Abstract
This study was conducted as a part of the Chromosome-Centric Human Proteome Project (C-HPP) of the Human Proteome Organization. The United States team of C-HPP is focused on characterizing the protein-coding genes in chromosome 17. Despite its small size, chromosome 17 is rich in protein-coding genes, it contains many cancer-associated genes, including BRCA1, ERBB2 (Her2/neu), and TP53. The goal of this study was to examine the splice variants expressed in three ERBB2 expressed breast cancer cell line models of hormone receptor negative breast cancers by integrating RNA-Seq and proteomic mass spectrometry data. The cell-lines represent distinct phenotypic variations subtype: SKBR3 (ERBB2+ (over-expression)/ ER−/PR−; adenocarcinoma), SUM190 (ERBB2+ (over-expression)/ER−/PR−; inflammatory breast cancer) and SUM149 (ERBB2 (low expression) ER−/PR −; inflammatory breast cancer). We identified more than one splice variant for 1167 genes expressed in at least one of the three cancer cell lines. We found multiple variants of genes that are in the signaling pathways downstream of ERBB2 along with variants specific to one cancer cell line compared to the other two cancer cell lines and to normal mammary cells. The overall transcript profiles based on read counts indicated more similarities between SKBR3 and SUM190. The top-ranking Gene Ontology and BioCarta pathways for the cell-line specific variants pointed to distinct key mechanisms including: amino sugar metabolism, caspase activity, and endocytosis in SKBR3; different aspects of metabolism, especially of lipids in SUM190; cell- to-cell adhesion, integrin and ERK1/ERK2 signaling, and translational control in SUM149. The analyses indicated an enrichment in the electron transport chain processes in the ERBB2 over-expressed cell line models; and an association of nucleotide binding, RNA splicing and translation processes with the IBC models, SUM190 and SUM149. Detailed experimental studies on the distinct variants identified from each of these three breast cancer cell line models may open opportunities for drug target discovery and help unveil their specific roles in cancer progression and metastasis.
Keywords: Splice variants (SpV), splice variant protein (SpP), splice variant transcript (SpT), ERBB2+ (Her2/neu), EGFR, proteotypic peptide, I-TASSER, breast cancer subtypes
Introduction
In Ensembl database version 70, 82% of the protein-coding genes have more than one transcript produced through exon skipping, exon swapping, intronic retention, alternative promoters or alternative polyadenylation sites, and alternatively spliced exons. Moreover, genes produce different splicing events in different cell types including tumor cells1, and splicing results in protein isoforms with different biological activities2. Splice variants of a gene may have opposite functions2-4. For example, two alternatively-spliced transcripts of the osr2 gene, which encode osr2-L (312 aa) and osr2-S (276 aa) have opposite transcriptional activities, activation and repression, respectively 4; we have inferred this functional difference from three-dimensional structural comparison5. Certain splice variants are cancer specific 6-7; for example, Nek2C, a splice variant of Nek2 is involved in breast cancer progression and the inhibition of Nek2C is a potential selective therapy for ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC) 6. It appears then, that some of the diversity of phenotypic behavior of cancer cells derives from alternative splicing of key signaling genes.
This study was conducted by the Chromosome 17 team of the Chromosome-centric Human Proteome Project (C-HPP) of the Human Proteome Organization (HUPO)8-10. HPP analyses involve integration of proteomics data into a genomic framework that will promote a better understanding of the relationship of the transcriptome to the proteome and of the pathways and biological networks involved in the phenotype11. Despite its relatively small size, chromosome 17 is rich in protein-coding genes, ranking second in gene density; it contains many cancer-associated genes, including BRCA1, ERBB2 (Her2/neu), TP53, and genes of the ERBB2 amplicon. Recent studies have shown the significant role of activation of ERBB2 receptor signaling pathways in affecting or driving metastasis-associated properties12, 13. ERBB2 (Her2/neu) and EGFR (ERBB1) are members of the human epidermal growth factor receptor Erbb protein family.
Although ERBB2 overexpression is associated with aggressive breast cancers, little is known about the repertoire of downstream pathways and network interactions that bring about the vast array of cellular phenotypes generated by ERBB2 overexpression in different breast cancers. The purpose of this study is to characterize comprehensively the splice variants (SpVs) expressed in aggressive ERBB2+ breast cancers which have poor prognosis due to high rates of recurrence and metastasis14 and to postulate likely pathways modulated by these variants to refine the pathobiology of ERBB2-induced breast cancers.
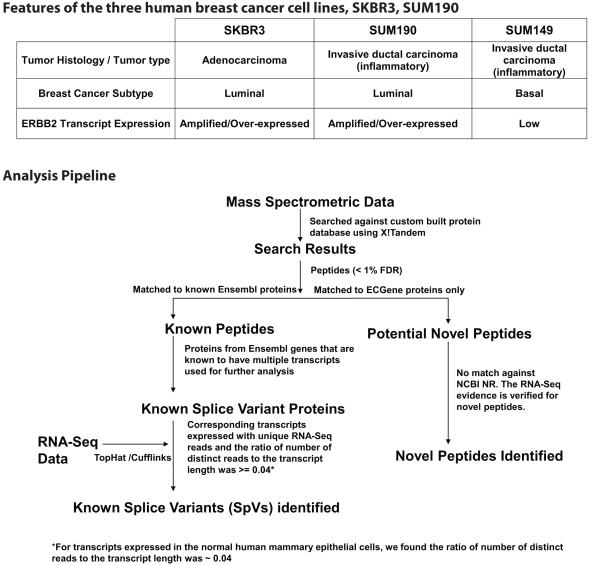
Tumors that over-express ERBB2 account for 15-20% of breast cancers in the US15. Breast cancer cell lines have been used widely to investigate breast cancer progression mechanisms and to develop new therapeutic approaches. SKBR3, which is ER –, PR - with ERBB2 (HER2) amplification has been used successfully as a preclinical model to screen for therapeutic agents targeting ERBB2 and to delineate mechanisms of resistance to ERBB2-based therapies16. SUM190 and SUM149 serve as models for inflammatory breast cancer, the most lethal form of breast cancer 17. Both these cell lines are ER –, PR – and clinically very aggressive, but ERBB2 is amplified in SUM190 and expressed at low levels in SUM149. The table in Figure 1 shows the different features of these three cell lines.
Figure 1.
Table shows the features of SKBR3, SUM190 and SUM149, the three breast cancer cell lines used in this study. The figure shows the analysis pipeline showing the identifications of known splice variants and novel peptides by integrating proteomic and RNA-Seq data.
The hetero-dimerization of ERBB2 with other ERBB proteins (ERBB1/EGFR, ERBB3, ERBB4) activates distinct signaling pathways that result in tumor cell survival; thus, the ERBB2 expression levels have an impact on the pathways that are activated 18. The contrasting ERBB2 expression observed in the similar clinical IBC phenotype represented by SUM149 and SUM190 and over-expression of ERBB2 in SKBR3, representing a non IBC epithelial adenocarcinoma tumor type, makes this group of cell lines useful for the comparisons that we set out to produce. Our goal was to comprehensively define the splice variants expressed in the three breast cancer cell models and to compare the enriched biological pathways involving these splice variants.
We integrated the information from RNA-Seq and proteomic mass spectrometry studies from the cell line models to identify both known splice variants and novel peptides. We identified multiple variants in a total of 1167 distinct genes, including ERBB2 and EGFR, which were expressed at different levels in the three breast cancer cell lines. The transcript expression profiles of the cell lines clustered differently for different pathways. Moreover, we found cell-line specific splice variants. The distinct splice variants identified from the three cell line models may represent new targets for drug development.
Materials and Methods
Cell lines
The human breast cancer cell line SKBR3, was obtained from the American Type Culture Collection (Manassas, VA) and maintained in culture with DMEM/F-12 medium supplemented with 10% FBS (Tissue Culture Biologicals, Seal Beach, CA) and 1% of Antibiotic- Antimycotic 100X (Gibco, Carlsbad, CA). SUM149 and SUM190 cells were obtained from Dr. Stephen Ethier (Kramanos Institute, Detroit, MI) and are commercially available (Asterand, Detroit, MI).Both human IBC cell lines were maintained in culture with Ham’s/F-12 medium supplemented with 10% FBS (Tissue Culture Biologicals, Seal Beach, CA), 5 μg/mL of insulin, 1 μg/mL of hydrocortisone and 1% of Antibiotic-Antimycotic 100X (Gibco, Carlsbad, CA).
Mass spectrometry
Cell lysis and in-gel digestion
Cells were washed 3 times in ice-cold PBS and then collected, using a cell scraper, in 20 μL lysis buffer (2% SDS in 50 mM NH4CO3). Cells were solubilized by sonication using 20 s bursts, followed by ice cooling for 20 s, repeated 10 times. The entire extract was concentrated in a speed vacuum to about 15 μL, and then loaded on a SDS-PAGE gel (4–12% gradient) to separate proteins by molecular weight. After staining with Coomassie blue, each gel lane was cut into five individual sections, which were minced into small pieces, washed with 600 mL water for 15 min, and centrifuged. 50% ACN was added to the pellet (1 mL), tubes were shaken to remove Coomassie stain, and the proteins were reduced with 250 μL of 10 mM DTT in 0.1 M NH4CO3 incubated for 30 min at 56°C. Samples were subsequently alkylated at room temperature in the dark for 80 min with 250 μL of 55mM iodoacetamide in 0.1 M NH4CO3. Trypsin digestion reagent (200 μL; 10 ng/mL of trypsin in 50 mM NH4CO3, pH 8.0) was added, and incubated for 30 min at 47°C and then overnight at 37°C. The supernatant was removed and saved. Gel pieces were further extracted with 5% formic acid (30 μL) and ACN (400 μL) at 37°C for 10 min and then twice with 5% formic acid (30 μL) and ACN (200 μL). The formic acid solution containing tryptic peptides was combined with the supernatant, concentrated to 5-10 μL, and subjected to LC-MS analysis. Three biological replicates were performed for each analysis.
