Abstract
The relaxivity of a magnetically responsive Gd complex can be controlled by non-covalent molecular recognition with a water-soluble deep cavitand. Lowered relaxivity is conferred by a self-assembled micellar “off state”, and the contrast can be regenerated by addition of a superior guest.
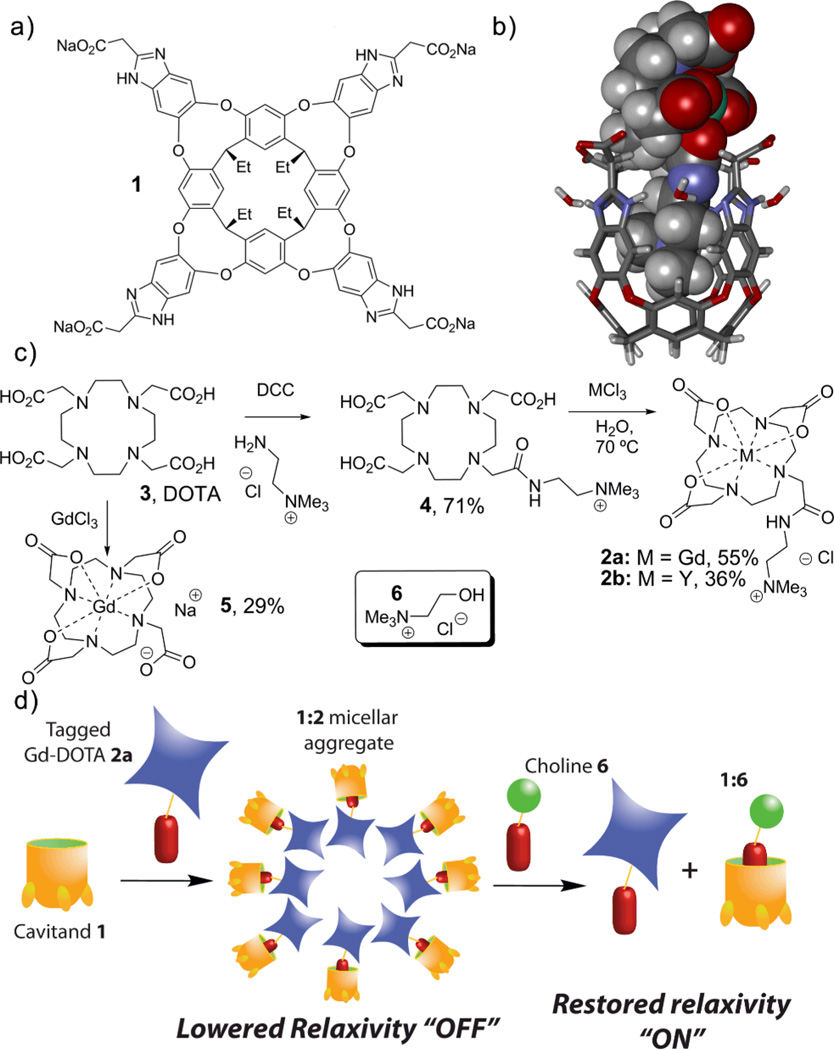
Shape-based molecular recognition has a number of applications in sensing for a variety of analytes.1 The most common applications report directly on the sensor:substrate interaction, but an alternate technique is based on allostery: the recognition event causes a second assembly event that can be detected by an external method.2 This has application for the detection of species that are difficult to complex via direct recognition. An example lies in the creation of magnetically responsive complexes that are based upon paramagnetic chelates of Gadolinium. Responsive agents that change contrast properties upon exposure to effectors3 such as metal ions,3a–e pH,3f–g enzymes3h and light3i are known, but they often rely on a direct interaction with the Gd center, most often with water, to modulate the response. An alternate strategy would be to “hide” the Gd center from water in a particle4 or via non-covalent, multicomponent assembly. Unfortunately, self-assembly in water is dominated by hydrophobic effects and mainly controlled by concentration, a property not amenable to external control. If the self-assembly could be triggered by a selective agent, simple control of the visualization properties would be accessible. Here, we report an example of this: a magnetically responsive Gd complex based on controlled, non-covalent assembly. The system is based on a shape-selective dimerization of a functionalized Gd chelate by a water-soluble host molecule. This reversible non-covalent binding of the relaxive complex by the receptor allows displacement of the complex by a stronger binder, and activation of the released guest. The relaxive species (2a, Fig 1c) is provided by addition of an R-NMe3+ “binding handle” to the known Gd ligand DOTA. This allows non-covalent recognition by the water-soluble deep cavitand 1. Cavitand 1 is capable of selective recognition of guests displaying an R-NMe3+ binding handle in pure water,5 lipid micelles,6a and living cells.6b
Fig. 1.
