Abstract
Telomere is a nucleoprotein structure at the ends of chromosomes that helps to protect the ends of chromosomes from being fused with other chromosomes. Knockout of histone methyltransferases Suv39h1 and Suv39h2 increases the telomere length in murine cells, whereas downregulation of SUV39H1 and SUV39H2 genes decreases the telomere length in human cells, suggesting that telomere biology is different among mammalian species. However, epigenetic regulation of the telomere has not been studied in mammals other than the human and mouse. In the present study, the effect of knockdown of SUV39H1 and SUV39H2 genes on telomere length was examined in porcine embryonic stem-like cells (pESLCs) and porcine embryonic fibroblasts (PEFs). The telomeres in SUV39H1 and SUV39H2 knockdown (SUV39KD) pESLCs (37.1 ± 0.9 kb) were longer (P<0.05) compared with those of the control (33.0 ± 0.7 kb). Similarly, SUV39KD PEFs had longer telomeres (22.1 ± 0.4 kb; P<0.05) compared with the control (17.8 ± 1.1 kb). Telomerase activities were not different between SUV39KD pESLCs (10.4 ± 1.7) and the control (10.1 ± 1.7) or between SUV39KD PEFs (1.0 ± 0.3) and the control (1.0 ± 0.4), suggesting that telomerase activities did not contribute to the telomere elongation in SUV39KD pESLCs and SUV39KD PEFs. Relative levels of trimethylation of histone H3 lysine 9 and expressions of DNMT1, DNMT3A and DNMT3B were decreased in SUV39KD cells, suggesting that telomere lengthening in SUV39KD pESLCs and SUV39KD PEFs might be not only related to the loss of histone modification marks but also linked to the decrease in DNA methyltransferase in pigs.
Keywords: Embryonic stem-like cells, Fibroblasts, Knockdown, Pig, Telomere length
Telomere is a nucleoprotein structure at the ends of chromosomes that helps to protect the ends of chromosomes from being fused with others [1]. Although the telomere DNA sequence varies among species, all telomere DNA sequences are composed of large arrays of short guanine-rich sequences and do not contain genes [2, 3]. In vertebrates, the telomere sequence consists of TTAGGG repeats and is highly conserved [3]. The telomere is bound by a complex of six proteins called shelterin or telosome [1, 4]. Shelterin plays an important part in maintenance of telomere length and protection of the ends of chromosomes [4]. Telomere length differs among individuals and species [5]. Telomere length is maintained by telomerase, a reverse transcriptase that adds telomeric repeats in every cell division. Alternatively, telomere length can be maintained by homologous recombination between telomeric sequences called alternative lengthening of telomeres (ALT) [6]. However, most normal cells have low telomerase activities and a low incidence of ALT [5]. The telomere length in normal cells, therefore, is shortened after each cell division. When telomeres in a population of cells become abnormally short and are unable to protect their chromosomes, cells would enter an irreversible growth arrest state called replicative senescence [1, 7].
Recent studies have shown that telomere length is regulated by epigenetic modifications such as DNA methylation and histone tail modifications [8,9,10,11,12]. Although telomeric repeats cannot be methylated, the subtelomeric regions located adjacent to telomeres are enriched with repetitive DNA and highly methylated [9, 11]. Mouse embryonic stem (ES) cells deficient in DNA methyltransferase Dnmt1 or both Dnmt3a and 3b show decreased DNA methylation in the subtelomeric region, resulting in dramatically elongated telomeres [9]. Human cells knocked-out for both DNMT1 and DNMT3B, however, have overall shorter telomeres than parental cells, although hypomethylation was also observed [12]. SUV39h1 and SUV39h2 are histone H3 lysine 9 (H3K9)-specific histone methyltransferases (HMTs) in heterochromatic regions in mammals [13]. Cells lacking the Suv39h1 and Suv39h2 HMTs show reduction of trimethylation (3Me) of H3K9 in telomeres and abnormal telomere elongation [8]. Knockdown of SUV39H1 in human cells, however, causes telomere shortening [14].
These data suggest that telomere biology is rather different among mammalian species. However, no study on epigenetic regulation of telomeres has been performed in mammals other than the mouse and human. In the present study, we investigated the impact of SUV39H1 and SUV39H2 on telomere length in porcine ES-like cells (pESLCs) and porcine embryonic fibroblasts (PEFs).