LTQ-FT MS
The in-gel digested peptides were analyzed with an online Dionex nano-LC instrument (Ultimate 3000, Sunnyvale, CA) and a 75 mm i.d. × 15 cm C-18 capillary column packed with Magic C18 (3 mm, 200 Å pore size) (Michrom Bioresources, Auburn, CA). The LC was coupled to a Fourier transfer mass spectrometer (LTQ-FT MS, Thermo Electron, San Jose, CA) operated in the data-dependent mode to switch automatically between MS and MS2 acquisition. Full-scan MS spectra with two microscans (m/z 400-2000) were acquired in the FT ion cyclotron resonance cell with a mass resolution of 100000 at m/z 400 (after accumulation to a target value of 2 × 106 ions in the linear IT), followed by ten sequential LTQ-MS/MS scans throughout the 90 min separation. The analytical separation was carried out using a three-step linear gradient, starting from 2% B to 40% B in 40 min (A: water with 0.1% formic acid; B: ACN with 0.1% formic acid), increased to 60% B in 10 min, and then to 80% B in 5 min. The column flow rate was maintained at 200 nL/min.
RNA-Seq Data
From total RNA of SKBR3, SUM149and SUM190 cells, strand-specific RNA-Seq libraries were prepared according to Illumina TruSeq standard procedures. Each library was sequenced (101 bases, paired end) on 1-3 HiSeq 2000 lanes to obtain an average of 120 million uniquely mapped reads19. The reads were aligned to the human genome (Ensembl GRCh37) using Tophat (v.2.0.5) embedded with Bowtie (v.2.2.0) with a maximum number of 2 mismatches. We assembled the alignments into gene transcripts (Ensembl) using Cufflinks (v. 2.0.2).
To find reads that are unique to a transcript, the non-redundant RNA-Seq reads from the SKBR3, SUM149 and SUM190 fastq files were aligned against the Ensembl cDNA sequences using NCBI blastn 20. For each dataset, a non redundant reads file was made from the paired end reads. Six bases from both ends of the reads were removed. Sequence alignments with > 95% identity for the full length of the trimmed reads with no gaps were considered true matches. We estimated the total number of distinct reads for the Ensembl protein coding transcripts; only the transcripts identified with unique reads were further analyzed.
Human Mammary Epithelial Cells (HMEC)
To compare the splice variants identified in the breast cancer cells against a normal breast cell, we downloaded the RNA-Seq dataset for HMEC (normal human mammary epithelial cells) from the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra). The accession number for HMEC is SRX061998 containing data from two runs. RNA-Seq analysis and blast search for unique reads were conducted as described above. We downloaded the tryptic peptides (FDR < 1%) identified in HMEC by Geiger et al 21 and integrated with the RNA-Seq data to produce the list of splice variant proteins.
Known Splice Variant (SpV) Identifications
In our previous publications22, 23 we have annotated spliced proteins as ‘Alternative Splice Variant’ (ASV). However, with the utilization of RNA-Seq data, the ASV abbreviation may lead to confusion between spliced protein and spliced transcript. Hence, we now annotate spliced transcripts as ‘SpTs’ and spliced proteins as “SpPs”; “SpVs” refer to both.
The mzXML data from the mass spectrometric analyses were searched against a custom-built ECGene22, 23 database using X! Tandem. Briefly, the database was created as follows: mRNA sequences of the predicted models from the ECgene (downloaded previously from http://genome.ewha.ac.kr/ECgene) and Ensembl transcripts (version 70) were translated in three reading frames. Within each dataset, the first instance of each protein sequence longer than 14 amino acids was recorded. The resulting proteins from both database translations were then combined and filtered for redundancy. For this filtering, proteins derived from Ensembl transcripts were preferentially recorded over those generated from ECgene records. A collection of common protein contaminant sequences was added to this set (115 sequences; ftp://ftp.thegpm.org/fasta/cRAP/). Lastly, all sequences were reversed and appended to the set of forward sequences as an internal control for false identifications.
The mass spectra search parameters included trypsin specificity up to three missed cleavages, carbamidomethyl as a complete modification and oxidation of methionine and threonine as variable modifications. Peptide identification was determined using a parent ion mass error of 50ppm and fragment ion tolerance of 0.8 Da. The use of the concatenated target-decoy database enabled us to calculate peptide spectral match FDR based on the X! Tandem expect value. We used empirical level peptide FDR (PSM-level FDR) over factual level peptide FDR as studies have shown that in most cases the empirical peptide-level FDR reliably estimates the peptide-level FDR24. Peptides identified with PSM-level FDR < 1% were considered for further analyses.
Splice variant proteins share high sequence identities; most peptides identified from a mass spectrometric analysis are shared by multiple variants. Hence, our approach was to use the corresponding transcript expression as the confirmation of a variant that is identified only by non-unique peptides from mass spectrometric analyses. Due to high sequence coverage in RNA-Seq data, reads from unique regions are more likely to be found by this method; moreover, the RNA-Seq reads from the UTR regions allow us to identify the transcripts of smaller proteins that share their entire peptide sequences with that of the corresponding canonical variants. However, we do recognize that our RNA-Seq and proteomic datasets come from different preparations of the same cell lines.
The expected number of distinct reads mapped to transcript is proportional to its length. Supplementary document part 1 shows the average number of distinct reads for the transcripts identified in HMEC grouped according to the transcript length. The ratio of the average number of distinct reads per average transcript length for each group in HMEC was ~ 0.04.
The analysis pipeline for identification of known splice variants (SpVs) and novel peptides is shown in Figure 1. Splice variants (SpVs) are considered as expressed in the sample studied if:
Peptides identified with PSM-level FDR < 1% from the X! Tandem search matched to the known Ensembl proteins that are derived from genes known to have multiple protein coding transcripts.
Corresponding protein-coding transcripts for these Ensembl proteins are found in the RNASeq data of the cell line types.
At least one unique read mapped to these transcripts by blast analyses. 4. The ratio of the number of distinct reads to the transcript length was >= 0.04.
Biological Annotations
Enrichment analysis for Gene Ontology (GO) was done using the R package topGO (http://www.bioconductor.org/packages/2.11/bioc/html/topGO.html). Using GSEA (Gene set enrichment analysis) with MsigDB 25 (http://www.broadinstitute.org/gsea/msigdb/annotate.jsp) we computed the overlap of the SpVs expressed in the breast cancer cell lines with the gene sets derived from the BioCarta pathways. BioCarta integrates proteomic information for pathway annotations. DAVID bioinformatics resources were used to do the functional annotation clustering (http://david.abcc.ncifcrf.gov/home.jsp). We used a cutoff of p < 0.05 for the enrichment analysis. DAVID provides a comprehensive set of functional annotation tools to understand biological meanings behind large lists of genes.
We have previously benchmarked the I-TASSER pipeline for structure modeling of pairs of protein isoforms which are known to have experimentally-solved structures in PDB5. The average RMSD between the experimentally-determined structure and the model predicted by I-TASSER was 1.72 Å. Generally, a structure model within 4-6 Å has a similar fold/topology to the native. We used I-TASSER to predict the structures of ERBB2 and EGFR splice variants expressed in breast cancer cell lines.
Novel Peptide Identifications
For distinct peptides (< 1% FDR) identified in searching the proteomics data against the custom built ECGene database that did not match to any known proteins (Figure 1), we sought mRNA evidence in the RNA-Seq reads. This was accomplished by querying the reverse translated cDNA sequence of the peptide against the non-redundant list of reads identified from each cell line.
Results
Known Splice Variant Protein (SpP) Identifications in Breast Cancer Cells
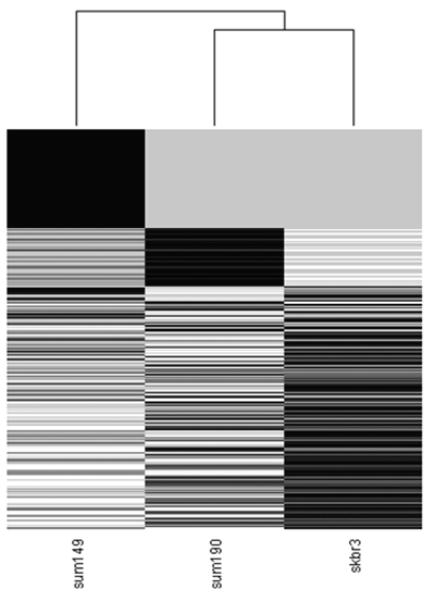
As shown in the Figure 1 pipeline, the integration of proteomics with RNA-Seq data enabled us to identify splice variant proteins expressed in the breast cancer cells. We identified 2684 (1362 genes), 1886 (894 genes) and 3124 (1435 genes) splice variant proteins in the SKBR3, SUM190 and SUM149 cell lines (Table 1a, Figure 2, Supplementary Tables 1-3) respectively. In total of 4406 distinct transcripts were identified from the three breast cancer cell lines, with 1052 splice variants in common. The heat map of the relative expressions of the transcripts in the three breast cancer cells shows SUM190 and SKBR3 as more similar to each other than to SUM149 (Figure 2).
Table 1a.
Summary of the known splice variant proteins identified from SKBR3, SUM190 and SUM149 analyses
| Cell line studied |
No. of total distinct peptides identified * |
No. of distinct known splice variant proteins with peptide evidence |
Total no. of distinct known splice variant proteins identified with peptide and transcript evidence |
No. of distinct genes with at least one of its splice variant protein identified with peptide and transcript evidence |
No. of distinct genes with more than one splice variant proteins identified with peptide and transcript evidence |
|---|---|---|---|---|---|
| SKBR3 | 8565 | 4478 | 2684 | 1362 | 712 (2034)** |
| SUM190 | 4361 | 3161 | 1886 | 894 | 508 (1500) ** |
| SUM149 | 8838 | 4598 | 3124 | 1435 | 831 (2520)** |
Only peptides with < 1% FDR used in the analyses
Number in parentheses indicate the total number of distinct splice variant proteins identified from these genes
Figure 2.
Heat map showing the total distinct RNA Seq reads for the transcripts identified in SKBR3, SUM190 and SUM149. SUM190 and SKBR3 are clustered together when compared to SUM149.
With regard to more than one isoform of a gene expressed in a cell line: 2034 variants from 712 genes were identified in SKBR3, 1500 from 508 genes in SUM190 and 2520 from 831 genes were expressed (Figure 2). From all three breast cancer cell lines, we found 1167 genes with more than one transcript expressed (3609 transcripts).
