Abstract
Inhibitor resistant (IR) class A β-lactamases pose a significant threat to many current antibiotic combinations. The K234R substitution in the SHV β-lactamase, from Klebsiella pneumoniae, results in resistance to ampicillin/clavulanate. After site-saturation mutagenesis of Lys-234 in SHV, microbiological and biochemical characterization of the resulting β-lactamases revealed that only –Arg conferred resistance to ampicillin/clavulanate. X-ray crystallography revealed two conformations of Arg-234 and Ser-130 in SHV K234R. The movement of Ser-130 is the principal cause of the observed clavulanate resistance. A panel of boronic acid inhibitors was designed and tested against SHV-1 and SHV K234R. A chiral ampicillin analogue was discovered to have a 2.4 ± 0.2 nM Ki for SHV K234R; the chiral ampicillin analogue formed a more complex hydrogen-bonding network in SHV K234R vs SHV-1. Consideration of the spatial position of Ser-130 and Lys-234 and this hydrogen-bonding network will be important in the design of novel antibiotics targeting IR β-lactamases.
INTRODUCTION
Nearly 1.7 million healthcare-associated infections in the United States each year result in almost 100,000 deaths.1,2 β-Lactam antibiotics are often given to treat such infections. In general, β-lactams act by attacking the penicillin-binding proteins (PBPs) of the bacterial cell wall, leading to disruption of the peptidoglycan polymer and bacterial lysis.2–6 Unfortunately, bacteria have evolved mechanisms to elude these antibiotics, one of which is the production of β-lactamase enzymes (EC 3.5.2.6).
Four classes of β-lactamases exist; three of these classes (class A, C, D) use serine as the active site nucleophile and one class comprises metallo-enzymes requiring Zn2+ for activity (class B).1,2 Class A enzymes are the most prevalent enzymes in Enterobacteriaceae (e.g., Escherichia coli and Klebsiella pneumoniae), where they are typically plasmid-encoded.1,2,7,8 TEM (Temoneira) and SHV are two of the enzymes most commonly found in Enterobacteriaceae.1,2,7
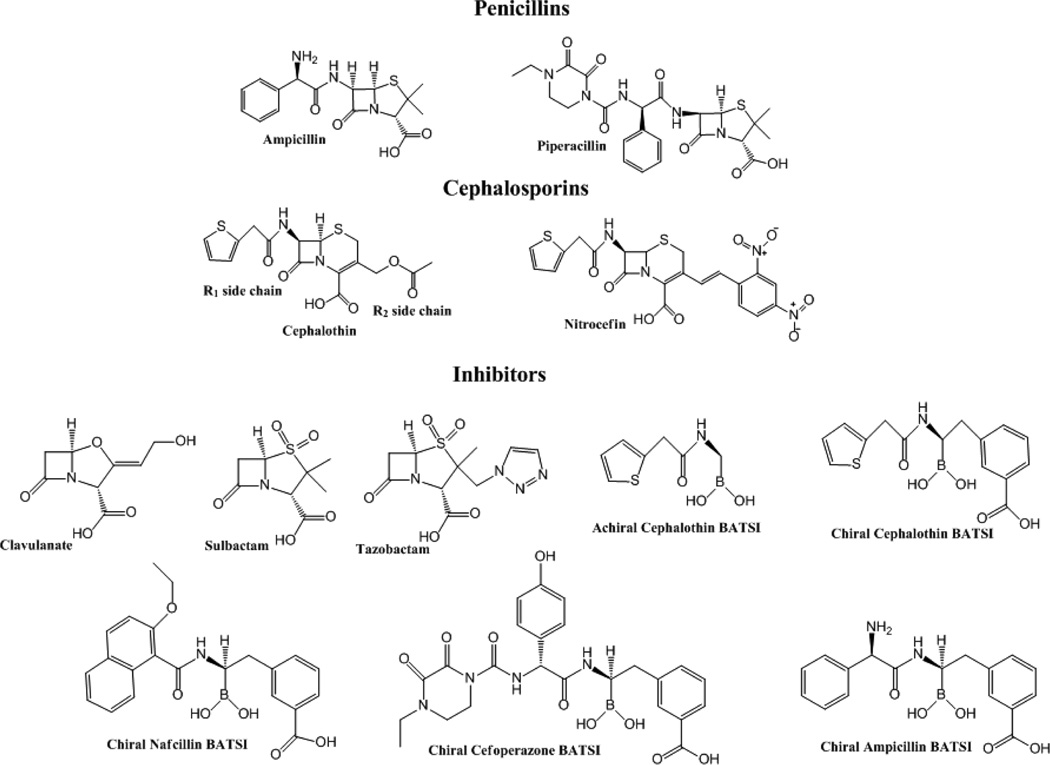
One means of combating the activity of β-lactamase enzymes has been the design of mechanism-based inhibitors, which subvert the activity of β-lactamases against antibiotics.2,9 The inhibitors are given with β-lactam antibiotics to protect against β-lactamases. There are three β-lactamase inhibitors currently on the market: clavulanate, sulbactam, and tazobactam (Figure 1), which are generally effective mostly against class A β-lactamases.2,9 β-Lactamase inhibitors form stable covalent complexes with the β-lactamase enzymes, which lead to their inactivation (Scheme 1).2 Unfortunately, β-lactamases have also evolved resistance to inhibitors; point mutations in blaTEM and blaSHV give rise to amino acid substitutions in the parent enzyme that result in inhibitor resistant (IR) β-lactamases that manifest as elevated MICs and Kis to β-lactam/β-lactamase inhibitor combinations.2,9
Figure 1.
Chemical structures of the compounds tested in this paper.
Scheme 1.
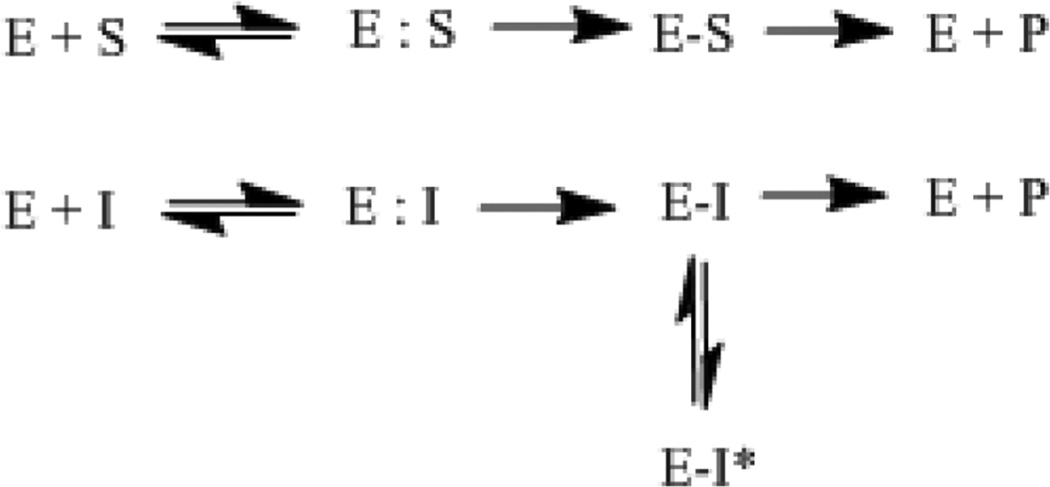
Scheme Showing the Breakdown of a β-Lactam (S) by the Enzyme (E) to Form the Inactive β-Lactam (P) and the Interaction of an Inhibitor (I) with the Enzyme to Either Form an Inactivated Inhibitor (P) and Active Enzyme or Inactivation of the Enzyme through a Long-Lived Inhibitor Complex (E–I*)
In the SHV family of β-lactamase enzymes, amino acid substitutions are found that lead to IR variants.8,10–21 The first IR SHV variant was detected in E. coli and designated SHV-10.13 The substitution contributing to clavulanate resistance in this variant was S130G (Ambler numbering22). S130G in the SHV β-lactamase has been studied extensively.16,19,21 Subsequently, substitutions at amino acid position Met-69 (SHV-49, M69I) and amino acid position Thr-235 (SHV-107, T235A) were also discovered that result in amoxicillin/clavulanate resistance.10,15
Three different IR SHV isolates were recently reported containing a K234R substitution (SHV-56, SHV-72, and SHV-84).11,12,14 Studies of these clinical isolates reported that position 234 is a key residue leading to IR in the SHV β-lactamase. Interestingly, Lys-234 is also part of the conserved KTG motif in class A β-lactamases.22,23 In SHV, Lys-234 is on the β3 strand, which forms a wall of the active site cavity.14,23 The proximity of Lys-234 to the active site of SHV and the frequent appearance of the K234R substitution among IR SHV enzymes lead us to postulate that this residue is critically important in the evolution of the IR phenotype in SHV.
On the basis of this reasoning, position 234 was explored to determine if additional amino acid substitutions would maintain the IR phenotype. To achieve this goal, site-saturation and site-directed mutagenesis were conducted to evaluate all 19 amino acid substitutions. Additionally, we probed the flexibility of the active site and enzyme–inhibitor or enzyme–substrate interactions in the K234R variant with the goal of obtaining insights that could aid in the design of novel antibiotics and inhibitors. To achieve this second objective, we synthesized a series of boronic acid transition state inhibitors (BATSIs). We were next compelled to determine changes in the structure of the K234R variant through X-ray crystallography that lead to the IR phenotype. From these results and previous studies, we propose a novel mechanism underlying inhibitor resistance involving Lys-234 and displacement of Ser-130, which prevents or slows proton transfer and inactivation.
RESULTS
Construction of Variants at Ambler Position 234; Mutagenesis and Immunoblotting
Using degenerate oligonucleotides, we first performed site-saturation mutagenesis on the blaSHV-1 gene at Ambler position 234. Thirteen of the 19 amino acid substitutions were obtained in the initial sequencing screen (100 blaSHV genes selected from E. coli ElectroMAX DH10B transformants and sequenced). blaSHV K234L, blaSHV K234Q, blaSHV K234G, blaSHV K234C, blaSHV K234Y, and blaSHV K234F were constructed by site-directed mutagenesis. Expression of the β-lactamase proteins were confirmed by immunoblotting (Supporting Information Figure 1). We observed that in E. coli there was lower expression of K234W, -Y, -F, -V, and -P substituted proteins.
Antimicrobial Susceptibility Testing
To assess the impact of the single amino acid substitutions at Lys234 on β-lactam and inhibitor susceptibility, minimum inhibitory concentrations (MICs) for all 19 variants against a panel of antibiotics and inhibitor combinations (ampicillin (amp), piperacillin (pip), cephalothin (thin), ampicillin/clavulanate (amp/clav), ampicillin/sulbactam (amp/sul), and ampicillin/tazobactam (amp/tazo) were determined in a uniform E. coli DH10B background. Results are shown in Table 1.
Table 1.
MIC Values (µg/mL) of E. coli DH10B Expressing SHV-1 and Lys-234 Variantsa
| amp | pip | thin | amp/ clav |
amp/ sul |
amp/ tazo |
|
|---|---|---|---|---|---|---|
| E. coli DH10B | 1 | 1 | 2 | <0.06 | <0.06 | <0.06 |
| E. coli blaSHV-1 | >16384 | >4096 | 256 | 4 | 512 | 128 |
| E. coli blaSHV K234R | 16384 | 2048 | 16 | 16 | 64 | 16 |
| E. coli blaSHV K234A | 32 | 8 | 8 | <0.06 | <0.06 | <0.06 |
| E. coli blaSHV K234Xb | 4 | 4 | 8 | <0.06 | <0.06 | <0.06 |
Inhibitors were calculated in the presence of 50 µg/mL ampicillin.
Includes other 16 amino acid substitutions.