Materials and Methods
Cell culture
Two pESLC lines were produced from in vitro fertilization blastocysts according to Haraguchi et al. [15]. The cells were subcultured at 37 C in a 5% CO2 atmosphere on mouse embryonic fibroblasts as feeder cells in ES medium consisting of KnockOut DMEM (KO-DMEM, Gibco, Grand Island, NY, USA) supplemented with 2 mM GlutaMAX, 1% MEM nonessential amino acids, 20% KnockOut Serum Replacement (KSR), 1% antibiotic-antimycotic liquid (all from Invitrogen, Carlsbad, CA, USA), 20 ng/ml porcine recombinant LIF (pLIF) (made in-house) and 0.1 mM ß-mercaptoethanol (Sigma, St. Louis, MO, USA), 6 µM CHIR99021 (Axon Medchem, Groningen, Netherlands); and 10 µM Y-27632 (Wako, Osaka, Japan). pESLCs were passaged using StemPro Accutase (Invitrogen) every two days. Three PEF cultures (passages 5–8) collected from Day 46 embryos (given by Transgenic Pig Research Unit, National Institute of Agrobiological Sciences, Tsukuba, Japan) were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Sigma) with 10% fetal bovine serum (FBS; Tissue Culture Biologicals, Tulare, CA, USA) and 1% antibiotic-antimycotic liquid at 37 C in a 5% CO2 atmosphere. PEFs were passaged using 0.05% trypsin-EDTA (Gibco) in phosphate-buffered saline (PBS; Nissui Pharmaceutical, Tokyo, Japan) when cells reached confluence.
RNA interference
Four sets of siRNA were designed by Cosmo Bio (Tokyo, Japan) for double knockdown of SUV39H1 and SUV39H2 genes (SUV39KD). The sequences of the two siRNA sets designed to target porcine SUV39H1 were 1) sense 5'-ggaaucagcuccaggaccutt-3' and antisense 5'-agguccuggagcugauucctt-3 and 2) sense 5'-gucgaguaccugugcgauutt-3 and antisense 5'-aaucgcacagguacucgactt-3'. The sequences of the two siRNA sets designed to target porcine SUV39H2 were 1) sense 5'-ggaauauguuggagaggua-3' and antisense 5'-uaccucuccaacauauucctt-3' and 2) sense 5-ggaagaaucacaaaggaautt-3 and antisense 5-auuccuuugugauucuucctt-3. One day prior to transfection, pESLCs and PEFs were passaged, and the cell concentration was adjusted to a density of 30–50% confluency. Before transfection, 80 nM of each siRNAs were mixed and incubated together with 0.4% (v/v) X-tremeGENE siRNA Transfection Reagent (Roche, Mannheim, Germany) in culture media without serum and antibiotics (transfection media) for 15–20 min at room temperature. The transfection media were then supplemented with either 10% FBS for PEFs or 20% KSR for pESLCs, and transfection was then carried out in culture media for 24 h or 48 h for pESLCs or PEFs, respectively. The transfections were performed for three successive passages, and the cells were collected after the third transfection. Non-siRNA-treated pESLCs and PEFs (at the same passage as SUV39KD pESLCs and SUV39KD PEFs, respectively) were also cultured in parallel with SUV39KD pESLCs and SUV39KD PEFs to serve as controls.
Extraction of total RNA and real-time PCR
Total RNA was isolated by using an RNeasy Micro kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. cDNA synthesis was performed using a PrimeScript RT reagent kit (TaKaRa, Tokyo, Japan) by following the manufacturer's protocol. PCR reactions contained approximately 5 ng cDNA, 0.25 μM of forward and reverse primers for each transcript and 10 μl of LightCycler 480 SYBR Green I Master (Roche Applied Science, Penzberg, Germany). The reference gene was α-tubulin (TUBA). The primer sequences for TUBA, SUV39H1, SUV39H2, DNMT1, DNMT3A, DNMT3B, ATP5A1 and CMOS are shown in Table 1. The forward and reverse primers for target genes were designed using the open source Primer3 ver0.4.0 software (http://frodo.wi.mit.edu/primer3/). The primer sets for SUV39H1 and SUV39H2 were designed so that they amplified the regions that contain the sites siRNAs targeted. Amplifications of cDNAs were performed twice and in triplicate on each plate. The enzyme was activated at 95 C for 5 min, and this was followed by 45 cycles of 95 C for 10 sec, 60 C for 5 sec, and 72 C for 10 sec. Serial dilutions were prepared by serially diluting a reference cDNA sample (from porcine cumulus cells) with PCR-graded water by 2-fold per dilution to produce five concentrations of cDNA. The relative quantifications were obtained with a LightCycler 480 Instrument (Roche Applied Science) and normalized to the relative quantification of the reference gene by the standard curve method.
Table 1. Primer sequences used for gene expression or chromatin immunoprecipitation (ChiP) assays.