Supplementary Table 4 shows the top 100 enriched Gene Ontology Biological process terms for the 4406 splice variants identified from the three breast cancer cell lines. Terms related to apoptosis, cell division, glucose metabolism, protein synthesis, signal transduction and splicing were among the top 100 enriched terms.
Known Variant Identifications in HMEC
Peptides (46909 distinct peptides) downloaded from the HMEC proteomic study were aligned to 31323 known Ensembl proteins. The RNA-Seq data integration and unique read analyses yielded 7186 distinct splice variants as expressed in HMEC (Supplementary Table 5). Due to the large-scale high-resolution proteomic analyses, the total number of SpVs identified in HMEC is larger than that of the breast cancer cell lines. However, the average number of splice variants per gene expressed in the three breast cancer cell types was higher than that of HMEC (2.0 versus 1.5).
Epidermal Growth Factor Receptor (EGFR) and ERBB2 Variants
Due to the recognized interacting roles of EGFR and ERBB2 in breast cancers 26, we examined the different splice variants of these genes expressed in the three breast cancer cell types. Table 1b shows the list of the variants identified from the cell lines studied. For the sake of readability we have given abbreviated symbols for these variants that specify their protein lengths.
Table 1b.
Splice variants of ERBB2 and EGFR identified by our integrated analyses in SKBR3, SUM149 and SUM190. √ indicates that the variant was expressed in the cell line.
| ERBB2 variants | |||||
|---|---|---|---|---|---|
| Ensembl Transcript ID | Ensembl Protein ID | Abbreviated symbol * |
SKBR3 | SUM190 | SUM149 |
| ENST00000578709 | ENSP00000463719 | ERBB2-102 | √ | ||
| ENST00000578199 | ENSP00000462808 | ERBB2-605 | √ | √ | |
| ENST00000584450 | ENSP00000463714 | ERBB2-1055 | √ | √ | |
| ENST00000540147 | ENSP00000443562 | ERBB2-1225-1 | √ | √ | √ |
| ENST00000584601 | ENSP00000462438 | ERBB2-1225-2 | √ | √ | √ |
| EGFR variants | |||||
| ENST00000450046 | ENSP00000413354 | EGFR-128 | √ | ||
| ENST00000420316 | ENSP00000413843 | EGFR-405 | √ | √ | |
| ENST00000342916 | ENSP00000342376 | EGFR-628 | √ | ||
| ENST00000442591 | ENSP00000410031 | EGFR-657 | √ | √ | |
| ENST00000344576 | ENSP00000345973 | EGFR-705 | √ | √ | |
| ENST00000455089 | ENSP00000415559 | EGFR-1091 | √ | ||
| ENST00000275493 | ENSP00000275493 | EGFR-1210 | √ | √ | |
For the sake of readability, we have given abbreviated symbols for the multiple variants of ERBB2 and EGFR. The number following the gene symbol denotes the length of the protein (number of amino acids)
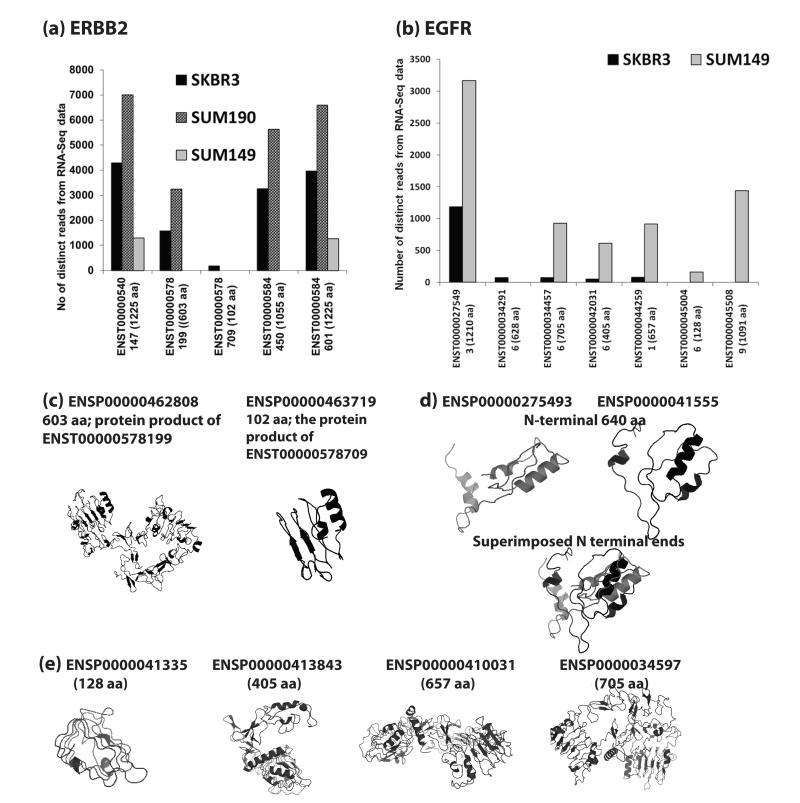
The integrative analyses indicate more than one ERBB2 isoform is expressed in all three breast cancer cell lines (Figure 3a). According to our analyses, five ERBB2 transcripts were expressed in SKBR3, 4 of the 5 variants were expressed in SUM190, and 2 were expressed in SUM149. Figure 3a shows the number of distinct reads mapped to the transcripts expressed in the cell lines. In SUM149, the two transcripts expressed (ERBB2-1225-1, ERBB2-1225-2) translated to the same protein sequence of 1225 amino acids (aa) length. In SKBR3, we found three other transcripts, ERBB2-102, ERBB2-603 and ERBB2-1055. In SUM190, ERBB2-603, ERBB2-1055, ERBB2-1225-1 and ERBB2-1225-2 were expressed. The variant ERBB2-1055 was highly expressed in the ERBB2 amplified cell lines, SKBR3 and SUM190, but was absent in SUM149. The comparison of the protein sequence of the variant ERBB2-1225-1 to that of ERBB2-1055 revealed that the shorter variant is missing the translated sequence of exon 27 at the C-terminal end. MotifScan27 analysis of the 170 amino acids missing in the truncated translated product of ERBB2-1055 showed a proline-rich region. The variant ERBB2-603 expressed both in SKBR3 and SUM190 matched to the N-terminal extracellular domains of ERBB2-1225-1 and ERBB2-1225-2. A short protein, ERBB2-102, was expressed only in SKBR3. The sequence matched to N terminal 102 amino acids of the long ERBB2-1225 variants and contains one L-Receptor domain. Figure 3c shows the I-TASSER predicted three-dimensional structures of the protein products of ERBB2-603 and ERBB2-102 with TM-scores 0.89 and 0.83, respectively; a TM-score >0.5 indicates a model of correct topology.
Figure 3.
(a) The relative transcript expression levels based on total distinct RNA-Seq reads for the ERBB2 variants expressed in SKBR3, SUM190 and SUM149. The numbers in parentheses next to the transcript ID is the length of the translated product. The variants ENST00000540147 and ENST00000584601 translated to the same protein product of 1225 amino acids (aa) length. (b) The relative transcript expression levels based on total distinct RNA-Seq reads for the EGFR variants expressed in SKBR3 and SUM149. No EGFR variants were identified by our analysis in SUM190. (c) The three dimensional structures predicted by I-TASSER for the translated products of ERBB2 splice variants, ENST00000578199 and ENST00000578709. The TM-scores of the models were 0.89 and 0.83. (d) The three dimensional structures predicted by I-TASSER for the N-terminal 640 amino acid regions of the translated products of the splice variants ENST00000275493 (ENSP00000275493, 1210 aa) and ENST00000455089 (ENSP00000415559, 1091 aa). Even though both sequences contain all the conserved domains, due the absence of the translated sequence from exon 4 in ENSP00000415559, the relative positions of the extracellular domains are shifted compared to that of the canonical protein, ENSP00000275493. (e) The three dimensional structures predicted by I-TASSER for the translated products of EGFR splice variants, ENST00000450046 (ENSP00000413354, 128 aa), ENST00000420316 (ENSP00000413843, 405 aa), ENST00000442591 (ENSP00000410031, 657 aa) and ENST00000344576 (ENSP00000345973, 705 aa). The TM-scores of these structures were 0.88, 0.74, 0.7 and 0.64 respectively.
The integrated analyses did not find any variant of EGFR in SUM190. We identified six variants of EGFR in SUM149 and five in SKBR3 (Table 1b, Figure 3b). The long canonical variant EGFR-1210, was expressed both in SKBR3 and SUM149 (Figure 3b). The numbers of total distinct reads suggest that shorter variants that were expressed in both cell lines were more highly expressed in SUM149 compared to SKBR3 (Figure 3b). The variant EGFR-1091 was expressed only in SUM149 with a high number of distinct reads. The protein product of EGFR-1091 differs from the canonical protein EGFR-1210 by 119 amino acids. Sequence analysis indicated that EGFR-1091 is missing the translated protein sequences from exons 4 and 28 found in EGFR-1210. Figure 3d shows the predicted structures of the N-terminal 640 amino acids from EGFR-1091 and EGFR-1210. The absence of exon 4 in the N-terminal end of EGFR-1091 results in a shift in its structure compared to the canonical variant. Figure 3e shows the predicted three-dimensional protein structures of four other smaller variants expressed in SUM149. The TM-score values of the predicted structures of these 4 variants were 0.88, 0.74, 0.7 and 0.64, respectively. The variants that translated to the proteins that are 705 aa and 657 aa long contain all the conserved domains (two L Receptor domains and two Furin-like repeats) found in the extracellular region of the long canonical variant. The variant which translated to a 405 aa long protein contains one L-Receptor domain and one Furin-like repeat. The shortest protein with 128 amino acids contains one L Receptor domain. The L domains from these receptors make up the bilobal ligand binding site.