(a) Water-soluble deep cavitand 1; (b) minimized structure of the cavitand 1:Gd chelate 2a complex (SPARTAN, AM1 forcefield); (c) synthesis of the guests; (d) a representation of the responsive assembly/release process.
The binding generally occurs with high affinities (~104 M−1 in D2O), controlled by cation-π interactions.5 Cavitand 1 displays properties of lipids, including a hydrophobic body and a charged terminus, allowing self-assembly with external lipids and incorporation into larger aggregates without loss of host behavior.6a Synthesis of the complex 2a was performed using known methods7 in 39% overall yield. The same route furnished a diamagnetic yttrium chelate 2b. A minimized structure of the proposed 1•2a complex is shown in Figure 1b.
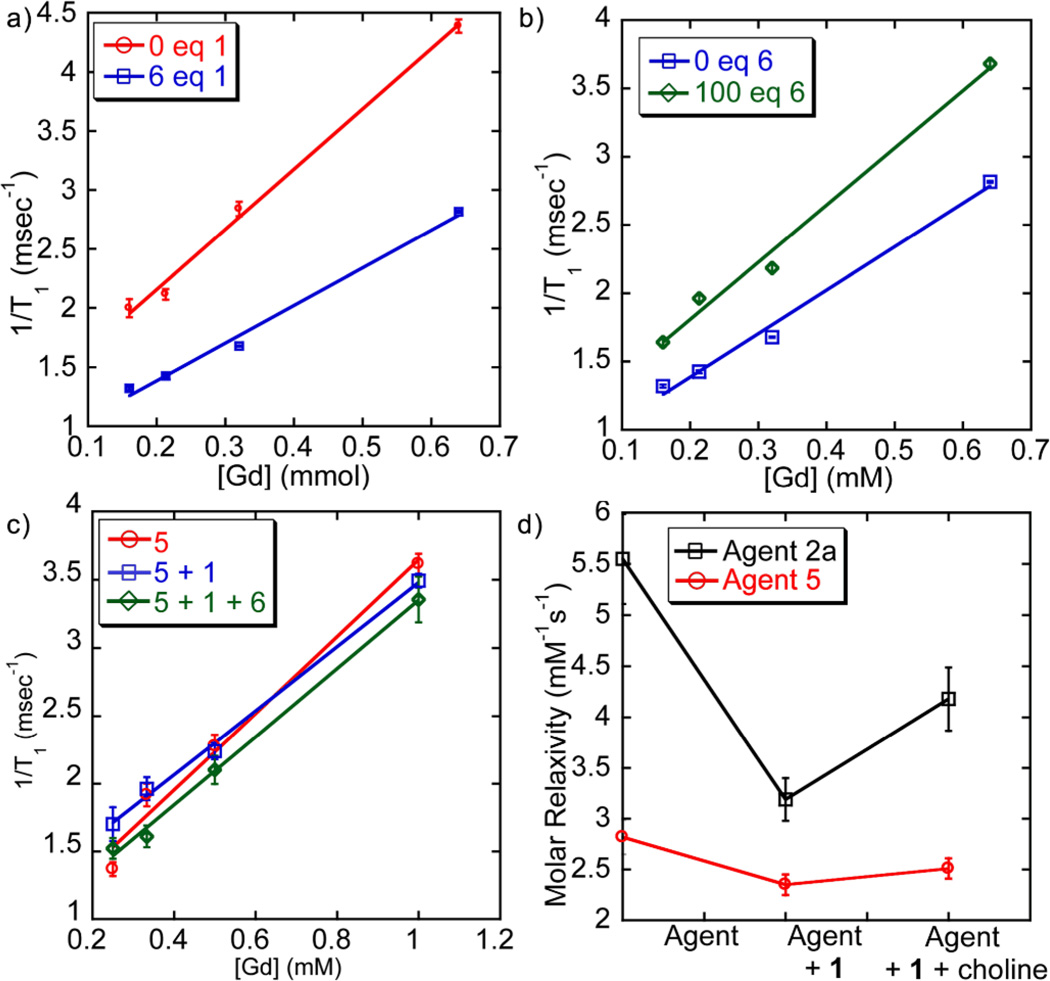
Upon addition of host 1 to an aqueous solution of complex 2a, significant attenuation of the relaxivity is observed. The T1 of water in a 0.64 mM solution of complex 2a increased from 228±3 ms to 301±1 ms upon introduction of one molar equiv. 1. As [1] increases, a continuous increase in T1 relaxation was observed. At 6 molar equiv. 1, the T1 344±1 ms and no further increase was observed (see ESI). The molar relaxivity decreased accordingly (Figure 2a): the relaxivity of 2a (5.6±0.4 mM−1s−1) is significantly lessened by the addition of excess 1 (3.2±0.2 mM−1s−1), a decrease of 43%.
Fig. 2.
Molar relaxivity plots (H2O, 298 K) of (a) Complex 2a + 0 or 6 mol.-eq. 1 ([2a] = 1 mM); (b) 2a + 6 mol.-eq. 1 + 0 or 100 mol.-eq. choline 6 ([2a] = 0.5 mM); (c) Control 5 + 1 + 0 or 100 mol.-eq. 6 (d) Molar relaxivities of complexes 2a and 5 with sequential addition of 1 (6 equiv) and 6 (100 equiv). Data acquired at 9.4T.