| Gene expression | ||
| Transcript | Forward primer | Reverse primer |
| SUV39H1 | aaggatgcagtgtgtgttgc | cctgttcgcggatcttttta |
| SUV39H2 | gcaggacgaactcaacagaa | caaccaaaggtggcttcatt |
| DNMT1 | ctgtgctgggatagata | agatgaccttcactttgct |
| DNMT3A | gaatcgctacagggcttctg | ctggatatgcttctgcgtga |
| DNMT3B | tgaagcacgaggggaatatc | tcagcaggtggtagtgttcg |
| ATP5A1 | gcttcaaatccagccaagaa | ggtgtcaggggctatcttga |
| CMOS | ttcaccccaaatgcttttgc | tggaactgcaaggaagcaca |
| ChiP assay | ||
| Locus | Forward primer | Reverse primer |
| Chr2start | tcctcagcactcaccttctg | gagctgcctctgcctttcta |
| Chr8start | atcagccgtaactgccttct | cactgcggccagtaagtatc |
| Chr12start | tcctggatgtaccagcttca | ctcctccgtgtagtccatca |
Extraction of genomic DNA and determination of telomere length by TRF assays
Genomic DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen) by following the manufacture's protocol. Mean telomere length was determined by mean terminal restriction fragment (TRF) length analysis using a TeloTAGGG Telomere Length Assay Kit (Roche) according to the manufacture's protocol. Briefly, the isolated genomic DNA (approximately 2 μg DNA per experimental sample) was digested with the restriction enzymes HinfI and RsaI for 2 h. Digested genomic DNA samples were fractionated by 0.8% agarose gel electrophoresis at 5 V/cm for approximately 3.5 h. Gels were denatured, neutralized and transferred to a positively charged nylon membrane (Roche) by capillary transfer at room temperature using 20× saline-sodium citrate (SSC) as a transfer buffer for overnight. The transferred DNA was fixed on the wet blotting membrane by UV cross-linking. After being washed twice with 2× SSC, the membrane was prehybridized in 20 ml of DIG Easy Hyb (Roche) for 6 h at 42 C and hybridized in 20 ml of DIG Easy Hyb containing 4 μl telomere probe for 18 h at 42 C. The signals were visualized by chemiluminescence using a DIG Luminescent Detection Kit (Roche) and exposed to Amersham Hyperfilm ECL (GE Healthcare, Buckinghamshire, UK). The assay was performed three times.
Telomerase activity assays
Telomerase activity assays were examined by using a TRAPEZE Telomerase Detection Kit (EMD Millipore, Billerica, MA, USA) following the manufacture's protocol. Briefly, samples were resuspended in 200 μl of 1× CHAPS Lysis Buffer. The suspensions were then incubated on ice for 30 min and centrifuged at 12,000 × g for 20 min at 4 C. About 160 μl of the supernatant was transferred into a fresh tube for determination and adjustment of protein concentration. The remaining extract was then quick-frozen on dry ice and stored at –80 C until use. The relative telomerase activities were determined using the SYBR Green real-time quantitative telomeric repeat amplification protocol (RQ-TRAP) assay according to Wege et al. [16] with some modifications. Briefly, the SYBR Green RQ-TRAP assay was conducted with 2 μl extracts, 0.1 μg of telomerase primer TS, 0.05 μg of anchored return primer ACX and 12.5 μl of LightCycler 480 SYBR Green I Master (Roche Applied Biosystems). Primer sequences were as described by Kim and Wu [17]. Inactivated (heat treated) samples and lysis buffer were also assayed on every plate as a negative control for each sample and primer-dimer/PCR contamination control, respectively. Samples were incubated for 20 min at 25 C and amplified in 35 PCR cycles of 30 sec at 95 C and 90 sec at 60 C (two-step PCR). All PCRs were performed twice and in triplicate on each plate with a LightCycler 480 Instrument (Roche Applied Science). Standard curves were generated from telomerase-positive cell extract (positive telomerase extract control supplied in the TRAPEZE Telomerase Detection Kit), which was serially diluted with PCR-graded water by 2-fold per dilution to produce five concentrations of DNA. The relative telomerase activities of cell samples were normalized to the relative quantification of non-siRNA-treated PEFs by the standard curve method.
Chromatin immunoprecipitation (ChIP) assay