Pathway Analysis
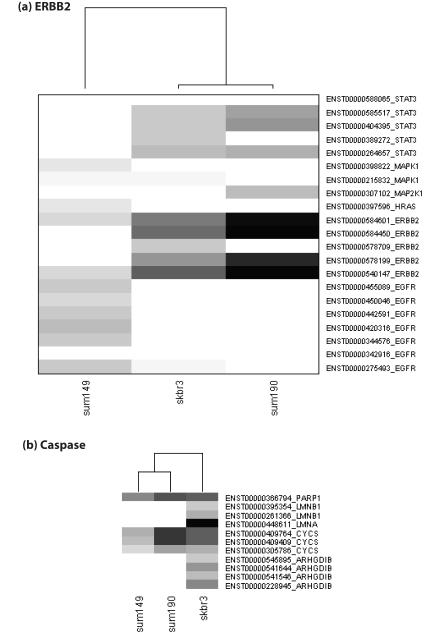
The top 100 BioCarta Genesets in MSigDb that overlapped significantly with the splice variants expressed in the three breast cancer cell lines are given in Supplementary Table 6. The heat map generated from the total distinct reads of the transcripts linked to BioCarta ERBB2 (Her2) signaling has SKBR3 and SUM190 clustered together (Figure 4a). In addition to ERBB2, multiple variants of STAT3 were expressed in SKBR3 and SUM190 but not in SUM149. Variants of MAPK1 were expressed in SUM149 and SUM190, but absent in SKBR3 (Figure 4a).
Figure 4.
Heat map showing the total distinct RNA Seq reads for the transcripts expressed in SKBR3, SUM190 and SUM149 linked to BioCarta pathways including (a) ERBB2 signaling (b) Caspase signaling.
The downstream ERBB receptor signaling affects biological mechanisms such as cell cycle, cell adhesion, cell motility, and apoptosis. Pathways linked to these mechanisms were found in the top 100 from the enrichment analyses (Figure 4, Supplementary document parts 2- 4, Supplementary Table 6). The caspase pathway transcript profile seems similar for SUM149 and SUM190 (Figure 4b). Variants of PARP1 and CYCS were expressed in these two cell lines. However, the transcript expression indicates variants of more genes including ARHGDIB, LMNA and LMNB1 involved in caspase pathway were expressed in SKBR3 compared to SUM149 and SUM190 (Figure 4b).
Supplementary documents part 2- 4 show heat maps for pathways including ucalpain cell motility, mcalpain cell motility, electron transport chain, G2/M check point in cell cycle, glycolysis and mRNA splicing. The expression profiles of these pathways show different clustering between the three breast cancer datasets. In electron transport chain, mcalpain, G2/M check point in cell cycle and ucalpain pathways, SKBR3 and SUM190 are clustered together. For the heat map of the transcripts involved in G2/M check point in cell cycle, two variants of ATR were expressed in SUM149 and absent in SKBR3 and SUM190. YWHAQ variants expressed at varying levels were found in all three breast cancer cell lines (Supplementary document part 3). SUM149 and SKBR3 are clustered together in glycolysis. For the heat map of the variants involved in the Rho cell motility signaling pathway, multiple variants of ARPC1B, CFL1, GSN, and PFN1 were expressed at different levels in the three cancer cell lines. For this pathway, the expression profiles of SKBR3 and SUM149 were similar. Variants of ROCK1 and SRC were identified only in SUM149. The heat map indicates that SUM190 and SUM149 are similar in mRNA splicing (Supplementary document part 4); however, we find multiple variants of splice factors including eftud2, nhp2l1, pcbp2, ptbp1, snpra, snrpd2, snrpe and ybx1 expressed at varying levels in the three breast cancer cell lines.
Similarities between the ERBB2 over-expressed SKBR3 and SUM190 cell lines
We found 138 transcripts from 92 genes identified with multiple unique reads in SKBR3 and SUM190 that were not in SUM149. Enrichment analyses using DAVID indicated mechanisms including Electron Transport Chain, intracellular transport and phosphate metabolic processes (Table 2, Supplementary Table 7). TMED proteins with GOLD domains were enriched.
Table 2.
Functional Annotation Clustering of the genes with one or more of its variants expressed only in SKBR3 and SUM190 compared to SUM149
| Term | PValue | Genes |
|---|---|---|
| Cluster 1 Enrichment Score: 8.42 | ||
| mitochondrial part | 0.0000 | CYB5R3, DLST, NDUFA5, NDUFA2, ALDH18A1, UQCRC1, NDUFA9, OGDH, NDUFA10, UQCRQ, IDH3A, HADHA, UQCRH, MCCC1, GSTK1, NDUFS8, NDUFV2, COX6B1, NDUFS1, ETFA |
| Cluster 2 Enrichment Score: 7.56 | ||
| electron transport chain |
0.0000 | NDUFA5, NDUFA2, UQCRC1, UQCRH, NDUFA9, NDUFV2, NDUFS8, NDUFA10, UQCRQ, NDUFS1, GLRX, ETFA |
| Cluster 3 Enrichment Score: 4.18 | ||
| vesicle-mediated transport |
0.0003 | COPB2, KDELR2, SEC31A, ARF3, COPZ1, PPT1, RAB6B, DOPEY2, GOSR1, CLTC, SAR1B, SAR1A |
| Cluster 4 Enrichment Score: 2.63 | ||
| domain:GOLD | 0.0028 | TMED4, TMED5, TMED9 |
| Cluster 5 Enrichment Score: 2.30 | ||
| Cardiac muscle contraction |
0.0047 | UQCRC1, UQCRH, COX6B1, TPM1, UQCRQ |
| Cluster 6 Enrichment Score: 2.24 | ||
| Citrate cycle (TCA cycle) |
0.0024 | DLST, IDH2, OGDH, IDH3A |
| Cluster 7 Enrichment Score: 1.91 | ||
| Golgi-associated vesicle |
0.0137 | COPB2, COPZ1, CLTC |
| Cluster 8 Enrichment Score: 1.66 | ||
| phosphate metabolic process |
0.0178 | NDUFA5, NDUFA2, NCEH1, UQCRC1, UQCRH, NDUFA9, ERBB2, NDUFV2, NDUFS8, NDUFA10, NDUFS1, PPA2 |
| Cluster 9 Enrichment Score: 1.63 | ||
| intracellular protein transport |
0.0174 | COPB2, KDELR2, COPZ1, GOSR1, CLTC, SAR1B, SAR1A |
| Cluster 10 Enrichment Score: 1.48 | ||
| membrane-bounded vesicle |
0.0237 | ANXA6, STOM, COPB2, LAMP2, SEC31A, COPZ1, PPT1, RAB6B, CLTC |
| Cluster 11 Enrichment Score: 1.12 | ||
| hsa04142: Lysosome |
0.0192 | LAMP2, GM2A, PSAP, PPT1, CLTC |
| Cluster 12 Enrichment Score: 1.08 | ||
| iron-sulfur cluster binding |
0.0329 | NDUFV2, NDUFS8, NDUFS1 |
| Cluster 13 Enrichment Score: 1.02 | ||
| GTPase activity | 0.1192 | ARF3, RALA, RAB6B, SAR1A |
Similarities between the IBC models SUM190 and SUM149 cell lines
We found 201 variants with multiple unique reads in SUM190 and SUM149 that were not in SKBR3. The enrichment analyses indicated terms related to multiple mechanisms including vesicle ATP binding, GTP binding, RNA binding, citrate cycle, Aminoacyl-tRNA synthesis, RNA translation, protein localization and Ras GTPase activity (Table 3, Supplementary Table 7). Heat repeat domains were enriched.
Table 3.
Functional Annotation Clustering of the genes with one or more of its variants expressed only in SUM190 and SUM149 compared to SKBR3
| Term | PValue | Genes |
|---|---|---|
| Cluster 1 Enrichment Score: 6.00 | ||
| ribonucleotide binding | 0.0000 | HSP90AB1, ADSS, ATL3, XRCC6, DTYMK, RAB1B, UBA6, ASNS, CCT3, KARS, WARS, ACTR2, LONP1, ACTR1A, TUBA1A, POTEF, HSPA8, HSPA9, RAB2A, ABCE1, YARS, RAB8B, EIF2S3, OLA1, MCM4, RECQL, ATP2A2, ARF1, ILF2, UBE2K, PSMC3, EIF4A1, CCT8, FARSB, TUBA4A, RAP1A, FARSA, ATP5A1 |
| Cluster 2 Enrichment Score: 5.87 | ||
| cytoplasmic vesicle | 0.0003 | RAB2A, HSP90AB1, YWHAZ, RAB8B, YWHAB, SLC3A2, NAP1L1, ACTN1, CANX, ANXA2, SLC1A5, PICALM, TFRC, TMEM33, CTSD, SEC23B, HSPA8 |
| Cluster 3 Enrichment Score: 3.28 | ||
| ATP binding | 0.0006 | HSP90AB1, XRCC6, DTYMK, UBA6, ASNS, CCT3, KARS, WARS, ACTR2, LONP1, ACTR1A, POTEF, HSPA8, HSPA9, ABCE1, YARS, OLA1, MCM4, RECQL, ILF2, ATP2A2, UBE2K, PSMC3, CCT8, EIF4A1, FARSB, ATP5A1, FARSA |
| Cluster 4 Enrichment Score: 3.04 | ||
| nicotinamide nucleotide metabolic process | 0.0004 | LDHB, KYNU, IDH1, DCXR, MDH1 |
| Cluster 5 Enrichment Score: 2.95 | ||
| Aminoacyl-tRNA biosynthesis | 0.0017 | WARS, YARS, FARSB, FARSA, KARS |
| Cluster 6 Enrichment Score: 2.88 | ||
| protein folding | 0.0213 | HSP90AB1, CCT8, CCT3, CANX, HSPA8, HSPA9 |
| Cluster 7 Enrichment Score: 2.77 | ||
| GTP binding | 0.0030 | RAB2A, ADSS, RAB8B, ARF1, ATL3, OLA1, TUBA4A, EIF2S3, RAB1B, RAP1A, TUBA1A |
| Cluster 8 Enrichment Score: 2.75 | ||
| microsome | 0.0067 | MGST3, ATP2A2, CYP51A1, SEC11A, LRRC59, SYNCRIP, SPCS2, MGST1 |
| Cluster 9 Enrichment Score: 2.47 | ||
| RNA recognition motif, RNP-1 | 0.0257 | HNRNPL, PTBP1, GRSF1, ESRP1, SYNCRIP, MATR3 |
| Cluster 10 Enrichment Score: 2.07 | ||
| membrane-enclosed lumen | 0.0109 | XPO1, HMGB2, MTDH, XRCC6, SYNCRIP, SERPINH1, KARS, CTNNB1, HNRNPL, RPA2, LONP1, NUMA1, RPS3A, LRRC59, MSN, HSPA9, SHMT2, RBBP4, PTBP1, CS, YWHAB, ACTN1, MCM4, PA2G4, ILF2, TXNDC5, ATP5A1, MATR3 |
| Cluster 11 Enrichment Score: 1.96 | ||
| Cell cycle | 0.2140 | YWHAZ, YWHAB, YWHAQ, MCM4 |
| Cluster 12 Enrichment Score: 1.92 | ||
| translation initiation factor activity | 0.0205 | EIF4G3, EIF4A1, EIF2S3, EIF3M |
| Cluster 13 Enrichment Score: 1.61 | ||
| Pyruvate metabolism | 0.0141 | LDHB, LDHA, ALDH7A1, MDH1 |
| Cluster 14 Enrichment Score: 1.54 | ||
| cellular protein localization | 0.0312 | XPO1, YWHAZ, IPO4, YWHAB, YWHAQ, SRP72, SEC23B, CTNNB1, HSPA9 |
| Cluster 15 Enrichment Score: 1.50 | ||
| Citrate cycle (TCA cycle) | 0.0590 | CS, IDH1, MDH1 |
| Cluster 16 Enrichment Score: 1.44 | ||
| purine nucleotide metabolic process | 0.0856 | ADSS, LONP1, ATP2A2, OLA1, ATP5A1 |
| Cluster 17 Enrichment Score: 1.35 | ||
| protein localization | 0.0516 | RAB2A, XPO1, YWHAZ, RAB8B, YWHAB, RAB1B, CANX, CTNNB1, ARF1, IPO4, YWHAQ, SRP72, SEC23B, HSPA9 |
| Cluster 18 Enrichment Score: 1.34 | ||
| repeat:HEAT | 0.0253 | XPO1, EIF4G3, IPO4 |
| Cluster 19 Enrichment Score: 1.30 | ||
| Ras GTPase | 0.0905 | RAB2A, RAB8B, RAB1B, RAP1A |
| Cluster 20 Enrichment Score: 1.26 | ||
| Pyruvate metabolism | 0.0141 | LDHB, LDHA, ALDH7A1, MDH1 |
Breast Cancer Cell-Line Specific Variants
We looked for splice variants expressed in only one breast cancer cell line compared to the other two cancer cell lines and in the normal HMEC. We found 396 distinct splice variants from 295 genes in SKBR3, 186 variants from 131 genes in SUM190, and 598 variants from 422 genes in SUM149 (Supplementary Table 8). Table 4 shows the table with the top 5 GO Biological Process terms and BioCarta Pathways associated with these cancer cell-line specific variants. Amino sugar metabolism, caspase activity, arrestin activation of MAP kinases, and endocytosis by NDK, phosphins and dynamins were among the top terms in SKBR3. Different aspects of metabolism, especially of lipids, were among the top terms in SUM190. In SUM149, cell-to-cell adhesion, integrin signaling, Erk1/Erk2 Mapk signaling, K48-linked ubiquination, and translational control by eIF4e and p70S6 were among the top enriched terms.
