methods has enabled developmental biologists to address fundamental questions of developmental gene regulation on a genome-wide scale. These efforts have demonstrated that transcription of developmental control genes by RNA polymerase II (Pol II) is commonly regulated at the transition to productive elongation, resulting in the promoter-proximal accumulation of transcriptionally engaged but paused Pol II prior to gene induction. Here we review the mechanisms and possible functions of Pol II pausing and their implications for development.
Keywords: Pol II pausing, Transcription elongation, Developmental control genes
Introduction
During metazoan development, pluripotent cells gradually acquire cell type-specific gene expression programs to give rise to a plethora of different lineages. Such programs are set up by sequence-specific transcription factors and co-regulators, traditionally thought to mediate recruitment of the transcription machinery to gene promoters. However, in recent years it has become clear that at a significant fraction of genes RNA polymerase II (Pol II) is also regulated after its recruitment to the promoter, at the level of transcription elongation. At these genes, Pol II initiates transcription and transitions to early elongation, before it pauses ∼30-50 bp downstream of the transcription start site (TSS). This promoter-proximal pausing is mediated by negative elongation factors, and positive transcription elongation factors are required to overcome this roadblock to productive transcription.
Although the phenomenon of Pol II pausing was first discovered several decades ago (see Adelman and Lis, 2012), it was only through genome-wide studies in mammalian embryonic stem cells (ESCs) and Drosophila that it became evident that Pol II pausing is not restricted to a few specialized genes but is a widespread phenomenon, affecting 10-40% of all genes (Guenther et al., 2007; Muse et al., 2007; Zeitlinger et al., 2007). Furthermore, these genome-wide studies revealed that, at least in Drosophila, paused Pol II is preferentially found at developmental control genes, including transcription factors and components of signal transduction cascades. This suggests that Pol II pausing has an important role in controlling gene expression during development.
In this review we focus on recent progress in understanding the relationship between Pol II pausing and development. This has been most extensively studied in Drosophila, and most of the data we present stem from this model organism. Yet, because of the broad conservation of the factors involved in this process, principles learned from fly development should also be applicable to all higher eukaryotes.
Molecular mechanisms of Pol II pausing
Pol II pausing in the transcription cycle
Pol II pausing is a late step during the initiation stage of transcription. It begins with the recruitment of Pol II and basal transcription factors to an accessible promoter to form the preinitiation complex (PIC) [for a recent review see Luse (Luse, 2013)]. After Pol II has initiated transcription and the nascent RNA reaches a length of 9-10 nt, Pol II escapes the promoter and the C-terminal heptapeptide repeat domain (CTD) of its largest subunit is phosphorylated on serine 5 residues. Once the transcript is 20-50 nt long, Pol II may pause and only enter into productive elongation after pause release (Zhou et al., 2012). Pol II pausing occurs both at highly transcribed genes as well as at those that are transcribed at very low levels and are poised for activation (e.g. heat shock genes).
Experimental evidence for Pol II pausing
Paused Pol II can be detected by several techniques. First, chromatin immunoprecipitation sequencing (ChIP-seq) studies find high levels of cross-linked Pol II ∼30-50 nt downstream of the TSS at many genes, consistent with the model that Pol II often has a long residence time at this position. Second, permanganate footprinting assays detect an open transcription bubble at the pausing position (Gilmour and Fan, 2009). Third, sequencing of short capped RNA species shows that short transcripts of 30-50 bp are produced from paused genes (Nechaev et al., 2010). Finally, global nuclear run-on sequencing assays (GRO-seq and PRO-seq) demonstrate that a large fraction of paused Pol II is still elongation competent (Core et al., 2008; Kwak et al., 2013). Since these different assays produce, overall, very similar results, the available evidence suggests that promoter-proximal paused Pol II is the most frequent form of non-elongating Pol II, and that the preinitiation or transcriptionally arrested forms are rare by comparison.
Establishment of Pol II pausing
Two key regulators of Pol II promoter-proximal pausing are the DRB sensitivity-inducing factor (DSIF) and the Negative elongation factor (NELF) complexes (for details, see Yamaguchi et al., 2013). Both associate with Pol II following promoter escape and transiently halt elongation, possibly through interaction with the newly emerging transcript. Moreover, a third Poll II-associated factor, Gdown1 (Polr2m), has recently been found to make Pol II pausing more stable in vitro (Guo and Price, 2013).