For ChIP analysis, 2×106 pESLCs or PEFs were used per condition. ChIP assays were performed based on Boyd and Farnham [18] with some modifications. In preparation for the chromatin preclear step, Pansorbin Cells (EMD Millipore) were washed with 2 mM EDTA and 50 mM Tris-Cl (pH 8.0), blocked with 10 mg/ml calf thymus (Sigma) and 10 mg/ml BSA and stored at 4 C before use. Cells were treated with 1% formaldehyde for 10 min to cross-link histones to DNA at room temperature on a shaking platform. The cross-linking reaction was stopped by adding glycine to a final concentration of 0.125 M. The cross-linked cells were washed twice with cold PBS and then lysed in 100 μl of 1% SDS (Sigma), 50 mM Tris-HCl (pH 8.0), 10 mM EDTA and 1% protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan) for 10 min on ice. The lysate was sonicated for 5 min (4 pulses for 15 sec with a 1-min rest interval) by a Bioruptor sonicator (Cosmo Bio) to obtain chromatin fragments <1 kb. The chromatin fragments were then centrifuged at 14,000 × g for 10 min at 4 C, and the supernatant was collected. About 10 μl of the chromatin fragments were cleaned using a QIAquick PCR Purification Kit (Qiagen) and run on a gel to check the sonication. The remaining chromatin fragments were precleared by adding 2 μl blocked/washed Pansorbin Cells and 1 mM phenylmethanesulfonyl fluoride (PMSF; Nacalai Tesque) and incubated on a rotating platform for 10 min at 4 C. The fragments were diluted 1:3 with 1.1% Triton-X100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl (pH 8.0), 0.01% SDS, 1 mM PMSF and 1% protease inhibitor cocktail. The fragments were then incubated with 5 μl of rabbit polyclonal antibody to 3MeH3K9 (EMD Millipore) or IgG from rabbit serum (for negative control; Sigma) at 4 C overnight on a rotating platform. After that, 2 μg of blocked/washed Pansorbin Cells and 1 mM PMSF were then added and the lysate was incubated on a rotating platform for 15 min at room temperature. The immunoprecipitated pellets were then washed twice with 0.2% SDS, 2 mM EDTA and 50 mM Tris-HCl (pH 8.0) and thrice with 0.5 M LiCl, 1% Nonidet P-40 (Sigma), 1% sodium deoxycholate, 100 mM Tris-HCl (pH 9.0) and 1 mM PMSF. The complexes of antibody-protein-chromatin were then eluted by incubation with 100 μl 1% SDS and 50 mM NaHCO3 for 15 min at room temperature with rotation. Cross-links were reversed by adding 4 μl of 5 M NaCl and incubating samples for 4 h at 65 C. Fragment DNAs were then purified using a QIAquick PCR Purification Kit (Qiagen) by following the manufacture's protocol. The presence of subtelomeric sequences of chromosomes 2, 8 and 12 in ChiP samples was detected using RT-PCR with the primer sets Chr2start, Chr8start and Chr12start shown in Table 1. The primer sequences were designed using the open source Primer3 ver0.4.0 software. The sequences of the “start” of chromosomes 2, 8 and 12 were obtained from the Sscrofa10.2 database, an assembly of the pig genome produced by the Swine Genome Sequencing Consortium (SGSC) (http://asia.ensembl.org/Sus_scrofa/Info/Index). RT-PCR reactions contained approximately 2 μl of each ChIP sample, 0.25 μM of forward and reverse primers and 10 μl of LightCycler 480 SYBR Green I Master (Roche Applied Science). Amplified reactions were performed twice and in quadruplicate on each plate. The enzyme was activated at 95 C for 5 min and this was followed by 45 cycles of 95 C for 10 sec, 60 C for 5 sec, and 72 C for 15 sec. The relative presence of subtelomeric sequences of chromosomes 2, 8 and 12, which represent the level of 3MeH3K9 at subtelomeres, was calculated by dividing the ChIP signals by the IgG negative control signals according to the fold enrichment method.
Statistical analysis
The data for gene expression, telomerase activities and level of 3MeH3K9 were analyzed by one-way ANOVA followed by Tukey's test using the Statview 5 software package (SAS Institute, Cary, NC, USA). Data are expressed as means ± SEM.
Results
Efficiency of siRNA knockdown of SUV39H1 and SUV39H2
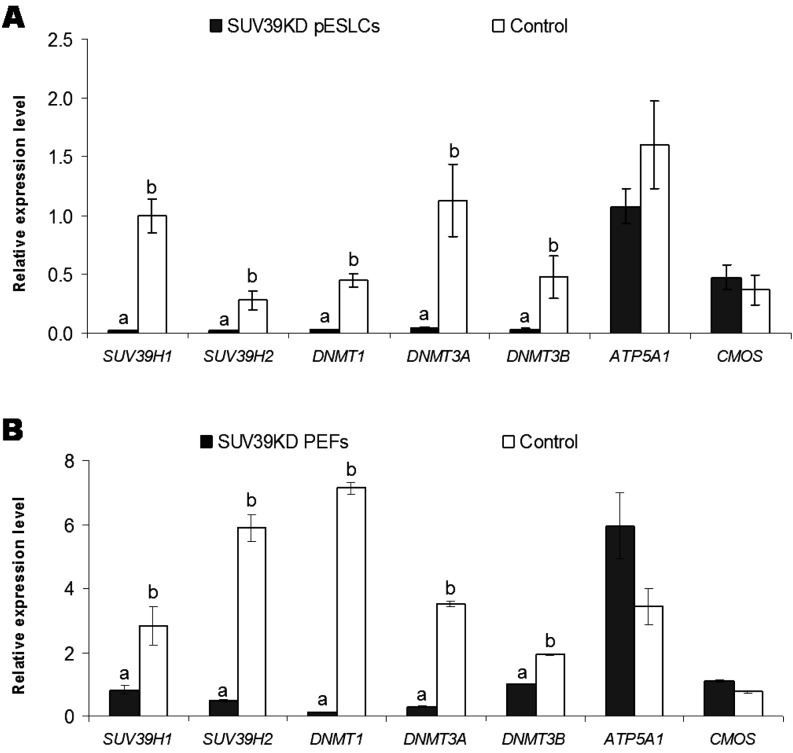
SUV39KD pESLCs and SUV39KD PEFs were obtained by siRNA knockdown. The efficiency of siRNA knockdown was evaluated by the expression levels of SUV39H1 and SUV39H2 genes in SUV39KD pESLCs and SUV39KD PEFs in comparison with their respective controls. The primer sets for SUV39H1 and SUV39H2 were designed so that they amplified the regions that contain the sites siRNAs targeted. Therefore, the expressions of these two genes in effectively knocked-down cells were expected to be remarkably decreased compared with controls. The expressions of SUV39H1 (0.02 ± 0.01) and SUV39H2 (0.02 ± 0.01) in SUV39KD pESLCs decreased (P<0.001) approximately 50 and 14 times compared with those of the control (1.00 ± 0.14 and 0.28 ± 0.08, respectively) (Fig. 1A). Likewise, the expressions of SUV39H1 (0.82 ± 0.14) and SUV39H2 (0.49 ± 0.03) in SUV39KD PEFs decreased (P<0.05) approximately 3.5 and 12 times compared with those of the control (2.82 ± 0.62 and 5.89 ± 0.43, respectively) (Fig. 1B).