All Lys-234 variants (here represented as K234X) with the exception of -R have lower MICs for the penicillins compared to SHV-1. The ampicillin and piperacillin MICs (16384 and 2048 µg/mL, respectively) for the K234R variant expressed in E. coli DH10B were preserved compared to the other Lys-234 variants, indicating that the Arg substitution maintains the ability to hydrolyze penicillins. Surprisingly, the E. coli K234A variant MICs (32 and 8 µg/mL for ampicillin and piperacillin, respectively) indicate that an alanine substitution at Ambler position 234 also allows some hydrolytic activity to be maintained for the penicillins relatively to the other amino acid substitutions (≤4 and ≤8 µg/mL for ampicillin and piperacillin, respectively). The K234A substitution was not further explored due to the lower level of ampicillin resistance relative to SHV-1 and K234R. When MICs were performed against cephalothin we found that all of the Lys-234 substitutions had increased susceptibility relative to wild-type SHV-1.
For the β-lactam/β-lactamase inhibitor combinations, only the K234R substitution in E. coli DH10B had a higher MIC than wild-type SHV-1 for ampicillin/clavulanic acid (16 vs 4 µg/mL, respectively, Table 1). The K234R MICs for ampicillin/sulbactam and ampicillin/tazobactam were lower than that of wild-type SHV-1 but significantly higher than for any other amino acid substitution at position 234. This indicates that this is a “restrictive position”, with only the K234R substitution maintaining inhibitor resistance.24 We have previously observed SHV enzymes that are resistant to ampicillin/clavulanate, and susceptible to ampicillin/sulbactam and ampicillin/tazobactam.17,18
Kinetics of K234R with Representative β-Lactam Substrates
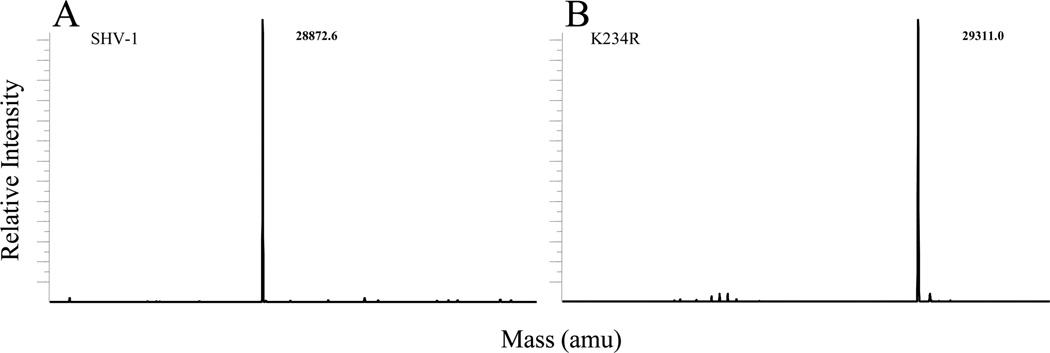
Both SHV-1 and K234R were purified as described and their identity was confirmed by mass spectrometry (Figure 2). Kinetic data collected for the K234R variant relative to values obtained for the wild-type SHV-1 enzyme are shown in Table 2. Overall, the kcat/Km values were lower for the K234R variant. A higher Km value for cephalosporins (nitrocefin and cephalothin) but a lower Km for piperacillin was demonstrated by the K234R variant. Except for piperacillin, we observed that the K234R variant has a lower kcat for all substrates tested.
Figure 2.
Deconvoluted ESI-MS spectra of (A) SHV-1 and (B) K234R β-lactamases. All measurements have an error of ±3amu.
Table 2.
Kinetic Properties of SHV-1 and K234R for Three Representative β-Lactams
| SHV-1 | K234R | |
|---|---|---|
| Piperacillin | ||
| Km (µM) | 74.6 ± 11.7 | 305 ± 47 |
| kcat (s−1) | 1179 ± 65 | 2736 ± 28 |
| kcat/Km (µM−1 s−1) | 15.8 ± 0.6 | 9 ± 0.2 |
| ΔΔGcat (kcal/mol) | +0.33 | |
| Nitrocefin | ||
| Km (µM) | 24.7 ± 5 | 10.4 ± 1.7 |
| kcat (s−1) | 347.8 ± 21.5 | 45.2 ± 0.5 |
| kcat/Km (µM−1 s−1) | 14.1 ± 0.22 | 4.3 ± 0.5 |
| ΔΔGcat (kcal/mol) | +0.70 | |
| Cephalothin | ||
| Km (µM) | 48.9 ± 11.2 | 29 ± 5 |
| kcat (s−1) | 42.6 ± 3.4 | 4.6 ± 1 |
| kcat/Km (µM−1 s−1) | 0.87 ± 0.3 | 0.16 ± 0.3 |
| ΔΔGcat (kcal/mol) | +1.00 | |
Kinetics of K234R with β-Lactamase Inhibitors
Table 3 summarizes the kinetics for SHV-1 and K234R when inactivated by various β-lactamase inhibitors. The K234R variant demonstrated a higher Ki than the wild-type SHV-1 for all three inhibitors (clavulanate, tazobactam, and sulbactam). The largest increase was for clavulanic acid. The partition ratios (kcat/kinact) were also determined for the three suicide inhibitors; the lower value for sulbactam and tazobactam for the K234R variant relative to SHV-1 is consistent with the increased susceptibility of the variant to these inhibitors (see MICs above).
Table 3.
Kinetic Properties of SHV-1 and K234R for Clavulanate, Tazobactam, Sulbactam, and BATSI Compounds
| SHV-1 | K234R | |
|---|---|---|
| Clavulanate | ||
| Ki (µM) | 1.3 ± 0.1 | 7.7 ± 0.7 |
| kcat/kinact (tn) | 25 | 50 |
| Tazobactam | ||
| Ki (µM) | 0.44 ± 0.04 | 0.9 ± 0.1 |
| kcat/kinact (tn) | 50 | 10 |
| Sulbactam | ||
| Ki (µM) | 1.6 ± 0.2 | 4.6 ± 0.5 |
| kcat/kinact (tn) | 12,000 | 60 |
| Achiral Cephalothin BATSI | ||
| Ki (µM) | 20 ±2 | 61 ± 6 |
| Chiral Cephalothin BATSI | ||
| Ki (µM) | 0.46 ± 0.05 | 0.012 ± 0.001 |
| Nafcillin BATSI | ||
| Ki (µM) | 0.037 ± 0.004 | 0.048 ± 0.005 |
| Chiral Ampicillin BATSI | ||
| Ki (µM) | 0.056 ± 0.005 | 0.0024 ± 0.0002 |
| Chiral Cefoperazone BATSI | ||
| Ki (µM) | 0.0093 ± 0.001 | 0.0135 ± 0.001 |
Different transition state analogues, BATSI compounds, were next investigated to determine interactions that may be responsible for the IR phenotype of the K234R substitution and also to elucidate chemical properties that may be important for novel inhibitor design (Figure 1). The BATSIs tested include: (1) an achiral compound with the R1 side chain of cephalothin, (2) a chiral compound with the R1 side chain of cephalothin and a R2 meta-carboxyphenyl group, (3) a nafcillin BATSI with the R1 side chain of nafcillin and a R2 methyl-meta-carboxyphenyl group, (4) a cefoperazone BATSI with the R1 side chain of cefoperazone and a R2 methyl-meta-carboxyphenyl group, and (5) a chiral ampicillin BATSI with the R1 side chain of ampicillin and a R2 methyl-meta-carboxyphenyl group.
The chiral cephalothin BATSI demonstrated a 44-fold lower Ki for SHV-1 than the achiral BATSI (0.46 ± 0.05 and 20 ± 2 µM, respectively). These observations suggested that the meta-carboxyphenyl group at the R2 position of the compound is important for interactions with the enzyme leading to its inhibition. The meta-carboxyphenyl group at the R2 position of the compound lowers the Ki of the K234R variant for the compound even more (61 ± 6 vs 0.012 ± 0.001 µM for the achiral vs chiral cephalothin BATSI, respectively) for a 5000-fold difference. The chiral nafcillin, chiral cefoperazone, and chiral ampicillin BATSI analogues have a Ki in the nM range for SHV-1 (37 ± 4, 9.3 ± 1, and 56 ± 6 nM, respectively) and for the K234R variant (48 ± 5, 13.5 ± 1.4, and 2.4 ± 0.2 nM, respectively). The values are comparable for SHV-1 and for SHVK234R for the chiral nafcillin and chiral cefoperazone BATSIs, but the chiral ampicillin BATSI has a 20-fold lower Ki for K234R relative to SHV-1.
Assessing the Secondary Structure and Stability of SHV-1 and K234R Using Circular Dichroism (CD)
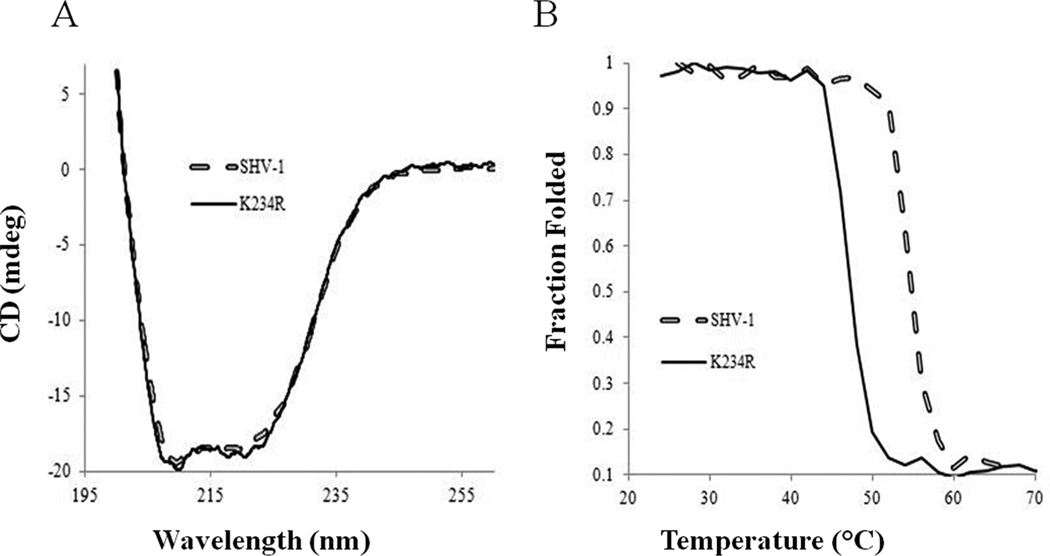
We explored K234R and SHV-1 through thermal stability measurements and CD because active site substitutions are known to destabilize proteins, and we were interested in overall structural changes of K234R that may be leading to the IR phenotype. The CD spectra and melting curves for SHV-1 and K234R are shown in Figures 3A and 33B, respectively. The wild-type SHV-1 required 6.2 °C more for thermal denaturation (a melting temperature of 54.0 °C vs 48.2 °C for K234R).
Figure 3.
(A) Far UV spectra of SHV-1 (dashed) and K234R (solid black) measured at 10 µM enzyme concentration. (B) Fraction of folded protein was calculated as described and is plotted here relative to the temperature in order to compute the Tm of each enzyme SHV-1 (dashed) has a Tm of 54.2 °C and K234R (solid black) has a melting temperature of 48.2 °C.