The complexation of complex 2a and cavitand 1 with subsequent lessening of T1 (the “off” state) can be reversed by expelling complex 2a from the host. This can be achieved by adding choline chloride (6), a strongly bound guest for 1. When choline chloride (6) was titrated into a solution of 2a (0.32 mM) with 6 molar equiv. 1, a continuous decrease in T1 was observed as [6] increased. The initial T1 relaxation of 2a (576±4 ms) decreased to 420±5 ms after the addition of 100 equiv. of choline, and molar relaxivities behave accordingly (Figure 2b). The r1 of the 1•2a construct (3.2±0.2 mM−1s−1) is increased to 4.2±0.3 mM−1s−1, an increase of 31% over the complex in its “off” state. We suspect that the incomplete recovery of relaxivity occurs because of a Coulombic attraction between cation 2a and the anionic 1•6 material. The R-NMe3+ binding handle of 2a is essential to the behavior of the system. The relaxivity-masking experiment above was repeated with Na[Gd(DOTA)] (5), which has little affinity for the cavitand. The molar relaxivity of 5 (2.8±0.2 mM−1s−1) was reduced upon addition of excess 1 by only 14% (r1 = 2.4±0.1 mM−1s−1). In addition, there was no change in relaxivity upon exposure of the 1•5 mixture to choline (r1 = 2.5±0.1 mM−1s−1). Without the selective binding of the complex, minimal reduction of molar relaxivity was observed, and no reversal is possible upon treatment with choline.