Fig. 1.
Gene expression in (A) SUV39H1 and SUV39H2 knockdown (SUV39KD) porcine embryonic stem-like cells (pESLCs) and the control and (B) SUV39KD porcine embryonic fibroblasts (PEFs) and the control. The efficiency of siRNA knockdown was evaluated by the expression levels of SUV39H1 and SUV39H2 genes in SUV39KD pESLCs and SUV39KD PEFs in comparison with their respective controls. The primer sets for SUV39H1 and SUV39H2 were designed so that they amplified the regions that contain the targeted sites of siRNAs. Therefore, the expression of SUV39H1 and SUV39H2 in effectively knocked down cells were expected to be dramatically decreased compared with controls. The expressions of these two genes in both SUV39KD pESLCs and SUV39KD PEFs significantly decreased compared with those of the controls. The expressions of DNMT1, DNMT3A and DNMT3B also decreased in SUV39KD cells compared with controls. In contrast, no significant down-regulation of the housekeeping genes ATP5A1 and CMOS was found in SUV39KD cells compared with the control, suggesting that siRNA knockdown specifically downregulated the expression of SUV39H1 and SUV39H2, and DNMT1, DNMT3A and DNMT3B but did not significantly alter the expressions of other essentials genes. Different letters denote statistically significant differences between groups.
Gene expression in SUV39KD cells
Following the downregulation of SUV39H1 and SUV39H2 genes, the expressions of DNMT1, DNMT3A and DNMT3B also decreased in SUV39KD cells. Significant decreases (P<0.05) in expressions of DNMT1 (0.03 ± 0.01), DNMT3A (0.05 ± 0.01), and DNMT3B (0.03 ± 0.01) in SUV39KD pESLCs was observed compared with the control (0.45 ± 0.06, 1.13 ± 0.3 and 0.48 ± 0.18, respectively) (Fig. 1A). In contrast, expression levels of ATP5A1 (1.08 ± 0.15) and CMOS (0.47 ± 0.11) in SUV39KD pESLCs did not differ from those of the control (1.60 ± 0.37 and 0.37 ± 0.13, respectively) (Fig. 1A). Similar to the case in pESLCs, significant decreases (P<0.05) in expressions of DNMT1 (0.12 ± 0.01), DNMT3A (0.30 ± 0.01) and DNMT3B (1.00 ± 0.01) were found in SUV39KD PEFs compared with the control (7.14 ± 0.19, 3.51 ± 0.07 and 1.93 ± 0.02, respectively) (Fig. 1B). In opposition to that, expression levels of ATP5A1 (5.94 ± 1.03) and CMOS (1.09 ± 0.04) in SUV39KD PEFs were similar to those of the control (3.42 ± 0.56 and 0.75 ± 0.03, respectively) (Fig. 1B).