Assessing the Changes in Secondary Structure and Stability of SHV-1 and K234R in the Presence of Inhibitors Using CD
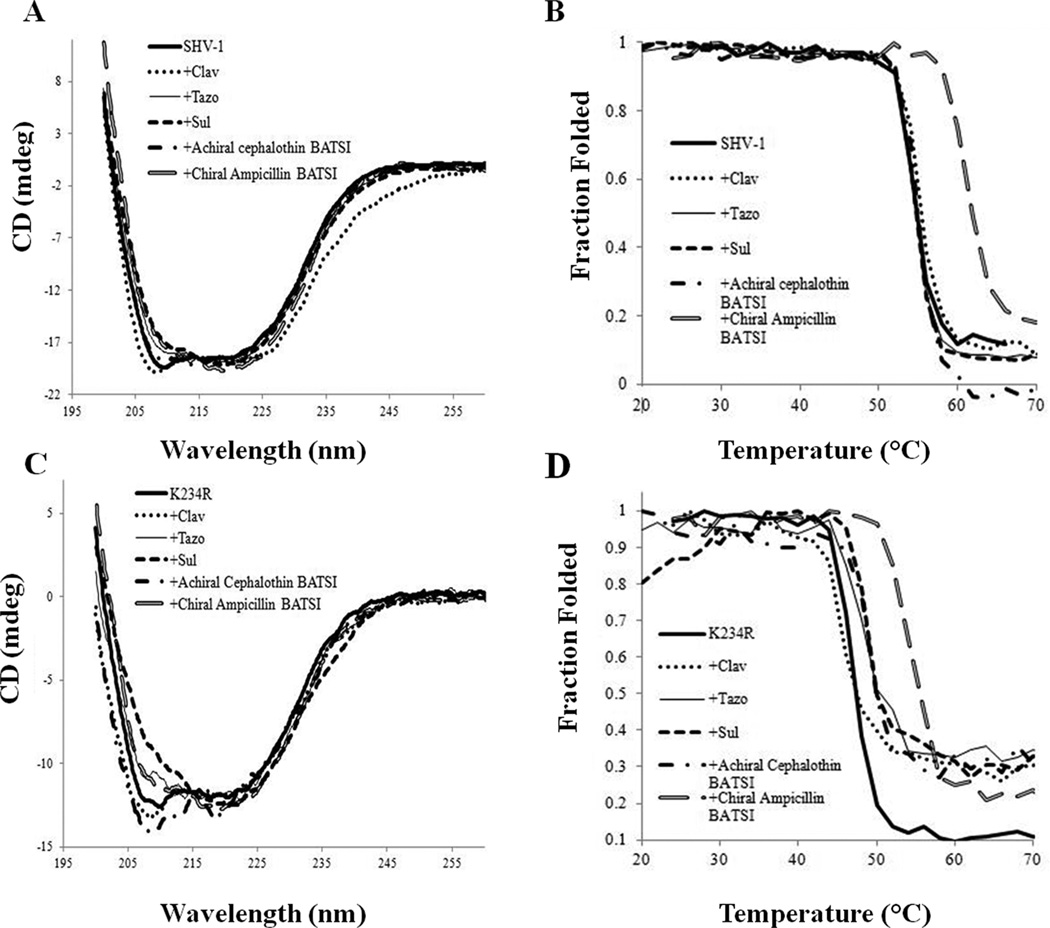
Far UV CD was also used to measure changes in the secondary structure and thermal stability of SHV-1 and the K234R variant in the presence of different inhibitors. These results are shown in Figure 4 and Table 4. From these measurements, only the binding of the chiral ampicillin boronate affects the melting temperature of either SHV-1 or K234R by increases of 7.2 °C for SHV-1 and of 6.6 °C for K234R (Figure 4B,D and Table 5). Tazobactam, sulbactam, and the achiral BATSI affect the secondary structure of K234R but not SHV-1 (Figure 4A,C). Clavulanate binding affects the secondary structure of both enzymes.
Figure 4.
(A) SHV-1 at 10 µM enzyme concentration in the presence of 50 µM of various inhibitors monitored by CD showing changes in the secondary structure upon the enzyme binding to the inhibitor. (B) Changes in the thermal denaturation curve of SHV-1 at 10 µM enzyme concentration in the presence of 50 µM of various inhibitors (the chiral ampicillin BATSI is the only compound to significantly shift the curve). (C) K234R at 10 µM enzyme concentration in the presence of 50 µM of various inhibitors monitored by CD showing changes in the secondary structure upon the enzyme binding to the inhibitor (large changes are seen with tazobactam and sulbactam, thin solid line and dashed black line, respectively). Additionally, clavulanate and the achiral cephalothin BATSI seem to increase the flexibility and movement of the protein (dotted and dashed-dotted black lines, respectively). (D) Changes in the thermal denaturation cruve of K234R at 10 µM enzyme concentration in the presence of 50 µM of various inhibitors (the chiral ampicillin BATSI is the only compound to greatly shift the curve). Sulbactam seems to cause an initial destabilization of the K234R variant, with stability recovered after the first few temperature determinations are made.
Table 4.
Melting Temperature of SHV-1 and K234R in the Presence of Various Inhibitors
| SHV-1 | K234R | |||||
|---|---|---|---|---|---|---|
| Tm (°C) | ΔΔGu (kcal/mol)a | ΔHVH (kcal/mol) | Tm (°C) | ΔΔGu (kcal/mol)a | ΔHVH (kcal/mol) | |
| 54.2 | 116 | 48.2 | 92 | |||
| Tazo | 54.4 | +0.07 | 149 | 49.0 | +0.23 | 114 |
| Sul | 54.4 | +0.07 | 183 | |||
| Clav | 55.0 | +0.28 | 113 | 46.1 | −0.6 | 68 |
| achiral cephalothin boronate | 55.1 | +0.32 | 160 | 48.9 | +0.2 | 100 |
| chiral Amp boronate | 61.4 | +2.56 | 143 | 54.8 | +1.88 | 119 |
Determined by ΔΔGu = ΔTm × ΔSapo
Table 5.
Data Collection and Refinement Statistics
| parameter | value for K234R |
|---|---|
| Data Collection Statistics | |
| space group | P212121 |
| cell dimensions | |
| a, b, c (Å) | 49.6, 55.4, 84.0 |
| α, β, γ (Å) | 90.0, 90.0, 90.0 |
| wavelength (Å) | 0.97 |
| resolution (Å)2 | 1.09 |
| Rsym | 0.03 (0.18) |
| I/σI | 44.7 (9.6) |
| completeness (%) | 84.3 (82.2) |
| redundancy | 4.6 (4.2) |
| Refinement Statistics | |
| resolution range (Å) | 23.8−1.08 |
| no. of reflections | 84309 |
| Rwork/Rfree | 0.12/0.15 |
| no. of atoms (protein/ligand/water) rmsd | 2210/49/229 |
| bond length (Å) | 0.015 |
| bond angle (deg) | 1.67 |
| av B-factors (Å2) | |
| protein (main chain/side chain/all) | 8.56/11.15/9.8 |
| HEPES/cymal-6 | 11.65/16.17 |
| water | 19.95 |
| Ramachandran plot statistics (%) | |
| preferred regions | 97.06 |
| allowed regions | 1.96 |
| outliers | 0.98 |
Crystal Structure of Apo K234R
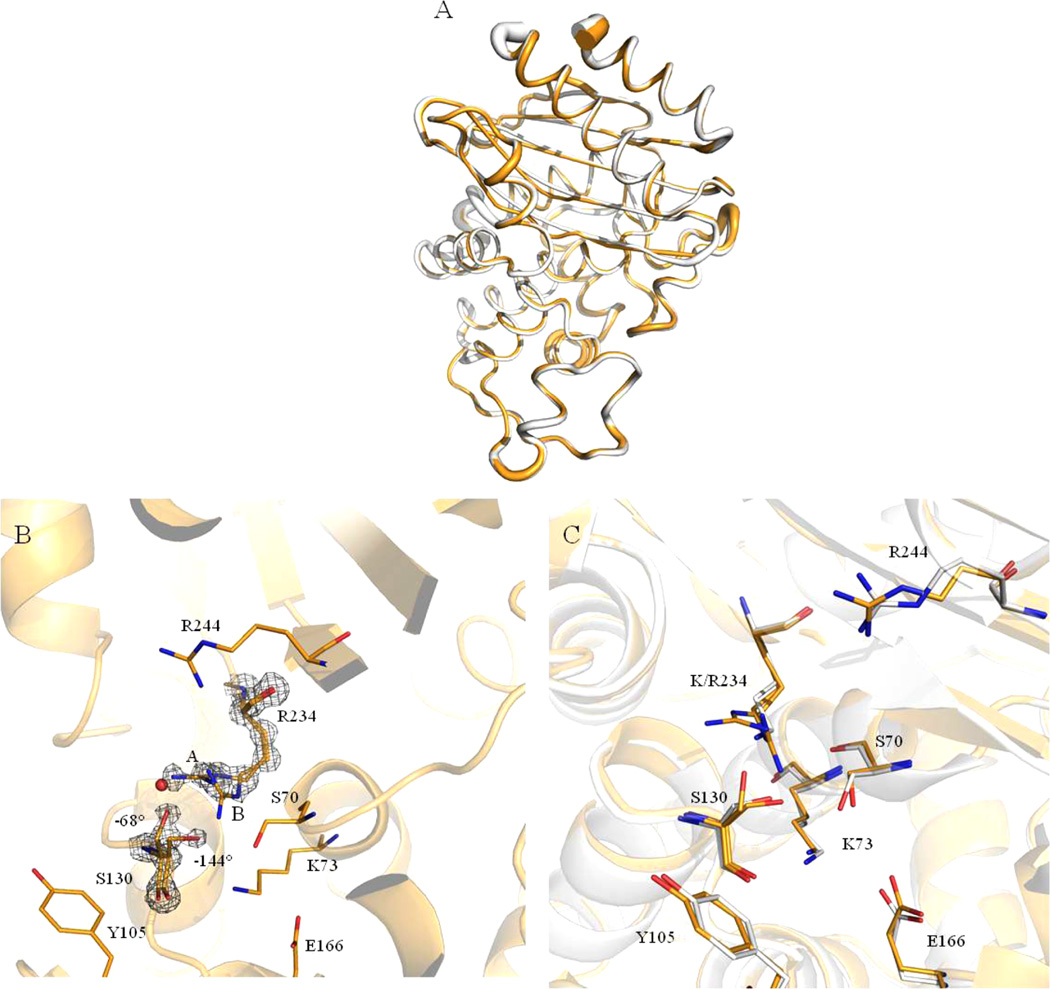
The K234R enzyme crystal structure was solved to high resolution, 1.09 Å, with R and Rfree values of 0.12 and 0.15, respectively (Table 5). The K234R structure is very similar to that of the wild-type SHV-1 enzyme structure (PDB ID: 1SHV, Figure 5A), as is evident by an overall Cα root mean square deviation (rmsd) of 0.32 Å. Ramachandran analysis indicates that 97% of residues reside in preferred regions, 2% reside in allowed regions, and 1% (two residues) are outliers. At this resolution, we could confidently assign alternative conformations for 23 residues. Two of these residues are in the active site, Ser-130 and Arg-234.
Figure 5.