Table 4.
Top 5 enriched Gene Ontology (GO) Biological Processes and BioCarta Pathways for the splice variants expressed only in one breast cancer cell type compared to the other two breast cancer cell lines and normal human epithelial cells
| Gene Ontology | BioCarta Pathway | |
|---|---|---|
| SKBR3 | ||
| GO:00 15986 |
ATP synthesis coupled proton transport |
Role of fl-arrestins in the activation and targeting of MAP kinases |
| GO:00 00272 |
polysaccharide catabolic process | Proteasome Complex |
| GO:00 06096 |
glycolysis | Eukaryotic protein translation |
| GO:19 01071 |
glucosamine-containing compound metabolism |
Caspase Cascade in Apoptosis |
| GO:00 06022 |
aminoglycan metabolic process | Endocytotic role of NDK, Phosphins and Dynamin |
| SUM190 | ||
| GO:00 51346 |
negative regulation of hydrolase activity |
Genes involved in Metabolism of amino acids and derivatives |
| GO:00 08610 |
lipid biosynthetic process | Amino sugar and nucleotide sugar metabolism |
| GO:00 51291 |
protein heterooligomerization | Genes involved in Metabolism of lipids and lipoproteins |
| GO:00 06690 |
icosanoid metabolic process | Pyruvate metabolism |
| GO:00 06767 |
water-soluble vitamin metabolic process |
Genes involved in Metabolism of vitamins and cofactors |
| SUM149 | ||
| GO:00 50900 |
leukocyte migration | Regulation of eIF4e and p70 S6 Kinase |
| GO:20 01236 |
regulation of extrinsic apoptotic signaling |
Cell to Cell Adhesion Signaling |
| GO:00 08612 |
peptidyl-lysine modification to hypusine |
Integrin Signaling Pathway |
| GO:00 34329 |
cell junction assembly | Role of Ran in mitotic spindle regulation |
| GO:00 70936 |
protein K48-linked ubiquitination | Erk1/Erk2 Mapk Signaling pathway |
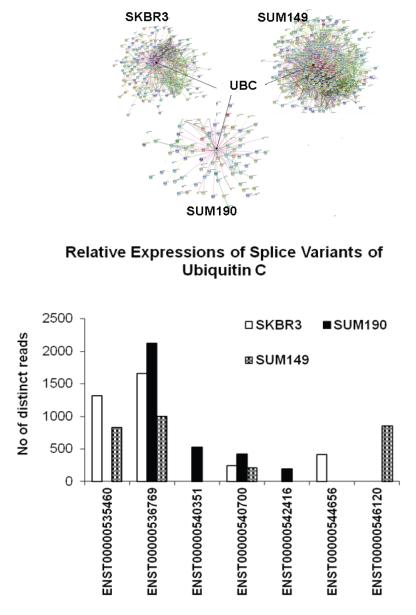
Using STRING (http://string-db.org/), we were able to visualize the protein interaction networks for these cell line-specific variants (Supplementary document parts 5-7) from the three cell line models. STRING is a database of known and predicted protein interactions. The interactions include physical and functional associations derived from sources including co-expression, literature, genomic context and high-throughput experiments. All three networks were enriched with protein interactions with Ubiquitin C (UBC) as the center for the networks. Identification of multiple variants of UBC with at least one unique variant in each breast cancer cell line is worthy of note (Figure 5). The protein sequences of these unique variants differ from the canonical long variant ENST00000536769 (685 aa, ENSP00000441543) which was found in all three cancer cells mainly by the number of ubiquitin domains; the canonical protein has nine ubiquitin domains while the unique variant specific to SKBR3 and SUM190 have two ubiquitin domains and the unique variant in SUM149 has eight.
Figure 5.
The interaction networks for the breast cancer cell line specific variants were created using STRING. We used only the sources with high confidence and direct interaction between the input genes. The protein interactions for all three networks were significantly enriched. Ubiquitin C (UBC) was found as the center of the network (See Supplementary document parts 5-7). Seven splice variants of UBC were identified. One variant each was unique to SKBR3 and to SUM149 and two were unique to SUM190. The canonical variant ENST00000536769, (ENSP00000441543, 685 aa) was expressed in all three cancer cell lines.
In order to substantiate the specificity of the breast cancer cell-line specific variants, we compared these variants to the proteins identified from three colorectal cancer (CRC) cell lines; Fanayan et al identified 4522 distinct proteins from proteogenomic analyses of three CRC cell-lines including LIM1215, LIM1899 and LIM240528. Interestingly, nearly 90% of the breast cancer cell-line variants we identified were not found in the CRC cell-lines (353 out of 396 in SKBR3, 168 out of 186 in SUM190 and 525 out of 598 in SUM149).
Novel Peptide Identifications
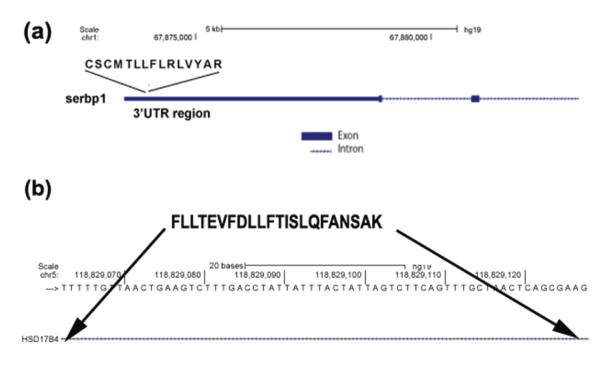
Table 5 shows the 10, 5, and 13 novel peptides (FDR < 1%) identified from SKBR3, SUM190 and SUM149 that did not match to any known protein sequences, but had evidence in RNA-Seq data (Supplementary document part 8 has the MS/MS spectra of the novel peptides). Since the UTR regions are part of the mRNA sequences, the occurrence of RNA-Seq reads that translated to the novel peptides identified from the 5′ and 3′ UTR regions may not exactly confirm these peptides. However, we were able to find mRNA evidence for the multiple peptides identified from intronic regions and for peptides resulting from alternate splice sites. Figure 6 shows the schematic diagrams of two novel peptides identified. Figure 6a shows the peptide ‘CSCMTLLFLRLVYAR’ identified from SKBR3 that aligned to the 3′UTR region of SERPINE1 mRNA binding protein 1 (SERBP1). Figure 6b shows the novel peptide ‘FLLTEVFDLLFTISLQFANSAK’ identified in SUM149 and SUM190 that matched to the intronic region of Hydroxysteroid (17-beta) dehydrogenase 4 (HSD17B4).
Table 5.