Telomere length in SUV39KD cells
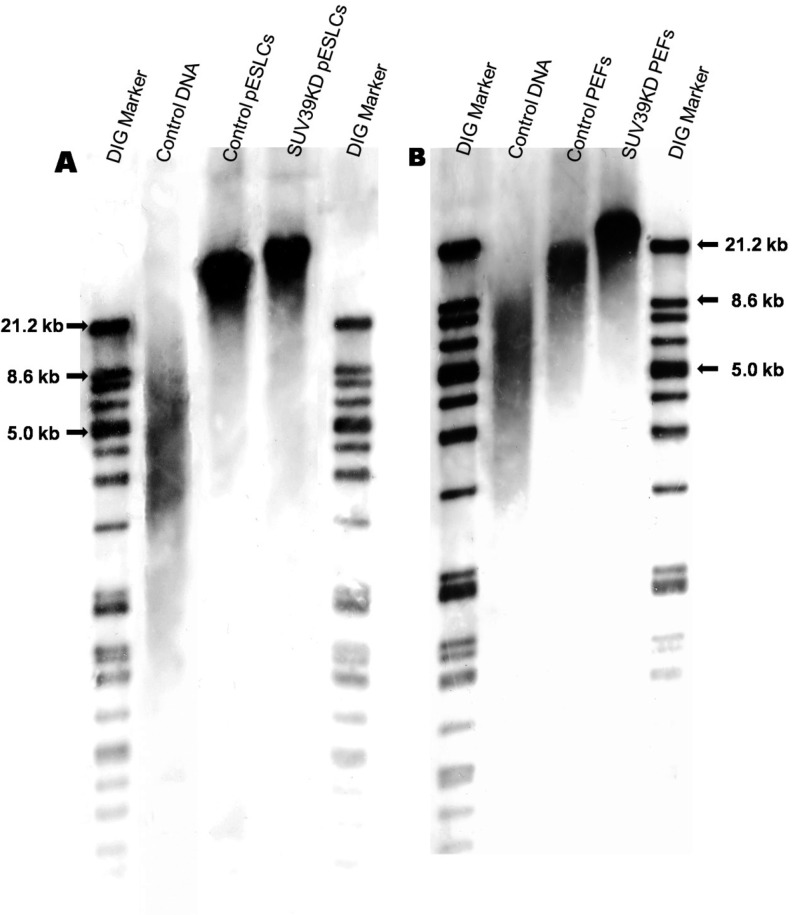
SUV39KD pESLCs (37.1 ± 0.9 kb) and SUV39KD PEFs (22.1 ± 0.4 kb) had longer telomeres (P<0.05) compared with their respective controls (33.0 ± 0.7 kb and 17.8 ± 1.1 kb, respectively) (Fig. 2A, B).
Fig. 2.
Telomere length in (A) SUV39H1 and SUV39H2 knockdown (SUV39KD) porcine embryonic stem like cells (pESLCs) compared with the control; and (B) SUV39KD porcine embryonic fibroblasts (PEFs) compared with the control. Telomere lengths were determined by using a TeloTAGGG Telomere Length Assay kit. SUV39KD pESLCs and SUV39KD PEFs had longer telomeres compared with controls. A: Control DNA, DNA from immortal human cell lines provided in the kit; Control pESLCs, non-siRNA-treated pESLCs. B: Control DNA, DNA from immortal human cell lines provided in the kit; Control PEFs, non-siRNA-treated PEFs.
Telomerase activity in SUV39KD cells
Relative telomerase activities were not different between SUV39KD pESLCs (10.4 ± 1.7) and the control (10.1 ± 1.7). Likewise, SUV39KD PEFs had similar relative telomerase activities (1.0 ± 0.3) compared with the control (1.0 ± 0.4).
Trimethylation of H3K9 in subtelomeric regions
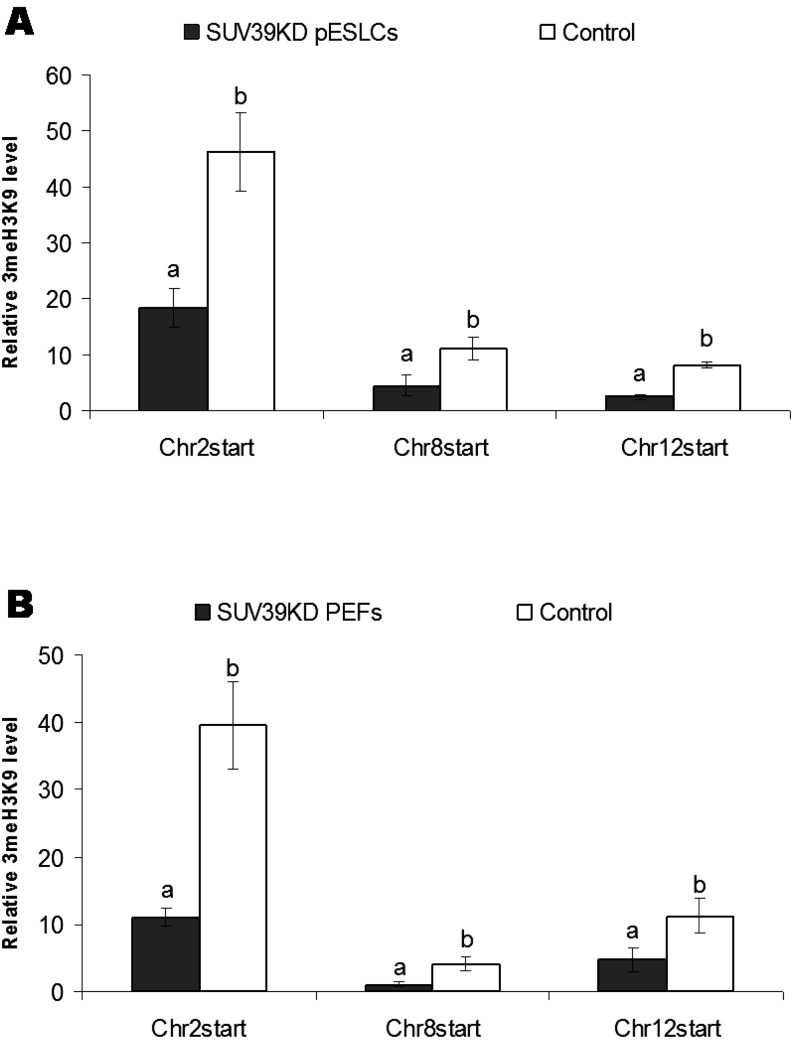
The relative levels of 3MeH3K9 at subtelomeric regions of chromosome 2 (18.3 ± 3.4), chromosome 8 (4.5 ± 1.8) and chromosome 12 (2.5 ± 0.4) in SUV39KD pESLCs were significantly lower (P<0.05) than those of the control (46.3 ± 6.9, 11.0 ± 2.0 and 8.0 ± 0.6, respectively) (Fig. 3A). Similar to the case in pESLCs, significant decreases (P<0.05) in the 3MeH3K9 levels at subtelomeres of chromosome 2 (11.0 ± 1.3), chromosome 8 (1.1 ± 0.3) and chromosome 12 (4.8 ± 1.8) were observed in SUV39KD PEFs compared with the control (39.6 ± 6.5, 4.2 ± 1.0 and 11.2 ± 2.5, respectively) (Fig. 3B).