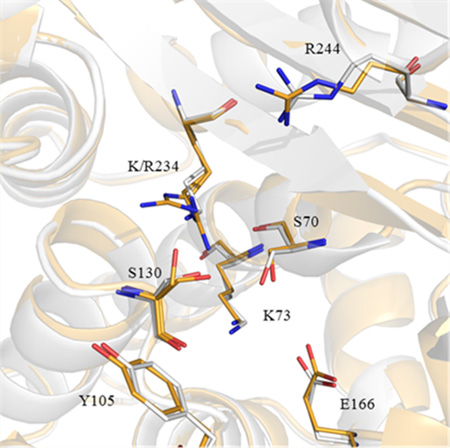
(A) Ribbon cartoon overlay of K234R (gold) and SHV-1 (PDB ID: 1SHV, white). Overall rmsd is 0.14 Å, all Cα superposition performed using SSM Superpose in COOT. Ribbon width is proportionate to B-factor, i.e., larger ribbon radius indicates higher B-factor. (B) SHVK234R active site. 2Fo–Fc density contoured at 1.5σ is shown for both R234 orientations. Two S130 orientations are also observed, with χ1 angles of −144 and −68°. χ1 angle defined as the angle between the N, CA, CB and CA, CB, Oγ planes as in Mendonca et. al.12 The red sphere represents the water molecule found in SHV-1. This space is filled by the A orientation of Arg-234 in the K234R structure, which leads the water molecule to be absent in the crystal structure of the K234R protein. (C) Active site overlay of SHV-1 (PDB ID: 1SHV, white) and K234R (gold). The change in position of K/R234, S130, and S70 is evident in this image.
The lysine to arginine substitution at position 234 is visualized in a 2Fo–Fc map contoured at 1.5σ (Figure 5B) with the arginine side chain having two conformations, A and B (occupancies of 65% and 35%, respectively). In the A conformation, the entire guanidium group is visible in the electron density, whereas this moiety is less clear in the minor B conformation, which is likely a result of increased flexibility and decreased occupancy. Most strikingly, Ser-130 has two conformations: one in the canonical wild-type SHV-1 orientation (−144°), toward Lys-73, which has an occupancy of 55%. In the other conformation, the hydroxyl of Ser-130 (−68°) is oriented toward Arg-234 and has an occupancy of 45% (Figures 5B,C). There is also a slight change in the position of Ser-70 in K234R relative to SHV-1, which likely contributes to the change in kinetic behavior and hydrolysis patterns of the variant. All of the other active site residues occupy almost identical positions as compared to wild-type SHV-1 (PDB ID: 1SHV), likewise no secondary structure changes are observed (Figure 5C). There is a HEPES buffer fragment in the active site of K234R as has been observed previously in a structure of SHV-2.25 The oxygens of the HEPES fragment are within hydrogen bonding distance of residues Thr-235 and Arg-244 as well as both conformations of Ser-130 and Arg-234 and the backbone nitrogen of Gly-236. Active site waters, including the deacylation water near Glu-166 and Asn-170, are conserved between the K234R and wild-type SHV structures, with the exception of a water near Arg-234. This water is present in the wild-type structure but is displaced by the second Arg-234 conformation (Figure 5B, red sphere).
Molecular Modeling/Molecular Dynamics Simulation (MM/MDS)
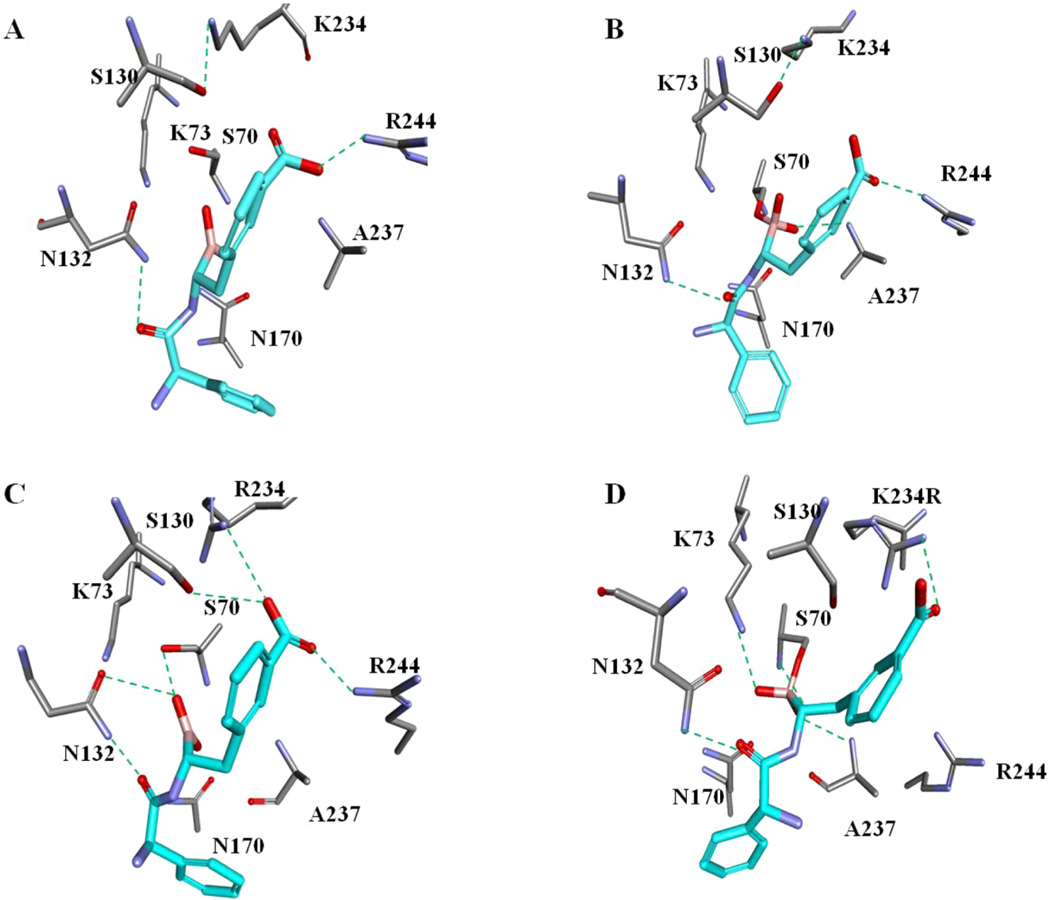
Molecular docking was performed with SHV-1 (Figure 6A,B) and K234R (Figure 6C,D) with the chiral ampicillin BATSI to understand the high affinity K234R shows for this inhibitor. To determine the role of Ser-130 in the inhibitor binding and to corroborate the X-ray structure findings, a 4 or 6 ps MDS was performed for SHV-1 and K234R in complex with the chiral ampicillin BATSI. During SHV-1 minimization and docking, the torsion angle of Ser-130 changes from −60° to −24° (Supporting Information Figure 2A), which shifts Ser-130 from pointing toward Lys-73 to Lys-234. After docking, the hydroxyl group of Ser-130 is oriented toward Lys-234 (Figure 6B). The 4 ps MDS of SHV-1 with the chiral ampicillin boronate shows a similar flexibility for the Ser-130 torsion angle. The model of SHV-1 and the chiral ampicillin BATSI show no interaction with the Lys-73 and Asn-132 residues. In the preacylation model of SHV-1 (Figure 6A), the hydrogen bonding pattern shows important hydrogen bonds forming with Ser-70, Asn-132, Asn-170, and Arg-244 for a total of four hydrogen bonds.
Figure 6.
Molecular docking of the chiral ampicillin BATSI (light blue) and SHV-1 in the preacylation state (before dative bond formation) (A) and after dative bond formation (B) and K234R before dative bond formation (C) and after dative bond formation (D).
During the K234R molecular dynamic simulation, the χ1 torsion angle of Ser-130 changes from −171° to −129° (Supporting Information Figure 2B) and the carboxyl of the chiral ampicillin BATSI makes/breaks hydrogen bonds with Arg-234 (Figure 6C,D). The preacylation model of K234R (Figure 6C) has a more extensive hydrogen bonding pattern with important hydrogen bonds occurring between the chiral ampicillin BATSI and amino acids Ser-70, Asn-132, Ser-130, Arg-234, and Arg-244 for a total of six hydrogen bonds. The hydrogen bonds with Ser-70, Asn-132, and Arg-244 are preserved in both models (Figure 6). The lack of interactions between the chiral ampicillin BATSI and Lys-234 in SHV-1 may be due to the difference in Ser-130 mobility angle in SHV-1 and K234R.
DISCUSSION
We performed an in-depth examination of position 234 in SHV to increase our understanding of the phenotype of this important residue in IR SHVs and to develop insight into novel compound design.
The Microbiological and Biochemical Impact of Amino Acid Substitutions at Ambler Position Lys-234 in SHV
Susceptibility testing of all 19 amino acid variants at position 234 revealed that only the K234R substituted enzyme was able to reduce susceptibility to ampicillin/clavulanate (Table 1). As observed by Drawz, et al. in studies on the N276D variant, the K234R variant retains a high level of resistance to ampicillin and piperacillin, which is not commonly observed in typical IR enzymes.8 Additionally, all of the K234 variants were less resistant to ampicillin/tazobactam and ampicillin/sulbactam than the wild-type SHV-1 in E. coli DH10B. This same susceptibility pattern was observed earlier in exploring the Asn-276 variants and in investigating the role of Arg-244 in SHV.8,17,18
Steady-State Kinetics
Steady-state kinetics assays were performed to find a biochemical basis for the IR phenotype. Overall, the K234R variant has impaired β-lactamase activity when compared with the wild-type SHV-1 β-lactamase (kcat/Km ratios are 18–57% of wild-type) (Table 2). The Km and kcat/Km were least affected for piperacillin but most affected for cephalosporins.
Resistance to clavulanate in K234R is manifested as a 7-to-8-fold higher Ki of the enzyme for the inhibitor; more importantly, K234R seems to turn over clavulanate better than SHV-1 (a kcat/kinact or partition ratio of 50 for K234R) (Table 3). Notably, the K234R variant also shows an increased Ki for tazobactam and sulbactam relative to SHV-1, which is not reflected in the MIC measurements (SHV-1 expressed in E. coli shows more resistance to these inhibitors than the K234R variant). However, the lower kcat/kinact for sulbactam and tazobactam for the K234R variant vs SHV-1 (60 vs 12000 and 10 vs 50, respectively) provide an explanation for the higher susceptibility of the K234R variant to sulbactam and tazobactam relative to SHV-1.
Using BATSIs to Probe SHV-1 and the Novel Variant K234R
BATSIs were used to probe kinetic relationships among the enzymes. These compounds replace the β-lactam with a boronic acid moiety and form reversible covalent bonds with β-lactamase enzymes, allowing measurement of binding energies and providing an ability to probe the constraints of the active site of the enzyme.26,27
As has been observed before, the meta-carboxyphenyl group seems to be important for the SHV β-lactamase to have a high affinity for the BATSI compounds (the achiral cephalothin analogue lacking this group has a higher Ki than any BATSI compounds with the m-carboxyphenyl group).26 The other notable observations are the 38-fold lower Ki of the chiral cephalothin BATSI for the K234R variant relative to SHV-1 and the 23-fold lower Ki of the chiral ampicillin BATSI for the K234R variant relative to SHV-1. This lowers the Ki for this compound against K234R to 2.4 nM. These changes in binding affinities can be translated into changes in activation energy (ΔΔGcat, Table 2). This measures the energy required to proceed from the free β-lactamase and compound to the transition state. The largest changes are therefore seen with the chiral ampicillin and chiral cephalothin BATSIs, which have a ΔΔGcat of −1.9 and −2.0 kcal/mol, indicating the formation of the transition state with the K234R variant is thermodynamically favored.
Thermal Stability and CD
SHV-1 and K234R were next studied using CD (Figure 3). The K234R substitution destabilizes the enzyme by 6.2 °C (Figure 3B). This large change in Tm in solution was not expected from a single amino acid substitution. One possible explanation for this observation is that the larger side chain of arginine is interfering with other residues in the protein and resulting in the disorder. Alternatively, the observed movement of Ser-130 and Arg-234 in the crystal structure may result in the destabilization of this protein. However, in accordance with our X-ray crystallography data, the secondary structure of both proteins is similar (Figure 3A). One thing that was observed in the X-ray crystallography image was the absence of a conserved water molecule near Arg-234 in the K234R structure that is present near Lys-234 in the SHV-1 structure (Figure 5B, red sphere). The absence of this water molecule may be a source of destabilization due to the lack of interactions with amino acid 234 in the K234R structure that are able to occur in SHV-1.