The novel peptides identified from SKBR3, SUM149 and SUM190 proteomic analyses with confirming RNA-Seq reads
| SKBR3 | |||
|---|---|---|---|
| Novel Peptide | Gene description | Gene Symbol | Location of the novel peptide or possible cause |
| ACISRGFLGSPGR | tripartite motif containing 39 | TRIM39 | 5′UTR |
| CSCMTLLFLRLVYAR | SERPINE1 mRNA binding protein 1 | SERBP1 | 3′UTR |
| GAPEPAQTQPQPQPQPAAPE GPEQPR |
ER degradation enhancer, mannosidase alpha-like 2 |
EDEM2 | intron |
| GGGRYWGDVEPTLLR | AF338194 | AF338194 | |
| HLFFVFSWALELK | interferon-related developmental regulator 1 |
IFRD1 | intron |
| NCSNCQTDSSFCPASR | cytochrome P450, family 20, subfamily A |
CYP20A1 | different frame |
| NDDIPEQDSLGLSNLQK | McKusick-Kaufman syndrome | MKKS | 5′UTR |
| RQEGQAVGAPTLLR | bromodomain containing 3 | BRD3 | different frame |
| SLTSLDTPLANSPSTAPQAATL SLGLR |
surfactant protein A1 | SFTPA1 | 3′ UTR |
| SRLSIAAGGVMDVNTALQEVL K |
ribosomal protein S12 | RPS12 | alternate 5′splice site |
| SUM190 | |||
| DPSQDGPDGCCSCMGFR | mechanistic target of rapamycin | MTOR | different frame |
| FLLTEVFDLLFTISLQFANSAK | hydroxysteroid (17-beta) dehydrogenase 4 |
HSD17B4 | intron |
| LPITITTIPTIGFNVETVEYK | ADP-ribosylation factor 1 | ARF1 | alternate 5′ splice site |
| LTQATFIILPLVLPQILLK | trafficking protein particle complex 2 | TRAPPC2 | intron |
| PGIKLWMSGNGTLCSPVHR | zinc finger protein 708 | ZNF708 | intron |
| SUM149 | |||
| AAQLTAFALLQAQLR | uncharacterized LOC152217 | LOC152217 | |
| AGTEEAEEGFQNWTKAGR | EST DA435764 | EST DA435764 |
|
| CGQCGSLEGPCTSGEDHR | EST DB501538 | EST DB501538 |
|
| ETSCDNCCLCPCCVK | ribonuclease P/MRP 25kDa subunit | RPP25 | 3′UTR |
| ELCVVPLHALLGPSGPVHSPG TVWQGRSR |
claudin 15 | CLDN15 | intron |
| FLLTEVFDLLFTISLQFANSAK | hydroxysteroid (17-beta) dehydrogenase 4 |
HSD17B4 | intron |
| HLAQPGPDLRAATTSSVCPLIK | chromosome 10 open reading frame 116 |
C10ORF116 | 3′UTR |
| LLAALLHSPQLVER | uncharacterized LOC152217 | LOC152217 | |
| MNINELIRSSSLFVAFQR | erythrocyte membrane protein band 4.1 like 5 |
EPB41L5 | 3′UTR |
| NDDIPEQDSLGLSNLQK | McKusick-Kaufman syndrome | MKKS | 5′UTR |
| TQTEPPTFLVELSR | importin b1 | KPNB1 | alternate 5′ splice site |
| VPGSLRILVSSETAVGILR | aspartate beta-hydroxylase | ASPH | intron |
| VQGLVASNLNLKPGECLR | lectin, galactoside-binding, soluble, 1 | LGALS1 | alternate 5′ splice site |
Figure 6.
(a) Schematic diagram of the novel peptide ‘CSCMTLLFLRLVYAR’ identified from SKBR3 analysis. This novel peptide matched to the 3′ UTR region of SERPINE1 mRNA binding protein 1 (serbp1). (b) Schematic diagram of the novel peptide ‘FLLTEVFDLLFTISLQFANSAK’ identified from SUM190 and SUM149 that matched to the intronic region of Hydroxysteroid (17-beta) dehydrogenase 4 (hsd17b4). We used UCSC Blat to map the peptide to the genome.
Discussion
The diversity of expressed proteins increases as cancer progresses; motility, survival in distinct niches, and metabolic adaptations regulate cellular homeostasis as the environment of the cancer changes. In order to understand the tumor evolution in different metastatic ecosystems, it is important to be able to assess the full spectrum of variability in expression of signaling and metabolic proteins. In this study, we combined high-throughput proteomic and RNA-sequencing technologies, along with bioinformatics, to identify known SpVs and novel peptides (Figure 1) expressed in the three hormone receptor negative breast cancer cell lines SKBR3, SUM190, and SUM149 and to annotate the mechanisms involving them. A similar approach has been used by Ning and Nesvizhiskii 29 to identify novel alternatively spliced isoforms.
The yield of SpVs unambiguously identified based on proteotypic peptides from mass spectrometry is low, as the majority of peptides identified are shared by the variants. In our analyses, the corresponding RNA-Seq transcript expression was used as a validation for the SpV with peptide evidence. The stringent analytic criteria and validations from both proteomic and transcriptomic data confirm the splice variant identifications as highly confident.
By our integrated analysis, we found many known SpVs (Table 1a), supporting the strength of the approach, as well as novel peptides (Table 5) expressed in these three breast cancer cell lines, indicating the sensitivity of our analyses. Moreover, we identified cancer cell-line specific variants that were found only in one cell type compared to the other two and the normal mammary epithelial cells. The top enriched GO biological processes for the variants expressed in the breast cancer lines include apoptosis, cell motility, and cell division that are the downstream effects of ERBB signaling pathways.
Even though the SpVs of a gene may be quite similar in their protein sequences, the differences resulting from alternative splicing may influence the function of these variants2-5. For example, alternative splicing could provide a mechanism for turning an activator into an effective inhibitor as in the case of the osr2 gene4. Moreover, the relative abundance of the splice isoforms can play a significant role in the normal functioning of a biological system30, 31.
The ERBB receptor proteins, EGFR and ERBB2 are involved in many cancers including breast cancers10, 32, 33. We found variants of these genes expressed at different levels in the three breast cancer cell types (Figure 3). The ERBB2-1055 is highly expressed in SKBR3 and SUM190; it is missing the proline-rich 170 amino acid region found at the C-terminal end of the longer protein ENSP00000385185 (1225 aa) whose transcript was expressed in all three breast cancer cell lines (and was over-expressed in SKBR3 and SUM190). There are no reports on the function of this proline-rich region of ERBB. Apart from this difference, ERBB2-1055 contains all the conserved domains: the extracellular (2 L Receptor domains, 3 Furin like cysteine rich regions), the transmembrane, and the intracellular catalytic tyrosine kinase domains. Marcotte et al 34 reported that the conserved amino acid motif surrounding tyrosine 877 (referred as EGFRYHAD) in ERBB2 is sufficient to confer binding to c-Src tyrosine kinase. c-Src specifically interacts with tyrosine-phosphorylated ERBB2 in ERBB2-induced mammary tumors and is a critical oncogene in signal transduction pathways associated with cancer. ERBB2-1055 may have similar functions as the canonical protein ERBB2-1225, as it is highly expressed in the ERBB2 amplified SKBR3 and SUM190, where ERRB2 signaling plays a significant role12.
If a splice variant is able to fold into a stable structure similar to that of the canonical variant, it may mimic the structural features and thus interact with interaction partners, with or without processing them further. We were able to reliably predict, with relatively high TM-scores, the three-dimensional structures for the smaller variants of EGFR and ERBB2 (Figure 3c and 3e) suggesting stable folding of these proteins. All of these smaller variants of ERBB2 and EGFR contain at least one of the conserved functional domains found in their long canonical counterparts. The EGFR variant EGFR-1091 was expressed only in SUM149. It is missing the translated protein sequences from exons 4 and 28 found in the canonical long protein EGFR-1210, but the translated shorter product contains all the known conserved domains of the canonical protein. Due to the splicing out of exon 4, there is a shift in the relative positions of the domains in the smaller variant compared to the canonical protein (Figure 3d).
The multiple variants of ERBB receptors expressed in the breast cancer cell lines can probably engage in distinct homo-dimerization or form heterodimers with other ERBB receptor variants that can trigger downstream signaling with distinctive patterns. Local densities of ERBB2 profoundly influence its association properties and biological function35.. Zhang et al reported the role of homo and heterodimers of ERBB receptors in different pathways in their genome wide analysis of ERBB2 and EGFR in inflammatory breast cancers 10. Hence, the different ERBB receptor variants identified in this analysis warrant further study, especially in relation to ERBB receptor-targeted drug therapies.
The heat maps of enriched pathways show multiple variants of genes expressed at different levels. For many of the pathways analyzed, SKBR3 and SUM190 were clustered together; this could be mainly driven by amplified ERBB2 expression (Figure 4, Supplementary document part 2). However, the expression profiles in glycolysis and Rho signaling were similar between SKBR3 and SUM149, but with regard to mRNA splicing, SUM149 and SUM190 were similar (Supplementary document part 2-4).
Rho signaling plays a major role in tumor cell motility36. For example, CAV1 is associated with integrins, Rho/ROCK, and SRC-dependent regulation of tumor cell motility and invasion; tyrosine phosphorylated CAV1 functions as an effector of Rho/ROCK signaling to promote late-stage tumor progression and metastasis37. Identification of multiple variants of CAV1, ITGB1, and SRC only from SUM149 (Supplementary Table 3) lends further support to the known significance of this adhesion/cell motility pathway in this cell line. Similar observation on CAV1 expression in SUM149 was shown by Zhang et al in their analyses10.
Calpains regulate biological functions like migration, adhesion, apoptosis, secretion, and autophagy, by modulating cleavage of specific substrates38. Since the calpain activation occurs in cell membranes, their substrates include actinins and integrins39, 40, proteins commonly implicated in ERBB2 over-expressed breast cancer metastases 41-43. Four variants of CAPN1 (ucalpain) were found in all three breast cancer cell lines, with highest expression in SKBR3 (Supplementary document part 3). The mcalpain pathway expression profile of SUM149 was different from SKBR3 and SUM190 mainly due to the expression of multiple variants of CAPSN1, ITGB1, and EGFR. The differential expression of the different variants of CAPN1, CAPN2, and CAPSN1 in the three breast cancer cell lines suggests a possible complex role of the calpain system in breast cancer mechanisms.
Gene Ontology terms enriched for the splice variants identified in the three breast cancer cell lines included splicing (Supplementary Table 4). Splicing could play a key role in determining the specific protein profile in each breast cancer sub-type. We identified multiple variants of genes involved in splicing including small nuclear ribonucleic proteins (snRNPs), DNA-directed RNA polymerases, splice factors and U2 auxiliary factors (u2afs)(Supplementary document part 4) that are differentially expressed in the three breast cancer cell lines. Pre-mRNA splicing is brought about by the Spliceosome, a large ribonucleoic protein complex composed of snRNPs and numerous non-snRNP proteins. The components of the Spliceosome facilitate a dynamic network of RNA–RNA interactions resulting in the two transesterification reactions required for intron removal and exon ligation44. The multiple variants of snRNPs and other splice factors identified may play distinct roles in the splicing patterns of each of these cell lines. The transcript profiles for mRNA splicing (Supplementary document part 4) that show SUM190 and SUM149 profiles clustered together suggesting similar splicing mechanisms may not be due to ERBB2 downstream signaling, since SUM149 does not over-express ERBB2.