Fig. 3.
Relative level of trimethylation of Histone H3 Lysine 9 (3MeH3K9) in the subtelomeres of chromosomes 2, 8 and 12 in (A) SUV39H1 and SUV39H2 knockdown (SUV39KD) porcine embryonic stem-like cells (pESLCs) compared with the control; and (B) SUV39KD porcine embryonic fibroblasts (PEFs) compared with the control. The relative levels of 3MeH3K9 in subtelomeric regions were determined by using a chromatin immunoprecipitation (ChIP) assay followed by RT-PCR analysis. A: The relative levels of 3MeH3K9 in the subtelomeric regions of chromosome 2, chromosome 8 and chromosome 12 in SUV39KD pESLCs were significantly lower than those of the control. B: Similar to this, significant decreases in 3MeH3K9 levels in the subtelomeres of chromosomes 2, 8 and 12 in SUV39KD PEFs were observed compared with the control. Different letters denote statistically significant differences between groups.
Discussion
In the present study, we successfully performed double knockdown of SUV39H1 and SUV39H2 by siRNA transfection in pESLCs and PEFs. In a preliminary experiment, we found that telomere length in SUV39KD pESLCs and SUV39KD PEFs did not significantly change compared with the control when the transfection was performed for only one passage (see Suppl Fig. 1: online only), although significant downregulations of SUV39H1 and SUV39H2 were found in SUV39KD cells compared with the controls. The possible cause for this observation is that although the siRNA treatment showed its effects on gene expressions in the very first trial, it might take a little longer to show effects on telomere length, since the cells were harvested only 24 h or 48 h after siRNA treatment for pESLCs and PEFs, respectively. However, SUV39KD PEFs started to show slow growth speed and changes in morphology after the third transfection. The siRNA knockdown of SUV39H1 and SUV39H2, therefore, was carried out for three consecutive passages. To check the efficiency of siRNA knockdown, we designed primers for SUV39H1 and SUV39H2 so that they amplified the regions that contain the targeted sites of designed siRNAs. Therefore, the expression of SUV39H1 and SUV39H2 genes in effectively knocked down cells were expected to be remarkably decreased compared with controls. The remarkably low expressions of these two genes in SUV39KD pESLCs and SUV39KD PEFs compared with controls suggested that SUV39H1 and SUV39H2 were effectively knocked down.
We found that the knockdown of SUV39H1 and SUV39H2 in pESLCs and PEFs significantly increased the length of the telomere in pigs. This result is in agreement with a previous study in the mouse [8] and inconsistent with a study in humans [14]. This might suggest that the telomere biology of the pig is more similar to that of the mouse in respect to epigenetic/genetic regulation of telomere length. In the present study, the telomere in PEF control was 17.8 kb in length. This length is comparable with that of the eGFP-transduced fetal fibroblast cell line in a previous report, which had a telomere length of 18.5 kb [19]. We found that, in comparison with the PEF control, the telomeres in SUV39KD PEFs increased approximately 24%. Meanwhile, an approximate 12% increment in telomere length was observed in SUV39KD pESLCs compared with the pESLC control. These increments are comparable to those in the previous study in the mouse (from 11% to 28%) [8].
SUV39h1 and SUV39h2 are 3MeH3K9-specific HMTs in mammals [13]. Thus, a decrease in the 3MeH3K9 level was expected as SUV39H1 and SUV39H2 genes were downregulated. We examined the relative level of 3MeH3K9 in the telomere-adjacent regions on chromosomes 2, 8 and 12. These are GC-rich regions based on the Sscrofa10.2 database. Significant reductions in the 3MeH3K9 level in these subtelomeric regions were observed in pESLCs and PEFs. The decreases in 3MeH3K9 at subtelomeric regions might be responsible for telomere lengthening, since this reduced chromobox proteins binding to the telomere [8] and Heterochromatin Protein 1, the chromobox homolog in Drosophila, plays an important role in telomere capping [20]. Further study on chromobox proteins is needed to confirm this hypothesis.