Release of Pol II pausing by P-TEFb
Pausing is overcome through P-TEFb (positive transcription elongation factor b), which comprises the cyclin-dependent kinase Cdk9 and Cyclin T. This complex phosphorylates NELF, DSIF and the Pol II CTD at serine 2 residues, causing NELF to dissociate while converting DSIF to a positive elongation factor (for details, see Zhou et al., 2012; Yamaguchi et al., 2013).
Inactive P-TEFb is sequestered in the 7SK small nuclear ribonucleoprotein complex, which is often localized to promoters (Ji et al., 2013). The release of P-TEFb from this complex and its activation can be stimulated by transcription factors through multiple mechanisms, either by direct contact (Barboric et al., 2001; Rahl et al., 2010) or indirectly via Mediator (Takahashi et al., 2011) or histone acetylation (Guo and Price, 2013). Active P-TEFb associates with super elongation complexes (SECs) (Guo and Price, 2013).
Dual role of transcription factors
Transcription factors have different abilities to stimulate Pol II initiation, Pol II elongation or both (Yankulov et al., 1994; Blau et al., 1996; Brown et al., 1998). Although long known, this became more appreciated when Pol II pausing was shown to be a widespread phenomenon. Tif1γ (Trim33), for example, appears to help overcome pausing at erythroid genes in zebrafish by recruiting P-TEFb and the transcription elongation factor FACT (Bai et al., 2010). Similarly, at highly transcribed genes in ESCs, the transcription factor c-Myc predominantly stimulates Pol II elongation through P-TEFb (Rahl et al., 2010).
Regulation of pause release by signaling pathways
Signaling pathways can affect Pol II pausing either via regulation of transcription factor activity or by more direct effects on the pausing machinery. For example, mitogen-activated protein (MAP) kinases may bind directly to target genes and phosphorylate histone H3 serine 10 (H3S10) at active promoters (Pokholok et al., 2006), which in turn could directly or indirectly promote P-TEFb recruitment (Ivaldi et al., 2007; Zippo et al., 2009). Moreover, PI3K/Akt signaling has been found to dissociate P-TEFb from the 7SK complex, freeing it for transcription stimulation (Contreras et al., 2007).
Pausing factors in evolution
Bona fide paused Pol II is absent in budding yeast, C. elegans and plants, and this correlates with a lack of NELF orthologs in these species. It is interesting to speculate that Pol II pausing might be particularly important for metazoans with complex development that require high transcriptional precision and synchrony (see below).
Intrinsic promoter differences
Pol II pausing is not found at all genes to the same extent. In the early Drosophila embryo, the most highly paused genes are developmental control genes (Zeitlinger et al., 2007), and an overall theme has been that paused genes tend to be highly regulated in response to extracellular stimuli.
This disposition for Pol II pausing among highly regulated genes is most likely due to intrinsic properties of the core promoter, as demonstrated by promoter-swapping experiments (Lagha et al., 2013) and the strong relationship between Pol II pausing and promoter sequence throughout Drosophila development (Gaertner et al., 2012; Chen et al., 2013). This suggests that transcription initiation varies between different promoter types. At promoters prone to pausing, the residence time of Pol II at the pause site may be intrinsically longer, and the transition to productive elongation might be more frequently subject to regulation.
Promoters of highly paused genes
In Drosophila, promoters with highly paused Pol II show very clear characteristics. They tend to undergo focused initiation within a very narrow window, often where an initiator (Inr) motif is found. These promoters are enriched for promoter elements such as the downstream promoter element (DPE), motif ten element (MTE) or the pause button (PB), which are located close to the pause site (Hendrix et al., 2008; Rach et al., 2011; Lenhard et al., 2012). They may regulate Pol II pausing through specific factors or, more indirectly, through their high CG content (Hendrix et al., 2008; Nechaev et al., 2010), where a G nucleotide may stabilize the pause position (Zhang et al., 2012).
Another characteristic of highly paused promoters in Drosophila is a strong tendency to be occupied by a nucleosome in the absence of Pol II (Gilchrist et al., 2010). This tendency can be predicted from DNA sequence and thus is also an intrinsic property of these promoters. Paused promoters are frequently bound by GAGA factor, which may antagonize the promoter nucleosome and help recruit NELF and paused Pol II (Adkins et al., 2006; Lee et al., 2008; Li et al., 2013).