The exact mechanism of the relaxivity control is not obvious. Immobilization of Gd chelates on scaffolds of larger size (e.g. macromolecules) tends to yield higher relaxivities due to reduced tumbling rate,8 but this system displays the opposite behavior: relaxivity lessens upon binding of the cavitand to 2a. Simple 1H NMR analysis is not practical with paramagnetic complex 2a, so the analogous diamagnetic yttrium complex 2b was employed. Upon addition of 2b to cavitand 1, no characteristic upfield peaks for the 1:R-NMe3+ complex were observed (see ESI). Instead, loss of signal was observed as the guest concentration increased. When cavitand 1 was titrated into 2b, similar results were obtained. Only after addition of 6 equiv. of 1 were sharp peaks observed, corresponding with those of free cavitand. The disappearance of signals for both host and guest and the lack of any observable precipitate from the solution suggest that an aggregation of the lipid-like cavitand 1 is occurring upon complexation with the amphiphilic guest 2b. This triggered self-assembly of cavitand 1 is rare. Hydrocarbon guests and small R-NMe3+ species such as choline or NMe4Br form simple 1:1 complexes.5 Longer R-NMe3+ species form micelles that incorporate the cavitand itself.6a Guest 2b, however, is of intermediate size: it is poorly capable of forming micelles itself, but a hydrophobic component of the guest protrudes from the cavity into the bulk water upon binding, lowering the solubility of the complex. When the host:guest binding event occurs, the 1:2b complex apparently initiates aggregation into a larger assembly that displays slow tumbling, and so the 1H signals are averaged. If the aggregate is disrupted by addition of MeCN-d3 to the solution, the individual peaks for both 1 and 2b are observed again (see ESI). The binding of 2b in 1 is relatively weak. While accurate calculation of the Ka (1:2b) is complicated by the aggregation phenomenon, it can be estimated as ca. 250 M−1, two orders of magnitude less than that of choline.5 This also explains why the relaxivity of 2a ceases to decrease after the addition of 6 molar equiv. 1: that is the amount of 1 required fully to bind all complexes in the cavitand. Lower cavitand concentration leads to free 2a molecules in the “on” state.
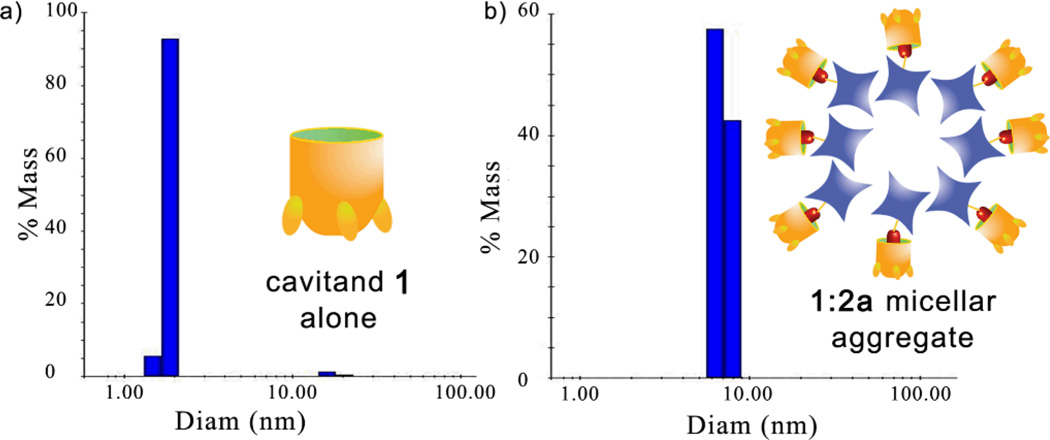
The assembly events were further characterized by dynamic light scattering (DLS, Figure 3). At the 1:2a molar ratio of 6:1, discrete nanoparticle aggregates with an average diameter of 7 nm were observed.9 The orientation of the 1•2a complex in this micellar aggregate is not obvious: one would expect the tetracarboxylate rim of 1 to be oriented towards the bulk solvent, but the Gd-DOTA complex 2a itself displays a lipophilic terminus. The relaxivity data provide a possible explanation: even though the assembly is larger and presumably tumbles at a slower rate than free 2a, the relaxivity of 2a is lowered upon binding to 1. In the aggregated form, the Gd center of 2a is most likely hidden and has limited access to water.10 This indicates that the micellar aggregate displays the characteristics of the cartoon shown in Figure 1d, wherein the Gd center is oriented to the aggregate’s interior. We expect that the aggregate does have a slower tumbling rate, but that the water-shielding properties of the system dominate.
Fig. 3.