Mitochondrial and electron transport chain (ETC) processes were enriched for the unique variants in ERBB2 over-expressed SKBR3 and SUM190. These observations concur with the report by Gupta and Srivastava, who reported a probable link between mitochondrial STAT3 and ETC complex in Her2 breast tumors32. We found multiple STAT3 variants only in SKBR3 and SUM190.
Another enriched process involving ANXA6 and TMED variants in SKBR3 and SUM190 was vesicular protein trafficking. The identification of the ANXA6 variants in SKBR3 and SUM190 supports our observations from previous studies on the HER2+ mouse model for human breast cancers 5, 23. The genes enriched for phosphate metabolism (Table 2) are also annotated to be involved in Alzheimer, Huntington and Parkinson diseases.
The enrichment analyses for the unique variants identified only in the IBC models, SUM190 and SUM149 indicated their roles in nucleotide binding, RNA processing, translation and protein localization (Table 3). Our annotation inferences of these unique variants suggest that the RNA splicing and translation processes in the two IBC models may be similar including intra-cellular transport via the HEAT repeat domains.
We identified splice variants unique to one cancer cell line compared to the other two and normal mammary epithelial cells. The Gene Ontology and BioCarta pathway annotations of these cell line-specific variants indicated distinct top-ranking terms (Table 4). The enriched terms suggest beta-arrestin and dynamin-dependent endocytosis followed by activation of MAP kinases, caspase activity and amino sugar metabolism in SKBR3; lipid synthesis and metabolism in SUM190, and cell-to-cell adhesion via integrin signaling in SUM149 as the key processes in these cell lines. It has been shown that ERBB2 overexpression increases translation of fatty acid synthase (FASN)45. The total distinct read counts of FASN in SUM190 and SKBR3 were higher compared to SUM149 (8055 and 5846 versus 4400). The distinctively high FASN read count in SUM190 suggests a more prominent role of lipid metabolism in SUM190 homeostasis.
The interactions between the cell line-specific variants showed UBC (Ubiquitin C) as the center of the network (Figure 5, Supplementary document parts 5-7). Ubiquitin regulation influences the half-life of most cellular proteins and their variants, thus regulating the relative abundance of the diverse alternatively spliced expressed variants. One unique UBC variant each was expressed in SKBR3 and SUM149 and two unique variants in SUM190. The absence of all ubiquitin domains in these unique shorter variants compared to that of the canonical protein may interfere with the normal functioning of the canonical protein or may act as an antagonist to the normal proteins that control cell growth and death46.
The identification of the likely mechanisms for the 10, 5, and 13 novel peptides with RNA-Seq read evidence from SKBR3, SUM190, and SUM149 (Table 5), points to complex alternative splicing mechanisms which lead to multiple transcripts from the same gene. Some of the genes including SERBP147, RPS1248, HSD17B49 and KPNB1 50 from which the novel peptides were identified, are known to be associated with breast cancers. Many of the abnormally spliced products may not be active. The fact that we found novel peptides from the non protein-coding regions by mass-spectrometric analyses searching a custom-built EST based protein database and then confirmed them with the corresponding mRNA sequences from RNA-Seq data, indicates either a deregulated splicing mechanism in tumor or incorrect annotation of coding and non-coding regions. The identification of the novel peptide ‘FLLTEVFDLLFTISLQFANSAK’ from both SUM149 and SUM190 and the clustering of the expression profiles of transcripts involved in mRNA splicing indicate similar splicing mechanisms in these two inflammatory cell line models (Supplementary document part 4).
Conclusion
Our integrated RNA-Seq and proteomics data analysis is the first of its kind where transcriptomic data are integrated with proteomic data to find known splice variants and novel peptides in a high throughput manner for breast cancer cell lines. The enriched pathways for which the ERBB2 amplified cell lines SKBR3 and SUM190 clustered together suggest the direct regulation of these processes by ERBB2 downstream signaling. Even though the transcript profiles for mRNA splicing mechanisms show similarities between the inflammatory models SUM190 and SUM149, the overall transcript profiles show more similarities between SKBR3 and SUM190. The identifications of more than one SpV of the same gene expressed in SKBR3, SUM190, and SUM149 imply possible distinct or cumulative roles of these variants in cancer processes. The cell line-specific variants suggest diverse biological processes in these cancer models. Identification of more than one variant of genes that are currently annotated as breast cancer oncogenes signifies the importance of knowing their expression levels in tumor samples when designing drugs targeting these genes, as they may interfere with the positive therapeutic outcome or may even be more specific targets. Detailed experimental studies on the distinct SpVs identified from each of these three breast cancer cell types may unveil their roles in cancer progression and metastasis. As we have shown for pairs of SpVs from the mouse model of Her2+ breast cancers, computational modeling of these protein variants can reveal important features of folding, conformation, and likely functional consequences5.
Supplementary Material
Acknowledgments
The authors thank Dr. Sofia Merajver, an internationally known breast cancer specialist at the University of Michigan, for her valuable comments and suggestions on the manuscript. This work was supported in part by NIH grants RM-08-029, P30 U54ES017885, U54DA021519, and UL1RR24986 (G.S.O.); NCI grant U01-CA128427 and Korean Research WCU grant R31-2008-000-10086-0 (W.S.H.); Centers of Excellence in Genomic Science from NIH 5P50HG00235711-3 and Johns Hopkins Sub-award (2001359289) from NIH 1U24CA160036.
Footnotes
From the HUPO Chromosome 17 C-HPP Team.
References
- 1.Germann S, Gratadou L, Dutertre M, Auboeuf D. Splicing Programs and Cancer. Journal of Nucleic Acids. 2012;2012:1–9. doi: 10.1155/2012/269570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Revil T, Toutant J, Shkreta L, Garneau D, Cloutier P, Chabot B. Protein kinase C-dependent control of bcl-x alternative splicing. Mol. Cell. Biol. 2007;27(24):8431–8441. doi: 10.1128/MCB.00565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Végran F, Boidot R, Oudin C, Riedinger J-M, Bonnetain F, Lizard-Nacol S. Overexpression of caspase-3s splice variant in locally advanced breast carcinoma is associated with poor response to neoadjuvant chemotherapy. Clinical Cancer Research. 2006;12(19):5794–5800. doi: 10.1158/1078-0432.CCR-06-0725. [DOI] [PubMed] [Google Scholar]
- 4.Kawai S, Kato T, Inaba H, Okahashi N, Amano A. Odd-skipped related 2 splicing variants show opposite transcriptional activity. Biochemical and Biophysical Research Communications. 2005;328(1):306–311. doi: 10.1016/j.bbrc.2004.12.173. [DOI] [PubMed] [Google Scholar]
- 5.Menon R, Roy A, Mukherjee S, Belkin S, Zhang Y, Omenn GS. Functional implications of structural predictions for alternative splice proteins expressed in Her2/neu-induced breast cancers. J Proteome Res. 2011;10(12):5503–11. doi: 10.1021/pr200772w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z, Wang Y, Wang S, Zhang J, Zhang F, Niu Y. Nek2C functions as a tumor promoter in human breast tumorigenesis. Int. J. Mol. Med. 2012;30(4):775–82. doi: 10.3892/ijmm.2012.1069. [DOI] [PubMed] [Google Scholar]
- 7.Choi JW, Kim DG, Lee AE, Kim HR, Lee JY, Kwon NH, Shin YK, Hwang SK, Chang SH, Cho MH, Choi YL, Kim J, Oh SH, Kim B, Kim SY, Jeon HS, Park JY, Kang HP, Park BJ, Han JM, Kim S. Cancer-associated splicing variant of tumor suppressor AIMP2/p38: pathological implication in tumorigenesis. PLoS Genet. 2011;7(3):1–13. doi: 10.1371/journal.pgen.1001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marko-Varga G, Omenn GS, Paik Y-K, Hancock WS. A First Step Toward Completion of a Genome-Wide Characterization of the Human Proteome. Journal of Proteome Research. 2013;12(1):1–5. doi: 10.1021/pr301183a. [DOI] [PubMed] [Google Scholar]
- 9.Liu S, Im H, Bairoch A, Cristofanilli M, Chen R, Deutsch EW, Dalton S, Fenyo D, Fanayan S, Gates C, Gaudet P, Hincapie M, Hanash S, Kim H, Jeong S-K, Lundberg E, Mias G, Menon R, Mu Z, Nice E, Paik Y-K, Uhlen M, Wells L, Wu S-L, Yan F, Zhang F, Zhang Y, Snyder M, Omenn GS, Beavis RC, Hancock WS. A Chromosome-centric Human Proteome Project (C-HPP) to Characterize the Sets of Proteins Encoded in Chromosome 17. J.Proteome Res. 2013;12(1):45–57. doi: 10.1021/pr300985j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang EY, Cristofanilli M, Robertson F, Reuben JM, Mu Z, Beavis RC, Im H, Snyder M, Hofree M, Ideker T, Omenn GS, Fanayan S, Jeong S-K, Paik Y.-k., Zhang AF, Wu S-L, Hancock WS. Genome Wide Proteomics of ERBB2 and EGFR and Other Oncogenic Pathways in Inflammatory Breast Cancer. Journal of Proteome Research. 2013;12(6):2805–17. doi: 10.1021/pr4001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aebersold R, Bader GD, Edwards AM, van Eyk JE, Kussmann M, Qin J, Omenn GS. The Biology/Disease-driven Human Proteome Project (B/D-HPP): Enabling Protein Research for the Life Sciences Community. Journal of Proteome Research. 2013;12(1):23–27. doi: 10.1021/pr301151m. [DOI] [PubMed] [Google Scholar]
- 12.Yu D, Hung MC. Overexpression of ErbB2 in cancer and ErbB2-targeting strategies. Oncogene. 2000;19(53):6115–21. doi: 10.1038/sj.onc.1203972. [DOI] [PubMed] [Google Scholar]