Promoters with dispersed initiation
At promoters with dispersed transcription, Pol II initiates from a broadly distributed range of start sites within ∼50-100 bp. Although these promoters are typically associated with so-called housekeeping genes (which are broadly expressed across tissues, regulating such as metabolism and the cytoskeleton), they are also found at highly regulated developmental genes in mammals (Lenhard et al., 2012). The promoters are strongly nucleosome depleted and have well-positioned downstream nucleosomes with high levels of histone modifications and H2A.Z (Rach et al., 2011; Gaertner et al., 2012; Chen et al., 2013). In Drosophila, these promoters are enriched for core promoter elements such as the DNA replication-related element (DRE), and the Ohler1, Ohler6 and Ohler7 promoter motifs (Ohler et al., 2002), whereas in mammals they mostly have CpG islands.
Pol II pausing at these promoters ranges from very low to moderately high (Zeitlinger et al., 2007; Rach et al., 2011; Kwak et al., 2013; Li and Gilmour, 2013) and often occurs further downstream (at +50-80 nt) compared with highly paused genes (Kwak et al., 2013). The mechanisms leading to pausing at this promoter type are not well understood but it is possible that the first downstream nucleosome (the ‘+1 nucleosome’) plays a crucial role (Kwak et al., 2013; Li and Gilmour, 2013).
TATA promoters
A third class of promoters in Drosophila and mammals is characterized by focused initiation, the presence of a TATA box and absence of pausing motifs. These promoters are typically associated with adult tissue-specific genes (Lenhard et al., 2012). In Drosophila, TATA promoters are also enriched among the earliest expressed developmental genes in the embryo (Chen et al., 2013) but are under-represented during the main patterning stages of embryonic development. Expression of tissue-specific TATA genes only increases again towards the end of embryogenesis (Gaertner et al., 2012).
Drosophila TATA genes are associated with a low degree of pausing and tend to be induced without prior poised Pol II (Gaertner et al., 2012; Chen et al., 2013). This suggests that, under some conditions, TATA prevents Pol II pausing and hence that Pol II initiation rather than elongation becomes rate limiting. Indeed, it has been shown in mammalian cells that TATA promotes P-TEFb activity, leading to more efficient elongation rates in vitro and in vivo (Amir-Zilberstein et al., 2007; Montanuy et al., 2008), suggesting that the properties of TATA promoters are conserved across metazoans (Dikstein, 2011).
Why regulate developmental gene expression by pausing?
Induction speed
Heat shock genes are induced rapidly to protect cells under high temperatures, and thus it is often assumed that Pol II primes heat shock genes for rapid induction. Recent evidence, however, suggests that paused Pol II is not absolutely required for rapid gene induction since genes lacking Pol II occupancy can be induced just as quickly as, and perhaps to even higher levels than, paused genes (Lin et al., 2011; Gilchrist et al., 2012; Chen et al., 2013). Furthermore, a genome-wide analysis of transcription rates by GRO-seq in mammalian cells revealed that induction of paused genes in response to TNF-alpha treatment is associated with a slight delay (Danko et al., 2013). Therefore, speedy induction is unlikely to be the primary purpose for paused Pol II.
The permissive state and its regulation
Pol II pausing is found broadly across tissues in the Drosophila embryo, frequently even in tissues in which the gene will never be induced (Levine, 2011; Gaertner et al., 2012). Here, the promoter lacks a nucleosome (i.e. is in an ‘open state’), but Pol II is paused and the levels of transcription are low. The simplest explanation is that such ‘poised Pol II’ is a permissive state that renders genes competent to respond to extracellular signals (Lis, 1998; Adelman and Lis, 2012).
This might be important for the interpretation of morphogen gradients, where all cells in a field must be prepared to respond to an extracellular signal in a concentration-dependent manner, even though many cells will not receive the signal. This is illustrated by the mesodermal transcription factor Snail, the expression of which on the ventral side of Drosophila embryos requires high levels of the Dorsal morphogen. Although cells on the dorsal side do not receive sufficient Dorsal to express snail, the snail gene is paused in these cells (Zeitlinger et al., 2007).
If poised Pol II constitutes a permissive state, one would expect the transition of genes into this state to be regulated during development. Thus, a promoter that harbors poised Pol II in one context may be in a ‘closed state’ in another, unresponsive to extracellular signals. Indeed, during Drosophila embryogenesis, many promoters are initially in a closed state with a strong promoter nucleosome and gradually open over time to acquire high levels of paused Pol II (Gaertner et al., 2012). Data so far indicate that the permissive state is established broadly across the entire embryo in a stage-specific fashion, although individual genes are expressed only in specific tissues. This suggests that the ‘opening’ of promoters is indeed a permissive step and prepares genes for activation by tissue-specific transcription factors (Gaertner et al., 2012).