CD was next performed with SHV-1 and K234R with a variety of inhibitors including the suicide inhibitors (clavulanate, sulbactam, and tazobactam) and BATSI compounds to observe changes in thermal denaturation and secondary structure of the enzymes in the presence of the inhibitors (Figure 4). For SHV-1, none of the compounds greatly affect the secondary structure of the enzyme although subtle changes can be seen in the presence of clavulanate (Figure 4A). Interestingly, the chiral ampicillin BATSI is able to stabilize the enzyme, increasing the melting temperature by 6.5 °C (Table 5 and Figure 4B). We suspect that this is a result of the added active site interactions of the meta-carboxyphenyl of the chiral ampicillin BATSI (56 ± 6 nM Ki) and not a function of the boronate specifically because the achiral boronate does not stabilize SHV-1.
The results seen when CD was performed with K234R and the various inhibitors are more intriguing (Table 5 and Figure 4C,D). There are changes observed in the secondary structure of the K234R variant in the presence of some of the substrates (sulbactam, tazobactam, and the chiral ampicillin BATSI) (Figure 4C). More subtle changes occur in the presence of clavulanate and the achiral cephalothin BATSI. Sulbactam, tazobactam, and the chiral ampicillin BATSI seem to decrease the α-helical content of K234R. Sulbactam has the greatest effect on these secondary structure changes. We note these inhibitors appear to change the structure of K234R with binding but do not change the structure of SHV-1. The lower stability of K234R may allow these compounds to more easily change the secondary structure of the enzyme.
One of the most unanticipated results is the melting curve of the K234R variant in the presence of sulbactam (Figure 4D). Sulbactam changes the melting curve of K234R so that it is no longer a single transition from folded to unfolded. Consequently, a melting point cannot be determined from this curve because the protein does not undergo a two-state transition. None of the other compounds show this effect with the K234R enzyme, and cause this type of change in the melting curve of SHV-1. Similar results are seen in the melting temperature measurements of the K234R variant in the presence of the different inhibitors relative to what was seen with SHV-1. Only the chiral ampicillin BATSI greatly increases the melting temperature of the K234R variant (6.6 °C). Again this seems to reflect the added active site interactions that resulted in the nanomolar Ki of this compound for the enzyme.
Overall, from the kinetics and CD studies, the chiral ampicillin boronate seems to be the most intriguing compound for combating IR variants of SHV with the K234R substitution. This compound shows a low Ki for the enzyme and seems to stabilize the β-lactamase as evident from the observed shifts in melting temperature.
Effect of the K234R Substitution on the Structure of SHV
Table 5 displays the refinement statistics obtained from the crystal of K234R. Overall, the structure of K234R is highly conserved with respect to wild-type SHV-1 (PDB ID: 1SHV) (Figure 5A,C). The variant active site bears striking resemblance to that of SHV-1. As mentioned earlier, the K234R substitution confers inhibitor resistance through changes in Ser-130.12 In that study, MDS suggested preferred χ1 torsion angles for Ser-130 in SHV-1 and in K234R.12 For SHV-1, the final torsion angle was −145 ± 11°, which orients the hydroxyl group toward Lys-73, whereas the K234R Ser-130 torsion angle is −61 ± 12°, which orients the hydroxyl group toward Arg-234.
In this reported structure, we observe both of these orientations (−68° and −144°, respectively) in a 65/35 occupancy ratio (Figure 5B). We also see two orientations of Arg-234 (A and B); this likely necessitates the two Ser-130 orientations because in the −144° Ser-130 orientation, the “B” Arg-234 orientation is too close to Ser-130 (1.76 Å). However, the “A” Arg-234 orientation is at a reasonable hydrogen bonding distance to Ser-130 (3.14 Å). This is in contrast to the wild-type enzyme structure where a single orientation of the shorter lysine side chain at position 234 would allow for both Ser-130 orientations (distances from Ser-130 to Lys-234 are 2.94 and 2.65 Å, respectively). Unlike in the K234R enzyme, when the carboxylic acid group of an inhibitor binds the SHV-1 active site, the Ser-130 residue is free to rotate to an optimal position for inhibitor binding. We see instability in both Ser-130 and Arg-234 in K234R; both of these residues participate in inhibitor binding as they are part of, or are in close proximity to, the carboxyl binding pocket. Small shifts are also observed for residues Arg-244 and Glu-166, but those could be due to the lower resolution of the SHV-1 structure.
On the basis of these structural observations, we suggest that inhibitor binding is affected, and thereby weakened, by this now mobile region of the active site. We do observe some local fluctuations in the Arg-234 and Ser-130 side chains, which may lead to the instability observed by CD. The lower catalytic efficiencies (Table 2) confirm the CD observations that there must be structural changes occurring in K234R. From the crystal structure, we would expect similar catalytic efficiencies because the active site configuration is relatively conserved between SHV-1 and K234R, but we are recording a significant loss in catalytic efficiency (2–5-fold loss). This supports our hypothesis that the crystal structure has captured a “snapshot” of the K234R variant, but other conformations of the β-lactamase exist in solution, leading to the change in catalytic properties by kinetic analysis and thermal denaturation by CD.
Molecular Modeling
As a result of the nanomolar affinity and stabilization of the protein by the chiral ampicillin BATSI, we performed molecular docking and MDS of SHV-1 and K234R with this compound. In accordance with our other data, the MDS revealed a greater flexibility and movement in K234R compared to SHV-1 (Supporting Information Figure 2A vs B). Before docking, the X-ray crystal structure of SHV-1 shows Ser-70 (χ = −114°) toward and hydrogen bonded with Lys-73. In the preacylation docking comparison of SHV-1 and K234R with the chiral ampicillin BATSI (Figure 6A vs C), a larger number of hydrogen bonds are seen between K234R and the chiral ampicillin BATSI compared to SHV-1 with this compound (Figure 6C vs A). Hydrogen bonds are seen with the chiral ampicillin BATSI and Ser-70, Ser-130, Asn-132, Arg-244, and Arg-234 in K234R (the hydrogen bonds with Lys/Arg-234 and Arg-244 are absent in SHV-1). The torsion angle of Ser-130 prevents the interaction of Lys-234 with the chiral ampicillin BATSI in SHV-1; interactions with Arg-244 are also not observed (Figure 6A). Additionally, the Ser-70 (χ1 = −125°) hydroxyl is positioned toward Ser-130 (which points toward Lys-234). Notably, the Ser-70 angle is different upon docking of the chiral ampicillin BATSI in K234R (χ1 = −100°, Figure 6C).
The preacylation conformation of K234R with the chiral ampicillin BATSI compound may allow the dative bond to form more easily, explaining the lower Ki of the chiral ampicillin boronate for K234R. This molecular modeling provides us with information that can be used to design drugs active against these IR enzymes. Lys-73 and Asn-132 may be important target sites for interactions between novel compounds and IR enzymes because two hydrogen bonds are present between the chiral ampicillin BATSI and these amino acids in K234R but not in SHV-1. Instead of focusing on interactions with Ser-70 and Ser-130 as sulbactam, tazobactam, and clavulanate do, it is important to utilize interactions with other important residues in order to combat enzymes that have already evolved inhibitor resistance.
Movement of Ser-130 Explains Variant Behavior
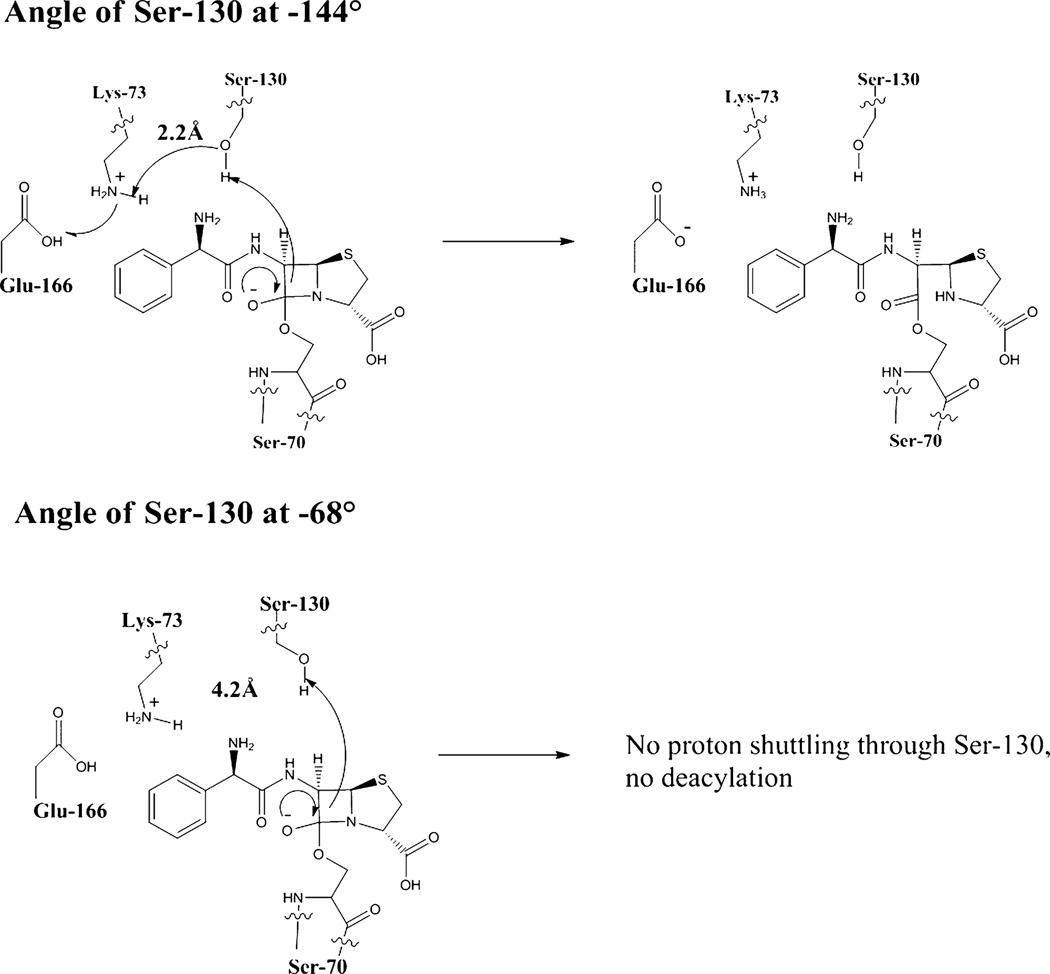
Quantum mechanical and molecular studies (QM and MM) have been performed with the TEM class A β-lactamase to understand the amino acids involved in the β-lactam breakdown reaction.28 These studies indicated that in TEM Ser-130 is involved in the tetrahedral collapse required for deacylation of the β-lactam substrate to occur. Ser-130 is involved in a proton relay, donating its proton to the β-lactam thiazolidine nitrogen while accepting a proton from Lys-73 (Figure 7). In K234R, one of the variations of Ser-130 moves the hydroxyl group from the −144° torsion angle to the −68° angle, which shifts the hydroxyl group away from Lys-73. In the −144° conformation of Ser-130, the oxygen atom of the hydroxyl group is 2.2 Å away from the Lys-73 hydrogen. However, in the −68° conformation, the oxygen is 4.2 Å away from the Lys-73 hydrogen. This greater distance likely prevents the observed proton shuttle from occurring between Lys-73, Ser-130, and the β-lactam. The inability of the proton shuttle to occur in one conformation of Ser-130 in K234R slows the catalytic breakdown of β-lactams by K234R and could underlie the kinetic results observed, i.e., the lower kcat and lower kcat/Km for the tested substrates.