- 13.Berchuck A, Kamel A, Whitaker R, Kerns B, Olt G, Kinney R, Soper JT, Dodge R, Clarke-Pearson DL, Marks P, et al. Overexpression of HER-2/neu is associated with poor survival in advanced epithelial ovarian cancer. Cancer Res. 1990;50(13):4087–91. [PubMed] [Google Scholar]
- 14.Vogel C, Chan A, Gril B, Kim SB, Kurebayashi J, Liu L, Lu YS, Moon H. Management of ErbB2-positive breast cancer: insights from preclinical and clinical studies with lapatinib. Jpn J Clin Oncol. 2010;40(11):999–1013. doi: 10.1093/jjco/hyq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jelovac D, Wolff AC. The adjuvant treatment of HER2-positive breast cancer. Curr Treat Options Oncol. 2012;13(2):230–9. doi: 10.1007/s11864-012-0186-4. [DOI] [PubMed] [Google Scholar]
- 16.Pohlmann PR, Mayer IA, Mernaugh R. Resistance to Trastuzumab in Breast Cancer. Clinical Cancer Research. 2009;15(24):7479–7491. doi: 10.1158/1078-0432.CCR-09-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nokes BT, Cunliffe HE, Lafleur B, Mount DW, Livingston RB, Futscher BW, Lang JE. In Vitro Assessment of the Inflammatory Breast Cancer Cell Line SUM 149: Discovery of 2 Single Nucleotide Polymorphisms in the RNase L Gene. J Cancer. 2013;4(2):104–16. doi: 10.7150/jca.5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olayioye MA. Update on HER-2 as a target for cancer therapy: intracellular signaling pathways of ErbB2/HER-2 and family members. Breast Cancer Res. 2001;3(6):385–9. doi: 10.1186/bcr327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, Clark MJ, Im H, Habegger L, Balasubramanian S, O’Huallachain M, Dudley JT, Hillenmeyer S, Haraksingh R, Sharon D, Euskirchen G, Lacroute P, Bettinger K, Boyle AP, Kasowski M, Grubert F, Seki S, Garcia M, Whirl-Carrillo M, Gallardo M, Blasco MA, Greenberg PL, Snyder P, Klein TE, Altman RB, Butte AJ, Ashley EA, Gerstein M, Nadeau KC, Tang H, Snyder M. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Geiger T, Cox J, Mann M. Proteomic changes resulting from gene copy number variations in cancer cells. PLoS Genet. 2010;6(9):1–15. doi: 10.1371/journal.pgen.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menon R, Zhang Q, Zhang Y, Fermin D, Bardeesy N, DePinho RA, Lu C, Hanash SM, Omenn GS, States DJ. Identification of novel alternative splice isoforms of circulating proteins in a mouse model of human pancreatic cancer. Cancer Res. 2009;69(1):300–9. doi: 10.1158/0008-5472.CAN-08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menon R, Omenn GS. Proteomic characterization of novel aternative splice variant proteins in Human epidermal growth factor receptor 2/neu induced breast cancers. Cancer Research. 2010;70(9):3440–3449. doi: 10.1158/0008-5472.CAN-09-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong K, Kim S, Bandeira N. False discovery rates in spectral identification. BMC Bioinformatics. 2012;13(Suppl 16):1–15. doi: 10.1186/1471-2105-13-S16-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henjes F, Bender C, von der Heyde S, Braun L, Mannsperger HA, Schmidt C, Wiemann S, Hasmann M, Aulmann S, Beissbarth T, Korf U. Strong EGFR signaling in cell line models of ERBB2-amplified breast cancer attenuates response towards ERBB2-targeting drugs. Oncogenesis. 1(7):1–14. doi: 10.1038/oncsis.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, Falquet L. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic acids research. 2007;35(suppl_2):W433–437. doi: 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fanayan S, Smith JT, Lee LY, Yan F, Snyder M, Hancock WS, Nice E. Proteogenomic Analysis of Human Colon Carcinoma Cell Lines LIM1215, LIM1899, and LIM2405. Journal of Proteome Research. 2013;12(4):1732–1742. doi: 10.1021/pr3010869. [DOI] [PubMed] [Google Scholar]
- 29.Ning K, Nesvizhskii AI. The utility of mass spectrometry-based proteomic data for validation of novel alternative splice forms reconstructed from RNA-Seq data: a preliminary assessment. BMC Bioinformatics. 2010;11(Suppl 11):S14. doi: 10.1186/1471-2105-11-S11-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goedert M, Jakes R. Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. Embo J. 1990;9(13):4225–30. doi: 10.1002/j.1460-2075.1990.tb07870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393(6686):702–5. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 32.Gupta P, Srivastava SK. Antitumor activity of phenethyl isothiocyanate in HER2-positive breast cancer models. BMC Med. 2012;10(80):1–18. doi: 10.1186/1741-7015-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henjes F, Bender C, von der Heyde S, Braun L, Mannsperger HA, Schmidt C, Wiemann S, Hasmann M, Aulmann S, Beissbarth T, Korf U. Strong EGFR signaling in cell line models of ERBB2-amplified breast cancer attenuates response towards ERBB2-targeting drugs. Oncogenesis. 2012;1:1–9. doi: 10.1038/oncsis.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcotte R, Zhou L, Kim H, Roskelly CD, Muller WJ. c-Src associates with ErbB2 through an interaction between catalytic domains and confers enhanced transforming potential. Mol Cell Biol. 2009;29(21):5858–71. doi: 10.1128/MCB.01731-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy P, Vereb G, Sebestyen Z, Horvath G, Lockett SJ, Damjanovich S, Park JW, Jovin TM, Szollosi J. Lipid rafts and the local density of ErbB proteins influence the biological role of homo- and heteroassociations of ErbB2. J. Cell Sci. 2002;115(22):4251–4262. doi: 10.1242/jcs.00118. [DOI] [PubMed] [Google Scholar]
- 36.Adesso L, Calabretta S, Barbagallo F, Capurso G, Pilozzi E, Geremia R, Delle Fave G, Sette C. Gemcitabine triggers a pro-survival response in pancreatic cancer cells through activation of the MNK2/eIF4E pathway. Oncogene. 2013;32(23):2848–57. doi: 10.1038/onc.2012.306. [DOI] [PubMed] [Google Scholar]
- 37.Unruh D, Turner K, Srinivasan R, Kocaturk B, Qi X, Chu Z, Aronow BJ, Plas DR, Gallo CA, Kalthoff H, Kirchhofer D, Ruf W, Ahmad SA, Lucas FV, Versteeg HH, Bogdanov VY. Alternatively spliced tissue factor contributes to tumor spread and activation of coagulation in pancreatic ductal adenocarcinoma. Int J Cancer. 2013 doi: 10.1002/ijc.28327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cataldo F, Peche LY, Klaric E, Brancolini C, Myers MP, Demarchi F, Schneider C. CAPNS1 Regulates USP1 Stability and Maintenance of Genome Integrity. Mol Cell Biol. 2013;33(12):2485–96. doi: 10.1128/MCB.01406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart MP, McDowall A, Hogg N. LFA-1-mediated adhesion is regulated by cytoskeletal restraint and by a Ca2+-dependent protease, calpain. J Cell Biol. 1998;140(3):699–707. doi: 10.1083/jcb.140.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao H, Chou J, Baty CJ, Burke NA, Watkins SC, Stolz DB, Wells A. Spatial localization of m-calpain to the plasma membrane by phosphoinositide biphosphate binding during epidermal growth factor receptor-mediated activation. Mol Cell Biol. 2006;26(14):5481–96. doi: 10.1128/MCB.02243-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ngan E, Northey JJ, Brown CM, Ursini-Siegel J, Siegel PM. A complex containing LPP and alpha-actinin mediates TGFbeta-induced migration and invasion of ErbB2-expressing breast cancer cells. J Cell Sci. 2013;126(Pt 9):1981–91. doi: 10.1242/jcs.118315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lahlou H, Muller WJ. beta1-integrins signaling and mammary tumor progression in transgenic mouse models: implications for human breast cancer. Breast Cancer Res. 2011;13(6):229. doi: 10.1186/bcr2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huck L, Pontier SM, Zuo DM, Muller WJ. beta1-integrin is dispensable for the induction of ErbB2 mammary tumors but plays a critical role in the metastatic phase of tumor progression. Proc Natl Acad Sci U S A. 2010;107(35):15559–64. doi: 10.1073/pnas.1003034107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Keefe RT. Mutations in U5 snRNA loop 1 influence the splicing of different genes in vivo. Nucleic Acids Res. 2002;30(24):5476–84. doi: 10.1093/nar/gkf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin Q, Yuan LX, Boulbes D, Baek JM, Wang YN, Gomez-Cabello D, Hawke DH, Yeung SC, Lee MH, Hortobagyi GN, Hung MC, Esteva FJ. Fatty acid synthase phosphorylation: a novel therapeutic target in HER2-overexpressing breast cancer cells. Breast Cancer Res. 2010;12(6):1–18. doi: 10.1186/bcr2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23(21):4776–89. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 47.Serce NB, Boesl A, Klaman I, von Serenyi S, Noetzel E, Press MF, Dimmler A, Hartmann A, Sehouli J, Knuechel R, Beckmann MW, Fasching PA, Dahl E. Overexpression of SERBP1 (Plasminogen activator inhibitor 1 RNA binding protein) in human breast cancer is correlated with favourable prognosis. BMC Cancer. 2012;12:597. doi: 10.1186/1471-2407-12-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng SS, Xing TY, Zhou HY, Xiong RH, Lu YG, Wen B, Liu SQ, Yang HJ. Comparative proteome analysis of breast cancer and adjacent normal breast tissues in human. Genomics Proteomics Bioinformatics. 2006;4(3):165–72. doi: 10.1016/S1672-0229(06)60029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhavani V, Srinivasulu M, Ahuja YR, Hasan Q. Role of BRCA1, HSD17B1 and HSD17B2 methylation in breast cancer tissue. Cancer Biomark. 2009;5(4):207–13. doi: 10.3233/CBM-2009-0105. [DOI] [PubMed] [Google Scholar]
- 50.Nordgard SH, Johansen FE, Alnaes GI, Bucher E, Syvanen AC, Naume B, Borresen-Dale AL, Kristensen VN. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosomes Cancer. 2008;47(8):680–96. doi: 10.1002/gcc.20569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.