Antagonistic repression
When cells do not receive an activation signal, paused genes are often kept silent by transcriptional repressors. Genome-wide binding of the zinc finger repressor Snail, for instance, was found to correlate with paused Pol II (Zeitlinger et al., 2007). This suggests that transcriptional repressors can block the release of Pol II (Bothma et al., 2011). To do so, repressors may act locally at enhancers to prevent adjacent activators from stimulating Pol II pause release at the promoter (also called ‘quenching’ or short-range repression).
Another mechanism that might keep the permissive state of paused Pol II in check is repression by Polycomb group (PcG) proteins. In both Drosophila and mammals, PcG proteins are frequently found at developmental control genes and prevent their inappropriate expression in a time- and tissue-specific fashion. Although the mechanisms of Pol II repression by PcG proteins may not be specific to paused Pol II and the biochemical characteristics of PcG-repressed paused genes remain to be clarified, it appears that the combination of both mechanisms is a powerful way to regulate transcription during development. In Drosophila, genes with both paused Pol II and PcG repression (the so-called ‘balanced state’) have particularly dynamic expression patterns during development (Schwartz et al., 2010; Gaertner et al., 2012). In mammals, PcG repression frequently coincides with histone H3 lysine 4 trimethylation (H3K4me3) (Bernstein et al., 2006), a marker for Pol II initiation. These ‘bivalent genes’ are also dynamically regulated during development, supporting the idea that a dynamic equilibrium between activation and repression keeps genes in a plastic, inducible state (Voigt et al., 2013).
Synchronicity of gene induction
Another reason why Pol II pausing is common at developmental genes might relate to the observation that paused genes have been shown to be induced more synchronously among a field of cells in the early Drosophila embryo (Boettiger and Levine, 2009). By contrast, genes without Pol II pausing appear to be induced more stochastically, creating a ‘salt-and-pepper’ pattern during gene activation. This difference in synchronicity depends on the core promoter (Lagha et al., 2013), indicating that differences in the transcription dynamics rather than regulation by transcription factors produce this effect.
In principle, there are two explanations for the more synchronous activation of paused genes. First, paused Pol II might reduce the stochasticity inherent to the stepwise assembly of the transcription machinery into a PIC and thus ensure that transcription begins synchronously in response to an extracellular signal. Second, Pol II pausing might serve to space out subsequent transcripts and thus attenuate transcription bursts. This might be particularly important when cells respond to low levels of an activating signal, as stochastic transcription bursts would lead to very high variation in the transcriptional response. Furthermore, longer time periods between subsequent rounds of transcription might give the transcription machinery more time for post-transcriptional processing and prevent Pol II crowding (Marbach-Bar et al., 2013). This could be important for developmental control genes, which tend to be long and frequently regulated by alternative splicing.
Whether a gene is activated in a more synchronous or stochastic fashion has important consequences for development. For example, the snail gene, which is essential for the coordinated invagination of cells during gastrulation, is expressed very synchronously. However, under the control of a promoter from a less paused gene, snail expression is more stochastic, resulting in variable invagination defects (Lagha et al., 2013). In the early Drosophila syncytium, by contrast, transcription variability may be averaged over space and time (Little et al., 2013), which could explain why Pol II pausing is rare at this stage (Chen et al., 2013). Taken together, Pol II pausing might be particularly important for pattern formation during development, when fields of cells have to respond in a precise and synchronous fashion to extracellular signals.
Perspectives
Although Pol II pausing has emerged as a key regulatory step of the transcription cycle, many mechanistic aspects of its regulation are still unclear and its consequences for development remain to be rigorously tested in vivo. How does Pol II pausing affect transcription dynamics at the different classes of promoter? How is paused Pol II recruited de novo to previously closed promoters during development? How does Pol II pausing vary across evolution and do other mechanisms exist in organisms that do not have NELF homologs? Future studies will undoubtedly reveal deeper insights into the role of paused Pol II during development, and perhaps uncover further surprises.
Acknowledgments
We apologize for not being able to cite many original papers, which we replaced by reviews owing to space limitations.
Footnotes
Competing interests
The authors declare no competing financial interests.