DLS data of a) cavitand 1 alone; b) after addition of 2a (H2O, 6:1 ratio of 1:2a)
Upon addition of choline (6), guest 2a is liberated from the assembled aggregate, leading to free 2a in solution that can resume its usual T1 contrast properties. Unfortunately, DLS analysis of the cavitand:choline complex was complicated by the appearance of larger aggregates that occur upon addition of salt and/or varying the concentration of 1. At higher [1], and in the presence of 6, larger aggregates ranging from 20–100 nm and 900–1000 nm were observed. The aggregation phenomenon is evidently complex, but the nature of the choline-containing aggregates is inconsequential to the T1 modulation behavior: the more strongly binding choline (6) occupies the cavitand aggregates and 2a is free to display strong contrast again.
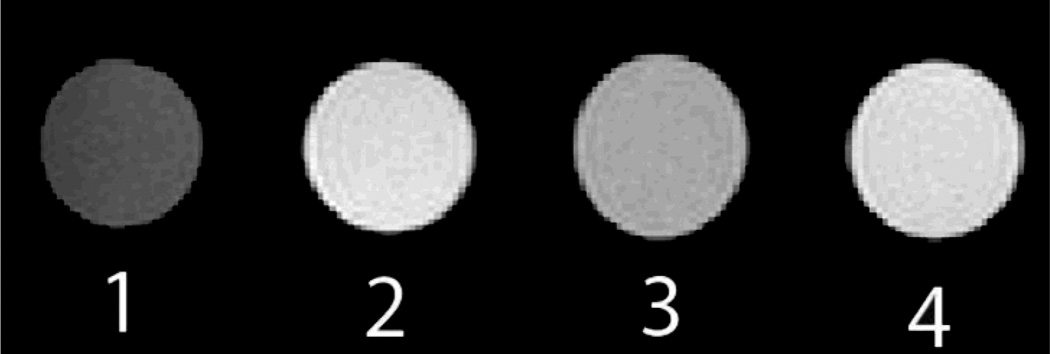
The responsiveness of the system can be most effectively demonstrated by a simple visualization of the contrast difference between the “on” and “off” states. To this end, phantoms were acquired in a 7 T MRI imager using a FLASH sequence (Figure 4). Consistent with the molar relaxivity data, the contrast of complex 2a was masked upon introduction of cavitand 1, which was returned with the introduction of choline chloride. A 30% difference in brightness was observed and quantified using ImageJ.
Fig. 4.
Phantoms acquired in a Pharmascan 7 T MRI using Bruker’s FLASH sequence, TR = 474 ms; TE = 6 ms. (1) H2O; (2) 0.64 mM 2a; (3) 0.64 mM 2a with 4 equivalents of cavitand 1; (4) 0.64 mM solution of Gd complex 2a with 4 equivalents of cavitand 1 upon exposure to 100 equivalents of choline.
In conclusion, we have developed a magnetically responsive Gd complex system that is controlled solely by non-covalent molecular recognition and self-assembly processes. The self-assembly cascade allows the relaxive complex to be turned “off” by shielding the Gd center in a self-assembled micellar aggregate with a suitable water-soluble cavitand host. The assembly is triggered by the binding event, so can be disrupted by addition of a superior guest molecule. This frees the complex, regenerating its contrast abilities. This process occurs under mild aqueous conditions: further research on the this system in biologically relevant environments is underway.
Supplementary Material
Acknowledgements
This work is sponsored by the USC Ming Hsieh Institute (TJW) and the National Science Foundation (CHE-1151773, to RJH). We are grateful to the NSF (DBI-0821671, CHE-0840366) and NIH (S10 RR25432) for their sponsorship of NMR spectrometers.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental details and characterization data. See DOI: 10.1039/c000000x/
Contributor Information
Richard J. Hooley, Email: richard.hooley@ucr.edu.
Travis J. Williams, Email: travisw@usc.edu.