Figure 7.
The top figure shows the wild-type enzyme with Ser-130 at a χ angle of −144° where proton shuffling can occur to allow breakdown of ampicillin by the β-lactamase. The bottom figure shows the movement of the hydroxyl group of Ser-130 away from Lys-73 when Ser-130 is at χ = −68°, which prevents the proton shuffling and ampicillin breakdown.
Additionally, Ser-130 is moved away from Ser-70 in the K234R structure relative to that of SHV-1. In the inactivation mechanism of class A β-lactamase enzymes by clavulanate, there is a cross-linking step between Ser-70, clavulanate, and Ser-130.2 The greater distance between Ser-70 and Ser-130 in K234R likely prevents this cross-linking from occurring in this enzyme. This explains the decreased resistance to clavulanate observed with this enzyme.
Mechanistic Implication
Our analysis of position 234 in SHV allows us to elucidate a novel pathway among IR variants of this enzyme involving the movement of Ser-130 and its relationship to K234R. Ser-130, like Lys-234, is involved in a conserved loop in class A β-lactamases, the SDN loop, which also includes Asp-131 and Asn-132.21 When a S130G substitution is present, resistance to clavulanate is observed.13,21 Ser-130 is involved in the permanent inactivation step of the inhibitor reaction, where cross-linking occurs between Ser-70, the inhibitor, and Ser-130.2 A glycine at position 130 prevents this cross-linking from occurring because the glycine does not have the hydroxyl group of serine, which allows cross-linking.
Other IR class A β-lactamase variants have been explored through crystal structures including TEM with a R244S substitution or M69I/V substitutions, which provide further information about the importance of Ser-130 in inhibitor resistance.29 These substitutions have also been observed in IR variants of SHV.15,17,18 In the crystal structures of the TEM Met-69 variants, Ser-130 was rotated, which is observed to diminish the ability of this amino acid to act as a site of cross-linking in inhibitor inactivation.29 In the R244S structure (TEM-30), there is a loss of a conserved water molecule which anchors the C3 carboxylate of the inhibitor.29 This affects the ability of Ser-130 to react with the inhibitor because it swings away from this amino acid.29 Thus, in the three previously studied IR class A β-lactamases (M69I/V, S130G, R244S), the common cause for the inhibitor resistance is a change in Ser-130.
Our work on the K234R variant indicates that the K234R substitution also leads to inhibitor resistance through a movement of Ser-130, which affects cross-linking with the inhibitor. A second hypothesis is that the presence of an arginine instead of a lysine at position 234 affects the electrostatic environment, which makes it easier for the breakdown of clavulanate and regeneration of the active enzyme (a higher partition ratio, kcat/kinact, is seen). Lastly, the altered interrelationships in the carboxyl binding pocket (as defined by Ser-130, Lys-234, Thr-235, and Arg-244) may be an additional cause for the inhibitor resistance and result in the higher Ki (Figure 5C).30 We have demonstrated that Ser-130 is the important residue underlying IR class A β-lactamases with a variety of different amino acid substitutions, (M69I/V, S130G, R244S, and now K234R) convening to lead to inhibitor resistance through an effect on the position of Ser-130.
EXPERIMENTAL METHODS
Mutagenesis, Sequencing, and Cloning
Using the template blaSHV-1 gene in the phagemid vector pBC SK(−) (Stratagene, La Jolla, CA), we used the Stratagene QuikChange Mutagenesis kit with degenerate oligonucleotides at Ambler position 234 to make the full complement of amino acid substitutions. After PCR mutagenesis, we electroporated the resulting plasmids into E. coli ElectroMAX DH10B cells (Invitrogen, Carlsbad, CA). These plasmids were isolated, and DNA sequencing of the blaSHV genes was performed. Variants not acquired after sequencing of 100 clones were created with specific mutagenic oligonucleotides that were designed as previously described.31
To aid in purification, the blaSHV K234R gene was cloned into the pGEX-6P-2 plasmid and transformed into E. coli OrigamiTM 2(DE3) competent cells (EMD Millipore, Billerica, MA). PCR was performed to obtain the blaSHV K234R gene from blaSHV K234R. Then restriction digests were performed on the plasmid and on the cloning vector. Ligation was performed and colonies were selected on ampicillin and sequenced.
Immunoblotting. E. coli
DH10B cells containing the blaSHV K234X plasmids were grown to OD600 = 0.8 and frozen at −20 °C for 48 h. A periplasmic extraction of the β-lactamases was performed by addition of 50 µL of 50 mM Tris-HCl (USB, Cleveland, OH) containing 5 µL of 40 µg/mL lysozyme (Sigma Aldrich), 5 µL of 10 mM MgSO4 (Teknova, Hollister, CA), and 1 µL of a 1000× dilution of benzonase (Novagen, Madison, WI), and tubes were allowed to stir for 25 min. Lastly, 1 µL of 50 mM EDTA was added to each tube followed by stirring for 5 min. Subsequently, debris was pelleted and 10 µL of sodium dodecyl sulfate (SDS) loading dye buffer was used to resuspend the pellet prior to incubation at 100 °C for 10 min. Immunoblots were performed with an anti-SHV polyclonal antibody to confirm periplasmic production of the SHV β-lactamases from all 19 constructs as previously described.32 Anti-DnaK (Assay Designs, Ann Arbor, MI) was used as a loading control as previously described.33
Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations (MICs) for E. coli DH10B cells expressing the blaSHV-1 or mutant blaSHV Lys234X genes cloned in phagemid pBC SK(−) were determined by Mueller–Hinton (MH) agar dilution according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.34 The MICs for various antibiotics were measured using a Steers replicator that delivered 10 µL of a diluted overnight culture containing 104 colony forming units. Ampicillin, piperacillin, and cephalothin were purchased from Sigma (St. Louis, MO). Lithium clavulanate was purchased from USP (Rockville, MD). Tazobactam was purchased from Chem-Impex International (Wood Dale, IL). Sulbactam (Pfizer, La Jolla, CA) was a kind gift. Tazobactam, sulbactam, and clavulanate were tested in combination with 50 µg/mL ampicillin.
β-Lactamase Expression and Purification
For kinetic assays and far-UV circular dichroism (CD), β-lactamase enzymes were expressed and purified as previously described35 using the pBC SK(−) plasmid or were expressed and purified from the pGEX-6P-2 plasmid (GE Healthcare, Piscataway, NJ). For SHV-1, E. coli DH10B cells containing the blaSHV-1 gene in pBC SK(−) were grown overnight in superoptimal broth (SOB) medium, harvested by centrifugation at 4 °C, and frozen at −20 °C. After thawing, the β-lactamase was liberated using stringent periplasmic fractionation with 40 µg/mL lysozyme (Sigma Aldrich) and 100 µL of 50 mM EDTA, pH 7.8. Preparative isoelectric focusing was performed with the lysate in a Sephadex granulated gel (GE Healthcare, Piscataway, NJ) using ampholines in the pH range 3.5–10 (Bio-Rad) and running the gel overnight at a constant power of 8 W on a Multiphor II isoelectric focusing apparatus (GE Healthcare). Preparative isoelectric focusing fractions were concentrated and purified using ÄKTA fast protein liquid chromatography (GE Healthcare, Piscataway, NJ). For SHVK234R, E. coli Origami 2(DE3) cells containing the blaSHV K234R gene in pGEX-6-P-2 were grown to OD600 = 0.6 in SOB medium at 37 °C, then 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added and the cultures were moved to a shaker at 16 °C overnight. The cells were harvested by centrifugation at 4 °C and frozen at −20 °C. After thawing, the β-lactamase was liberated by fractionation with 40 µg/mL lysozyme (Sigma Aldrich). The lysate was purified on the GSTrap FF HiTrap affinity column according to the manufacturer’s instructions (GE Healthcare, Piscataway, NJ). Residual TEM β-lactamase from the pGEX-6P-2 vector was removed using ÄKTA fast protein liquid chromatography (FPLC). The GST tag was cleaved using PreScission Protease (GE Healthcare, Piscataway, NJ). Then the protein was purified a second time with the GSTrap FF HiTrap affinity column to separate the SHVK234R and the cleaved GST tag. Purity was assessed by gel electrophoresis using a 5% stacking, 12% resolving SDS-polyacrylamide gelelectrophoresis (SDS-PAGE) and was found to be greater than 95%. Purity was confirmed with mass-spectrometry. β-Lactamase concentration was determined using a spectrophotometric assay (Biorad Laboratories, Hercules, CA) with ε = 33585 M−1 cm−1 for SHV-1 and ε = 32095 M−1 cm−1 for the K234R variant.
For crystallization, a different protein expression and purification procedure was followed. The K234R substitution was introduced into SHV-1 subcloned into the pET24a+ vector (Novagen, Madison, WI) without the leader sequence. Mutagenesis was performed using the QuikChange Lightning Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA) to change lysine (AAG) to arginine (CGC). Mutagenized DNA was confirmed by sequencing and transformed into OneShot BL21(DE3) Star chemically competent E. coli cells (Invitrogen, Carlsbad, CA), per manufacturer protocol, and plated overnight at 37 °C on lysogeny broth (LB) agar containing 50 µg/mL kanamycin. Then 100 mL of LB media supplemented with 50 µg/mL kanamycin was inoculated with the plated colonies. This culture was incubated overnight at 37 °C with 225 rpm shaking. This was used to inoculate 10 L of LB with 50 µg/mL of kanamycin, which was incubated until OD600 = 0.5–0.6 was reached. A final concentration of 0.5 mM of IPTG was added to induce protein expression. Then 3–4 h after induction, cells were harvested by centrifugation and lysed by microfluidization on a Microfluidics M-110Y microfluidizer processor using two passes at 90 psi. Cleared lysate was purified by pIEF as described above. Nitrocefin positive fractions were pooled, concentrated, and loaded on a QSepharose FF (GE Lifesciences, Piscataway, NJ) anion exchange column using a starting buffer consisting of 20 mM bis-Tris pH 6.5 and using an elution buffer consisting of 20 mM bis-Tris pH 6.5 and 1 M KCl. The K234R β-lactamase was present in the flow-through fractions of this column with the elution buffer, whereas the majority of the impurities were not. Nitrocefin positive peaks were pooled, concentrated, and loaded onto a Superdex75 gel filtration column. Protein purity was assessed by SDS-PAGE gel and was found to be greater than 95%. The protein was concentrated to 5 mg/mL using Vivaspin20 centrifugal concentrator (Sartorious Stedim, Goettingen, Germany).
Kinetic Measurements
Steady-state kinetic measurements were performed using continuous assays at room temperature in an Agilent 8453 diode array spectrophotometer (Agilent, Palo Alto, CA). Each assay was performed in 10 mM phosphate-buffered saline (PBS) at pH 7.4. Measurements were obtained using nitrocefin (NCF) (BD Biosciences, San Jose, CA) (Δε482 = 17400 M−1 cm−1), piperacillin (Δε235 = −820 M−1 cm−1), and cephalothin (Δε262 = −7660 M−1 cm−1). To investigate the role of position 234 in recognition of substrates and with changes in the binding pocket in a Lys to Arg substitution, we used a variety of BATSI compounds (achiral cephalothin,18 chiral cephalothin (synthesis described here), chiral nafcillin (synthesis described here), chiral cefoperazone (synthesis described here), and chiral ampicillin (synthesis described here)).
The kinetic parameters Vmax and Km were obtained with nonlinear least-squares fit of the data (Henri–Michaelis–Menten equation) using EnzFitter (Biosoft Corporation, Ferguson, MO) as described in eq 1.
| (1) |
The dissociation constants of the preacylation complex, Kis, were determined by direct competition assays. In our experiments, we use the term Ki to represent the Km of the inhibitor. The initial velocity was measured in the presence of a constant concentration of enzyme with increasing concentrations of inhibitor against the indicator substrate, NCF. Care was taken to initiate the reaction with the addition of both NCF and the inhibitor and to limit the initial velocity determination to the first 5 s.