Funding
Work in the authors’ laboratories is supported by the Stowers Institute for Medical Research and the National Institutes of Health.
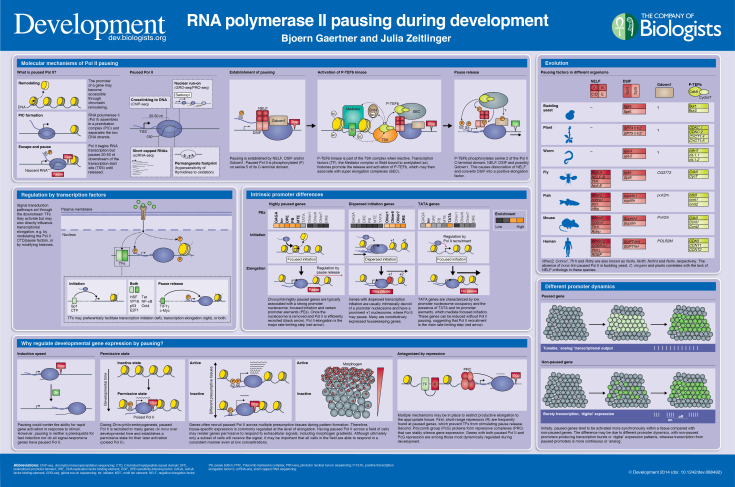
Development at a Glance
A high-resolution version of the poster is available for downloading in the online version of this article at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.088492/-/DC1
References
- Adelman K., Lis J. T. (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins N. L., Hagerman T. A., Georgel P. (2006). GAGA protein: a multi-faceted transcription factor. Biochem. Cell Biol. 84, 559–567 [DOI] [PubMed] [Google Scholar]
- Amir-Zilberstein L., Ainbinder E., Toube L., Yamaguchi Y., Handa H., Dikstein R. (2007). Differential regulation of NF-kappaB by elongation factors is determined by core promoter type. Mol. Cell. Biol. 27, 5246–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Kim J., Yang Z., Jurynec M. J., Akie T. E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A., et al. (2010). TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 142, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M., Nissen R. M., Kanazawa S., Jabrane-Ferrat N., Peterlin B. M. (2001). NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8, 327–337 [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- Blau J., Xiao H., McCracken S., O’Hare P., Greenblatt J., Bentley D. (1996). Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16, 2044–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger A. N., Levine M. (2009). Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325, 471–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma J. P., Magliocco J., Levine M. (2011) The snail repressor inhibits release, not elongation, of paused Pol II in the Drosophila embryo. Curr. Biol. 21, 1571–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. A., Weirich C. S., Newton E. M., Kingston R. E. (1998). Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 17, 3146–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Johnston J., Shao W., Meier S., Staber C., Zeitlinger J. (2013). A global change in RNA polymerase II pausing during the Drosophila midblastula transition. Elife 2, e00861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras X., Barboric M., Lenasi T., Peterlin B. M. (2007). HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3, e146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L. J., Waterfall J. J., Lis J. T. (2008). Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko C. G., Hah N., Luo X., Martins A. L., Core L., Lis J. T., Siepel A., Kraus W. L. (2013). Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell 50, 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikstein R. (2011). The unexpected traits associated with core promoter elements. Transcription 2, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B., Johnston J., Chen K., Wallaschek N., Paulson A., Garruss A. S., Gaudenz K., De Kumar B., Krumlauf R., Zeitlinger J. (2012). Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell Rep. 2, 1670–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist D. A., Dos Santos G., Fargo D. C., Xie B., Gao Y., Li L., Adelman K. (2010). Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist D. A., Fromm G., dos Santos G., Pham L. N., McDaniel I. E., Burkholder A., Fargo D. C., Adelman K. (2012). Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 26, 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Fan R. (2009). Detecting transcriptionally engaged RNA polymerase in eukaryotic cells with permanganate genomic footprinting. Methods 48, 368–374 [DOI] [PubMed] [Google Scholar]
- Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. (2007). A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Price D. H. (2013). RNA polymerase ii transcription elongation control. Chem. Rev. 113, 8583–8603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix D. A., Hong J. W., Zeitlinger J., Rokhsar D. S., Levine M. S. (2008). Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 105, 7762–7767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivaldi M. S., Karam C. S., Corces V. G. (2007). Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev. 21, 2818–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Zhou Y., Pandit S., Huang J., Li H., Lin C. Y., Xiao R., Burge C. B., Fu X. D. (2013). SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153, 855–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H., Fuda N. J., Core L. J., Lis J. T. (2013). Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M., Bothma J. P., Esposito E., Ng S., Stefanik L., Tsui C., Johnston J., Chen K., Gilmour D. S., Zeitlinger J., et al. (2013). Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153, 976–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Li X., Hechmer A., Eisen M., Biggin M. D., Venters B. J., Jiang C., Li J., Pugh B. F., Gilmour D. S. (2008). NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 28, 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B., Sandelin A., Carninci P. (2012). Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 13, 233–245 [DOI] [PubMed] [Google Scholar]