Notes and references
- 1.Pirondini L, Dalcanale E. Chem. Soc. Rev. 2007;36:695. doi: 10.1039/b516256b. [DOI] [PubMed] [Google Scholar]
- 2.Zhu L, Anslyn EV. Angew. Chem. Int. Ed. 2006;45:1190. doi: 10.1002/anie.200501476. [DOI] [PubMed] [Google Scholar]
- 3.(a) Rolla GA, Tei L, Fekete M, Arena F, Gianolio E, Botta M. Bioorg. Med. Chem. 2011;19:1115. doi: 10.1016/j.bmc.2010.07.064. [DOI] [PubMed] [Google Scholar]; (b) Que EL, Gianolio E, Baker SL, Wong AP, Aime S, Chang CJ. J. Am. Chem. Soc. 2009;131:8527. doi: 10.1021/ja900884j. [DOI] [PubMed] [Google Scholar]; (c) Luo J, Li W, Xu P, Zhang L, Chen Z. Inorg. Chem. 2012;51:9508. doi: 10.1021/ic301308z. [DOI] [PubMed] [Google Scholar]; (d) Li W, Fraser SE, Meade TJ. J. Am. Chem. Soc. 1999;121:1413. [Google Scholar]; (e) Major JL, Parigi G, Luchinat C, Meade TJ. Proc. Natl. Acad. Sci. USA. 2007;104:13881. doi: 10.1073/pnas.0706247104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Zhang S, Wu K, Sherry AD. Angew. Chem. Int. Ed. 1999;38:3192. [PubMed] [Google Scholar]; (g) Lowe MP, Parker D, Reany O, Aime S, Botta M, Castellano G, Gianolio E, Pagliarin R. J. Am. Chem. Soc. 2011;123:7601. doi: 10.1021/ja0103647. [DOI] [PubMed] [Google Scholar]; (h) Moats RA, Frasier SE, Meade TJ. Angew. Chem. Int. Ed. Engl. 1997;36:726. [Google Scholar]; (i) Li Y, Qian Y, Liu T, Zhang G, Liu S. Biomacromolecules. 2012;13:3877. doi: 10.1021/bm301425j. [DOI] [PubMed] [Google Scholar]
- 4.Huang W-Y, Davies G-L, Davis JJ. Chem. Commun. 2013;49:60. doi: 10.1039/c2cc37545a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biros SM, Ullrich EC, Hof F, Trembleau L, Rebek J., Jr J. Am. Chem. Soc. 2004;126:2870. doi: 10.1021/ja038823m. [DOI] [PubMed] [Google Scholar]
- 6.(a) Ghang Y-J, Schramm MP, Zhang F, Acey RA, David CN, Wilson EH, Wang Y, Cheng Q, Hooley RJ. J. Am. Chem. Soc. 2013;135:7090. doi: 10.1021/ja401273g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schramm MP, Hooley RJ, Rebek J., Jr J. Am. Chem. Soc. 2007;129:9773. doi: 10.1021/ja0723378. [DOI] [PubMed] [Google Scholar]
- 7.Wangler C, Wangler B, Eisenhut M, Haberkorn U, Mier W. Bioorg. Med. Chem. 2008;16:2606–2616. doi: 10.1016/j.bmc.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 8.Skinner PJ, Beeby A, Dickins RS, Parker D, Aime S, Botta M. J.C.S. Perkin Trans 2. 2000:1328. [Google Scholar]
- 9.Other cavitand micelles: Kubitschke J, Javor S, Rebek J., Jr Chem. Commun. 2012;48:9251. doi: 10.1039/c2cc34065h.
- 10.(a) Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Chem. Rev. 1999;99:2293. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]; (b) Takahara S, Sumiyama N, Kittaka S, Yamaguchi T, Bellissent-Funel MC. J. Phys. Chem. B. 2005;109:11231. doi: 10.1021/jp046036l. [DOI] [PubMed] [Google Scholar]; (c) Carnato F, Tei L, Dastrù W, Marchese L, Botta M. Chem Commun. 2009;45:1246. doi: 10.1039/b820591d. [DOI] [PubMed] [Google Scholar]; (d) Taylor KML, Kim JS, Rieter WJ, An H, Lin W, Lin W. J. Am. Chem. Soc. 2008;130:2154. doi: 10.1021/ja710193c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.