Ki values for the BATSIs were determined as described above. However, because of an incubation effect, SHV β-lactamase and the boronic acid compounds were preincubated for 5 min in PBS before initiating the reaction with the addition of substrate (NCF) as previously described.18,36
The Ki data was corrected to account for the affinity of NCF for SHV-1 and K234R according to eq 2.37
| (2) |
For the inhibitors, Ki was used in place of Km to obtain the energies needed to form the preacylation complex with either enzyme.
The activation energy required for bond breaking and formation (ΔΔGcat) was calculated using the kinetic constants and eq 3 as previously described.38,39
| (3) |
Mass Spectrometry
Electrospray ionization (ESI)-MS was performed on SHV-1 and the SHVK234R variant with an Applied Biosystems (Foster City, California) Q-STAR Elite quadrupole-time-of-flight mass spectrometer equipped with a TurboIon spray source (Cleveland State University). First, 10 µg of enzyme was desalted and concentrated for ESI-MS using a C18 ZipTip (Millipore, Billerica, Massachusetts) according to the manufacturer’s protocol. Then eluted protein samples were diluted with 50% acetonitrile and 0.2% formic acid to a concentration of 10 mM, infused at a rate of 0.5 µL per min, and data were collected for 2 min. Spectra were deconvoluted using the Applied Biosystems Analyst program.
Circular Dichroism (CD)
CD experiments were performed in a Jasco J-815 spectrometer (Easton, MD) with a Peltier effect temperature controller (GE Healthcare, Piscataway, NJ). Quartz cells with a 0.1 cm path length (Hellma, New York) were used for all experiments. Spectra were obtained with a protein concentration of 10 µM. Compounds were tested at a concentration of 50 µM to ensure that they did not interfere with the refraction of the light by the protein in the far UV spectrum. Thermal melting was performed between 22 and 72 °C with a heating rate of 2 °C/min. Raw equilibrium denaturation data, monitored by far-UV CD at 208, 214, and 220 nm, were normalized to the fraction of denatured protein (fu). The data at 208 nm was used for calculations of melting temperature (Tm), van’t Hoff enthalpy (ΔHVH), the entropy of unfolding (ΔSu), and the change in Gibbs free energy of unfolding (ΔΔGu). The Van’t Hoff plot (ln Keq vs 1/T) was used to calculate ΔHVH as the slope times −R where R is the ideal gas constant. The y-intercept is ΔS × R. Equations 4 and 5 were used to determine Tm and ΔHVH.
| (4) |
| (5) |
With the assumption of a reversible two-state transition, equilibrium constants (Keq) at any given temperature were calculated from eq 6.40
| (6) |
The ΔΔGu between SHV-1 and SHVK234R was calculated using the method of Schellman shown in eq 7.41,42
| (7) |
In the calculation of ΔΔGu between the apo-enzymes and the complexes, ΔSapo was substituted for ΔSWT in eq 7.
Molecular Modeling and Molecular Dynamics Simulation (MM/MDS)
The atomic coordinates of SHV-1 (PDB ID: 1SHV) and the crystal structure reported here were used to construct the acyl–enzyme complexes. Acyl–enzyme models of the chiral ampicillin BATSI with the wild-type SHV-1 β-lactamase and K234R were constructed as previously described using Discovery Studio 3.1 (Accelrys, Inc. San Diego, CA) molecular modeling software.43 The K234R variant and the WT molecules were solvated (in a box of water with 7 Å distance from the molecule to the box) and minimized using the conjugated gradient method. The chiral ampicillin BATSI structure was constructed using Fragment Builder tools and was minimized using a Standard Dynamics Cascade protocol of DS 3.1. The inhibitor was automatically docked into the active site of the SHV-1 and the K234R β-lactamase using the CDOCKER module of DS 3.1.44 This protocol used a CHARMm-based molecular dynamics (MD) scheme to dock ligands into a receptor binding site. Random conformations were generated using high-temperature MD. The conformations were then translated into the binding site. Candidate poses were then created using random rigid-body rotations followed by simulated annealing. A final minimization was used to refine the ligand poses. The resulting conformations were analyzed with the most favorable positioning of the chiral ampicillin BATSI chosen and the complexes between the enzyme and inhibitor created as previously described.45 The complex was energy minimized and 4–6 ps molecular dynamic simulation was performed as previously described.38 The MDS was performed in three steps: heating/cooling, equilibration, and production. The target temperature was 300 K, and the long-range electrostatics were treated with Particle Mesh Ewald. The pressure was kept constant (NPT) during the equilibration and production steps to better mimic the experimental conditions for explicitly solvated systems with periodic boundary conditions (PBC). The distance cutoff value used for counting nonbonded interaction pairs was 14 Å. During the MDS the time steps were 0.001 ps, and the atom velocities and positions were calculated at time points that differ by a half time step.46–48
BATSI Synthesis and Analysis
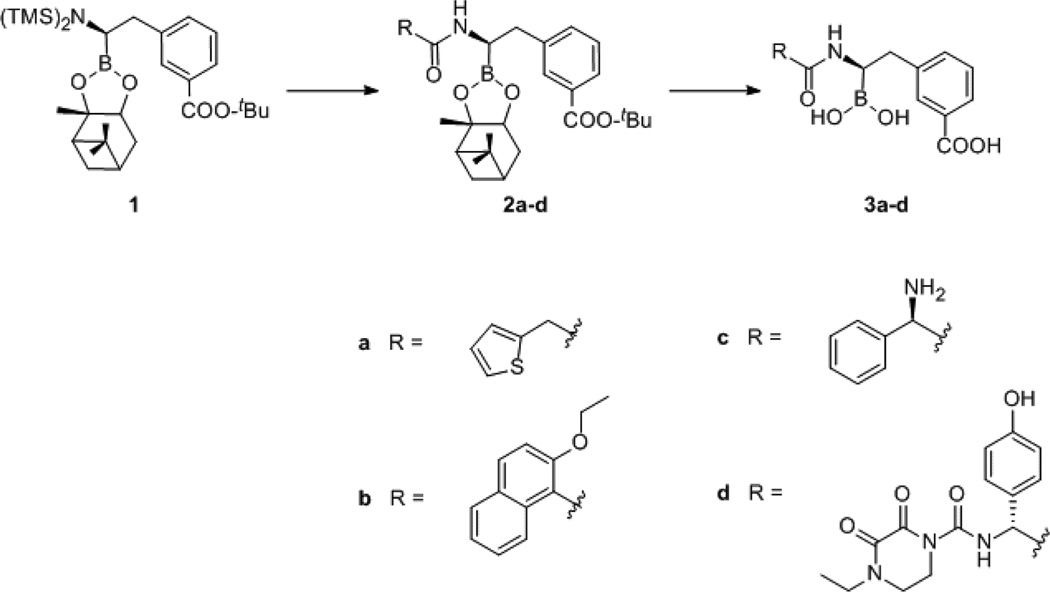
(Scheme 2) All reactions were performed under argon using oven-dried glassware and dry solvents. Anhydrous tetrahydrofuran (THF) and diethyl ether were obtained by standard methods and freshly distilled over sodium benzophenone ketyl prior to use under argon. All reagents were purchased from Sigma-Aldrich and Fluka. Reactions were monitored by thin layer chromatography (TLC), which were visualized by UV fluorescence and by Hanessian’s cerium molybdate stain. Chromatographic purification of the compounds was performed on silica gel (particle size 0.05–0.20 mm). Melting points were measured on a Büchi 510 apparatus. Optical rotations were recorded at 20 °C on a Perkin-Elmer 241 polarimeter and are expressed in 10−1 deg cm2 g−1. 1H and 13C NMR spectra were recorded on a Bruker DPX-200 or Avance-400 spectrometer; chemical shifts (δ) are reported in ppm downfield from tetramethylsilane (TMS) as internal standard (s singlet, d doublet, t triplet, q quartet, m multiplet, br broad signal). Two-dimensional NMR techniques (COSY, HMBC, HSQC) were used to aid in the assignment of signals in 1H and 13C spectra. Particularly, in the 13C spectra, the signal of the boron-bearing carbon atom tend to be broadened, often beyond the detection limit; however, its resonance was unambiguously determined by HSQC. Mass spectra were determined on a gas chromatograph HP 5890 with mass spectrometer detector HP 5972 (EI, 70 eV) or on an Agilent Technologies LC-MS(n) Ion Trap 6310A. Elemental analyses were performed on a Carlo Erba Elemental Analyzer 1110; elemental analyses for the compounds were within ±0.4% of the theoretical values. Elemental analyses of free boronic acids 3a–d were not obtainable because of the formation of dehydration products. Nevertheless, these boronic acids could be converted into analytically pure pinacol/pinanediol esters by simple exposure to equimolar amount of pinanediol in anhydrous THF. The starting (+)-pinanediol (1R)-1-(N-bis(trimethylsilyl)-amino)-2-phenylethaneboronate 1 was obtained as already described.49 Full synthesis of all compounds provided in Supporting Information.
Scheme 2.
Synthesis Scheme Leading to the Creation of the Various BATSI Compounds (a = Chiral Cephalothin, b = Chiral Nafcillin, c = Chiral Ampicillin, d = Chiral Cefoperazone)a
aSynthesis provided in Supporting Information.
Crystallization, Data Collection, and Refinement
The K234R protein was crystallized using the vapor diffusion method in 24-well sitting drop trays (Hampton Research, Aliso Viejo, CA). Well solution (500 µL) consisted of 21–30% PEG6K and 0.1 M HEPES pH 6.6–7.6. Five mg/mL K234R was combined with Cymal-6 (Hampton Research, Aliso Viejo, CA) at a final concentration of 0.56 mM and then mixed in a 1:1 ratio with well solution, yielding a final drop size of 5 µL. Crystals grew to full size within one week. Data was collected at Stanford Synchrotron Radiation Lightsource (SSRL), beamline BL9–2. Images were integrated and scaled using HKL2000.50 K234R crystallizes in the P212121 space group (Table 1 for data collection and refinement statistics). The structure was solved using isomorphous replacement with an existing apo wild-type SHV-1 structure (PDB ID: 1SHV). Refmac5 in the CCP4 suite51 was used to perform restrained anisotropic refinement and COOT52 was used for model building and manual refinement. Density for a partial HEPES buffer molecule was observed in the active site after initial refinement, and the fragment was included in subsequent refinement steps. Library files for the HEPES fragment were generated using PRODRG2.53 The coordinates for the apo K234R structure were deposited in the Protein Data Bank (PDB ID: 4FCF).