- Levine M. (2011). Paused RNA polymerase II as a developmental checkpoint. Cell 145, 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Gilmour D. S. (2013). Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J. 32, 1829–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu Y., Rhee H. S., Ghosh S. K., Bai L., Pugh B. F., Gilmour D. S. (2013). Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol. Cell 50, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Garrett A. S., De Kumar B., Smith E. R., Gogol M., Seidel C., Krumlauf R., Shilatifard A. (2011). Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 25, 1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. (1998). Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 63, 347–356 [DOI] [PubMed] [Google Scholar]
- Little S. C., Tikhonov M., Gregor T. (2013). Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell 154, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luse D. S. (2013). Promoter clearance by RNA polymerase II. Biochim. Biophys. Acta 1829, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach-Bar N., Ben-Noon A., Ashkenazi S., Tamarkin-Ben Harush A., Avnit-Sagi T., Walker M. D., Dikstein R. (2013). Disparity between microRNA levels and promoter strength is associated with initiation rate and Pol II pausing. Nat. Commun. 4, 2118 [DOI] [PubMed] [Google Scholar]
- Montanuy I., Torremocha R., Hernández-Munain C., Suñé C. (2008). Promoter influences transcription elongation: TATA-box element mediates the assembly of processive transcription complexes responsive to cyclin-dependent kinase 9. J. Biol. Chem. 283, 7368–7378 [DOI] [PubMed] [Google Scholar]
- Muse G. W., Gilchrist D. A., Nechaev S., Shah R., Parker J. S., Grissom S. F., Zeitlinger J., Adelman K. (2007). RNA polymerase is poised for activation across the genome. Nat. Genet. 39, 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S., Fargo D. C., dos Santos G., Liu L., Gao Y., Adelman K. (2010). Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327, 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U., Liao G. C., Niemann H., Rubin G. M. (2002). Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3, RESEARCH0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D. K., Zeitlinger J., Hannett N. M., Reynolds D. B., Young R. A. (2006). Activated signal transduction kinases frequently occupy target genes. Science 313, 533–536 [DOI] [PubMed] [Google Scholar]
- Rach E. A., Winter D. R., Benjamin A. M., Corcoran D. L., Ni T., Zhu J., Ohler U. (2011). Transcription initiation patterns indicate divergent strategies for gene regulation at the chromatin level. PLoS Genet. 7, e1001274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl P. B., Lin C. Y., Seila A. C., Flynn R. A., McCuine S., Burge C. B., Sharp P. A., Young R. A. (2010). c-Myc regulates transcriptional pause release. Cell 141, 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Kahn T. G., Stenberg P., Ohno K., Bourgon R., Pirrotta V. (2010). Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 6, e1000805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Parmely T. J., Sato S., Tomomori-Sato C., Banks C. A., Kong S. E., Szutorisz H., Swanson S. K., Martin-Brown S., Washburn M. P., et al. (2011). Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P., Tee W. W., Reinberg D. (2013). A double take on bivalent promoters. Genes Dev. 27, 1318–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Shibata H., Handa H. (2013). Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta 1829, 98–104 [DOI] [PubMed] [Google Scholar]
- Yankulov K., Blau J., Purton T., Roberts S., Bentley D. L. (1994). Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell 77, 749–759 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Stark A., Kellis M., Hong J. W., Nechaev S., Adelman K., Levine M., Young R. A. (2007). RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39, 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Feng Y., Chatterjee S., Tuske S., Ho M. X., Arnold E., Ebright R. H. (2012). Structural basis of transcription initiation. Science 338, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Li T., Price D. H. (2012). RNA polymerase II elongation control. Annu. Rev. Biochem. 81, 119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A., Serafini R., Rocchigiani M., Pennacchini S., Krepelova A., Oliviero S. (2009). Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138, 1122–1136 [DOI] [PubMed] [Google Scholar]