In mammals, DNA methylation requires 3MeH3K9 by SUV39h1 and SUV39h2 HMTs, suggesting that histone methylation directs DNA methylation at the pericentric chromatin [21, 22]. Downregulation of the genes SUV39H1 and SUV39H2, therefore, might also affect DNA methyltransferases. We revealed that the expressions of DNMT1, DNMT3A and DNMT3B were decreased in both SUV39KD pESLCs and SUV39KD PEFs compared with controls. The downregulation of DNMT1, DNMT3A and DNMT3B that occurred as a consequence of SUV39KD might be due to the fact that, at the enzyme levels, SUV39h1 and its interacting partner bind to and direct Dnmt1 and Dnmt3a [21]. The downregulations of DNMT1, DNMT3A and DNMT3B could lead to loss of DNA methylation at subtelomeric regions, which results in aberrant telomere elongation [9]. Therefore, a decrease in DNA methyltransferases might also be responsible for the abnormal lengthening of telomeres in SUV39KD cells in pigs. Further study on the protein levels of DNA methyltransferases and the changes in DNA methylation at subtelomeres in SUV39KD cells are needed to confirm this hypothesis. The possibility that telomerase activity also contributes to the abnormal elongation of telomeres is excluded, as we found that telomerase activities did not differ between SUV39KD pESLCs and the control or between SUV39KD PEFs and the control. It should be noted that no significant differences in expressions of the housekeeping genes ATP5A1 and CMOS were found in SUV39KD cells in comparison with controls. This result confirms that SUV39KD of pESLCs and PEFs seemed to specifically downregulate the expression of SUV39H1, SUV39H2, DNMT1, DNMT3A and DNMT3B but do not significantly alter the expressions of other essential genes in our study.
It also should be noted that we obtained similar results with another pESLC line and two other PEF cultures (data not shown). This may eliminate the possibility that all the observations might only apply to specific cell cultures.
In conclusion, double knockdown of SUV39H1 and SUV39H2 results in elongation of telomeres in pigs. This elongation might be not only related to the decrease of trimethylation of histone H3 lysine 9, but also linked to the reduction of DNA methyltransferases.
Supplementary Material
Acknowledgment
We would like to thank Dr A Onishi and the Transgenic Pig Research Unit (National Institute of Agrobiological Sciences) for giving us the pig embryonic fibroblasts and Ms Ando and Ms Suzuki for technical assistance.
References
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 2005; 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- 2.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA 1988; 85: 6622–6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA 1989; 86: 7049–7053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem 2004; 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- 5.Shay JW, Wright WE. Hallmarks of telomeres in ageing research. J Pathol 2007; 211: 114–123 [DOI] [PubMed] [Google Scholar]
- 6.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 2010; 11: 319–330 [DOI] [PubMed] [Google Scholar]
- 7.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet 2007; 8: 299–309 [DOI] [PubMed] [Google Scholar]
- 8.García-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 2004; 36: 94–99 [DOI] [PubMed] [Google Scholar]
- 9.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 2006; 8: 416–424 [DOI] [PubMed] [Google Scholar]
- 10.Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 2007; 178: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. Epigenetic regulation of telomeres in human cancer. Oncogene 2008; 27: 6817–6833 [DOI] [PubMed] [Google Scholar]
- 12.Farnung BO, Giulotto E, Azzalin CM. Promoting transcription of chromosome ends. Transcription 2010; 1: 140–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001; 107: 323–337 [DOI] [PubMed] [Google Scholar]
- 14.Kondo Y, Shen L, Ahmed S, Boumber Y, Sekido Y, Haddad BR, Issa JP. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One 2008; 3: e2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haraguchi S, Kikuchi K, Nakai M, Tokunaga T. Establishment of self-renewing porcine embryonic stem cell-like cells by signal inhibition. J Reprod Dev2012; [Epub ahead of print] DOI: http://dx.doi.org/10.1262/jrd.2012-008 [DOI] [PubMed]
- 16.Wege H, Chui MS, Le HT, Tran JM, Zern MA. SYBR Green real-time telomeric repeat amplification protocol for the rapid quantification of telomerase activity. Nucleic Acids Res 2003; 31: E3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim NW, Wu F. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplication protocol (TRAP). Nucleic Acids Res 1997; 25: 2595–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyd KE, Farnham PJ. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol Cell Biol 1999; 19: 8393–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang L, Carter DB, Xu J, Yang X, Prather RS, Tian XC. Telomere lengths in cloned transgenic pigs. Biol Reprod 2004; 70: 1589–1593 [DOI] [PubMed] [Google Scholar]
- 20.Raffa GD, Siriaco G, Cugusi S, Ciapponi L, Cenci G, Wojcik E, Gatti M. The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc Natl Acad Sci USA 2009; 106: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 2003; 31: 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 2003; 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.