Supplementary Material
ACKNOWLEDGMENTS
M.L.W. has been supported by Medical Scientist Training Program Training Grant, Case Western Reserve University-T32 GM07250. K.P.W. is funded by the Veterans Affairs Career Development Program. R.A.B. is funded by Veterans Affairs Merit Review Program, VISN 10 GRECC, and NIH grants 1R01-A1063517 and 1R01-A100560. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS USED
- SHV
sulfhydryl-variable
- IR
inhibitor resistant
- TEM
Temoneira
- MIC
minimum inhibitory concentration
- amp
ampicillin
- pip
piperacillin
- thin
cephalothin
- clav
clavulanate
- sul
sulbactam
- tazo
tazobactam
- MM/MDS
molecular modeling/molecular dynamics simulation
- BATSI
boronic acid transition state inhibitor
- LB
lysogeny broth
- CD
circular dichroism
- MH
Muller–Hinton
- SOB
superoptimal broth
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Tm
melting temperature
- fu
fraction of denatured protein
- ΔHVH
van’t Hoff enthalpy
- ΔSu
entropy of unfolding
- ΔΔGu
Gibbs free energy of unfolding
- DS3.1
Discovery Studio 3.1
- THF
tetrahydrofuran
- TLC
thin layer chromatography
- TMS
tetramethylsilane
- SSRL
Stanford Synchrotron Radiation Light Source
- QM
quantum mechanical
Footnotes
ASSOCIATED CONTENT
Supporting Information
Full synthesis of the boronate compounds, Western blot of the SHV 234 variants, and MD simulation of Ser-130 movement in SHV-1 and K234R. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
The crystal structure can be found as 4FCF in the Protein Data Bank. The coordinates have been deposited.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
REFERENCES
- 1.Klevens RM, Edwards JR, Richards CL, Horan TC, Gaynes RP, Pollock DA, Cardo DM. Estimating Health Care-Associated Infections and Deaths in U.S. Hospitals, 2002. Public Health Rep. 2007;122:160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drawz S, Bonomo R. Three Decades of β-Lactamase Inhibitors. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buynak JD. Cutting and Stitching: The Cross-linking of Peptidoglycan in the Assembly of the Bacterial Cell Wall. ACS Chem. Biol. 2007;2:602–605. doi: 10.1021/cb700182u. [DOI] [PubMed] [Google Scholar]
- 4.Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Three-Dmensional Structure of the Bacterial Cell Wall Peptidoglycan. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lakaye B, Dubus A, Lepage S, Groslambert S, Frere JM. When Drug Inactivation Renders the Target Irrelevant to Antibiotic Resistance: A Case Story with β-Lactams. Mol. Microbiol. 1999;31:89–101. doi: 10.1046/j.1365-2958.1999.01150.x. [DOI] [PubMed] [Google Scholar]
- 6.Lovering AL, Safadi SS, Strynadka NC. Structural Perspective of Peptidoglycan Biosynthesis and Assembly. Annu. Rev. Biochem. 81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 7.Paterson DL, Bonomo RA. Extended-Spectrum β-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drawz S, Bethel CR, Hujer KM, Hurless KN, Distler AM, Caselli E, Prati F, Bonomo RA. The Role of a Second-Shell Residue in Modifying Substrate and Inhibitor Interactions in the SHV β-Lactamase: A Study of Ambler Position Asn276. Biochemistry. 2009;48:4557–4566. doi: 10.1021/bi9003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Llarena FJ, Bou G. β-Lactamase inhibitors: the story so far. Curr. Med. Chem. 2009;16:3740–3765. doi: 10.2174/092986709789104957. [DOI] [PubMed] [Google Scholar]
- 10.Manageiro V, Ferreira E, Cougnoux A, Albuquerque L, Canica M, Bonnet R. Characterization of the Inhibitor-Resistant SHV β-Lactamase SHV-107 in a Clinical Klebsiella pneumoniae Strain Coproducing GES-7 Enzyme. Antimicrob. Agents Chemother. 2012;56:1042–1046. doi: 10.1128/AAC.01444-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manageiro V, Ferreira E, Albuquerque L, Bonnet R, Canica M. Biochemical Study of a New Inhibitor-Resistant β-Lactamase, SHV-84, Produced by a Clinical Escherichia coli Strain. Antimicrob. Agents Chemother. 2010;54:2271–2272. doi: 10.1128/AAC.01442-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendonca N, Manageiro V, Robin F, Salgado MJ, Ferreira E, Canica M, Bonnet R. The Lys234Arg Substitution in the Enzyme SHV-72 Is a Determinant for Resistance to Clavulanic Acid Inhibition. Antimicrob. Agents Chemother. 2008;52:1806–1811. doi: 10.1128/AAC.01381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prinarakis EE, Miriagou V, Tzelepi E, Gazouli M, Tzouvelekis LS. Emergence of an Inhibitor-Resistant β-Lactamase (SHV-10) Derived from an SHV-5 Variant. Antimicrob. Agents Chemother. 1997;41:838–840. doi: 10.1128/aac.41.4.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubois V, Poirel L, Demarthe F, Aprin C, Coulange L, Minarini LAR, Bezian M-C, Nordmann P, Quentin C. Molecular and Biochemical Characterization of SHV-56, a Novel Inhibitor-Resistant β-Lactamase from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2008;52:3792–3794. doi: 10.1128/AAC.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubois V, Poirel L, Arpin C, Coulange L, Bebear C, Nordmann P, Quentin C. SHV-49, a Novel Inhibitor-Resistant β-Lactamase in a Clinical Isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004;48:4466–4469. doi: 10.1128/AAC.48.11.4466-4469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sulton D, Pagan-Rodriguez D, Zhou X, Liu Y, Hujer AM, Bethel CR, Helfand MS, Thomson JM, Anderson AE, Buynak JD, Ng LM, Bonomo RA. Clavulanic Acid Inactivation of SHV-1 and the Inhibitor-Resistant S130G SHV-1 β-Lactamase. J. Biol. Chem. 2005;280:35528–35536. doi: 10.1074/jbc.M501251200. [DOI] [PubMed] [Google Scholar]
- 17.Thomson J, Distler AM, Bonomo RA. Overcoming Resistance to β-Lactamase Inhibitors: Comparing Sulbactam to Novel Inhibitors against Clavulanate Resistant SHV Enzymes with Substitutions at Ambler Position 244. Biochemistry. 2007;46:11361–11368. doi: 10.1021/bi700792a. [DOI] [PubMed] [Google Scholar]
- 18.Thomson J, Distler AM, Prati F, Bonomo RA. Probing Active Site Chemistry in SHV β-Lactamase Variants at Ambler Position 244. J. Biol. Chem. 2006;281:26734–26744. doi: 10.1074/jbc.M603222200. [DOI] [PubMed] [Google Scholar]
- 19.Sun T, Bethel CR, Bonomo RA, Knox JR. Inhibitor-Resistant Class A β-Lactamases: Consequences of the Ser130-to-Gly Mutation Seen in Apo and Tazobactam Structures of the SHV-1 Variant. Biochemistry. 2004;43:14111–14117. doi: 10.1021/bi0487903. [DOI] [PubMed] [Google Scholar]
- 20.Helfand MS, Taracila MA, Totir MA, Bonomo RA, Buynak JD, van den Akker F, Carey PR. Raman Crystallographic Studies of the Intermediates Formed by Ser130Gly SHV, a β-Lactamase that Confers Resistance to Clinical Inhibitors. Biochemistry. 2007;46:8689–8699. doi: 10.1021/bi700581q. [DOI] [PubMed] [Google Scholar]
- 21.Helfand MS, Bethel CR, Hujer AM, Hujer KM, Anderson VE, Bonomo RA. Understanding Resistance to β-Lactams and β-Lactamase Inhibitors in the SHV β-Lactamase: Lessons from the Mutagenesis of SER-130. J. Biol. Chem. 2003;278:52724–52729. doi: 10.1074/jbc.M306059200. [DOI] [PubMed] [Google Scholar]
- 22.Ambler RP, Coulson AFW, Frere J-M, Ghuysen J-M, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. A Standard Numbering Scheme for the Class A β-lactamases. Biochem. J. 1991;276:269–272. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghuysen J-M. Serine β-Lactamases and Penicillin-Binding Proteins. Annu. Rev. Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 24.Huang W, Petrosino J, Hirsch M, Shenkin PS, Palzkill T. Amino acid sequence determinants of β-lactamase structure and activity. J. Mol. Biol. 1996;258:688–703. doi: 10.1006/jmbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- 25.Nukaga M, Mayama K, Hujer AM, Bonomo RA, Knox JR. Ultrahigh Resolution Structure of a Class A β-Lactamase: On the Mechanism and Specificity of the Extended-Spectrum SHV-2 Enzyme. J. Mol. Biol. 2003;328:289–301. doi: 10.1016/s0022-2836(03)00210-9. [DOI] [PubMed] [Google Scholar]
- 26.Thomson JM, Prati F, Bethel CR, Bonomo RA. Use of Novel Boronic Acid Transition State Inhibitors to Probe Substrate Affinity in SHV-Type Extended Spectrum β-Lactamases. Antimicrob. Agents Chemother. 2007;51:1577–1579. doi: 10.1128/AAC.01293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crompton IE, Cuthbert BK, Lowe G, Waley SG. The Inhibition of Serine β-lactamases by Specific Boronic Acids. Biochem. J. 1988;251:453–459. doi: 10.1042/bj2510453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meroueh SO, Fisher JF, Schlegel HB, Mobashery S. Ab Initio QM/MM Study of Class A β-Lactamase Acylation: Dual Participation of Glu166 and Lys73 in a Concerted Base Promotion of Ser70. J. Am. Chem. Soc. 2005;127:15397–15407. doi: 10.1021/ja051592u. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Minasov G, Shoichet BK. The Structural Bases of Antibiotic Resistance in the Clinically Derived Mutant β-Lactamases TEM-30, TEM-32, and TEM-34. J. Biol. Chem. 2002;277:32149–32156. doi: 10.1074/jbc.M204212200. [DOI] [PubMed] [Google Scholar]
- 30.Zafaralla G, Manavathu EK, Lerner SA, Mobashery S. Elucidation of the Role of Arginine-244 in the Turnover Processes of Class A β-Lactamases. Biochemistry. 1992;31:3847–3852. doi: 10.1021/bi00130a016. [DOI] [PubMed] [Google Scholar]
- 31.Hujer AM, Hujer KM, Helfand MS, Anderson VE, Bonomo RA. Amino Acid Substitutions at Ambler Position Gly238 in the SHV-1 β-Lactamase: Exploring Sequence Requirements for Resistance to Penicillins and Cephalosporins. Antimicrob. Agents Chemother. 2002;46:3971–3977. doi: 10.1128/AAC.46.12.3971-3977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hujer AM, Bethel CR, Bonomo RA. Antibody Mapping of the Linear Epitopes of CMY-2 and SHV-1 β-Lactamases. Antimicrob. Agents Chemother. 2004;48:3980–3988. doi: 10.1128/AAC.48.10.3980-3988.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papp-Wallace KM, Taracila MA, Smith KM, Xu Y, Bonomo RA. Understanding the Molecular Determinants of Substrate and Inhibitor Specificities in the Carbapenemase KPC-2: Exploring the Roles of Arg220 and Glu276. Antimicrob. Agents Chemother. 2012;56:4428–4438. doi: 10.1128/AAC.05769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 1st ed. Wayne, PA: Approved Standard: Clinical and Laboratory Standards Institute; 2008. [Google Scholar]
- 35.Lin S, Thomas M, Shlaes DM, Rudin SD, Knox JR, Anderson V, Bonomo RA. Kinetic Analysis of an Inhibitor-Resistant Variant of the OHIO-1 β-Lactamase, An SHV-Family Class A Enzyme. Biochem. J. 1998;333:395–400. doi: 10.1042/bj3330395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morandi F, Caselli E, Morandi S, Focia PJ, Blazquez J, Shoichet BK, Prati F. Nanomolar Inhibitors of AmpC β-Lactamase. J. Am. Chem. Soc. 2003;125:685–695. doi: 10.1021/ja0288338. [DOI] [PubMed] [Google Scholar]
- 37.De Meester F, Joris B, Reckinger G, Bellefroid-Bourguignon C, Frere J-M, Waley SG. Automated Analysis of Enzyme Inactivation Phenomena: Application to β-Lactamases and DD-Peptidases. Biochem. Pharmacol. 1987;36:2393–2403. doi: 10.1016/0006-2952(87)90609-5. [DOI] [PubMed] [Google Scholar]
- 38.Papp-Wallace KM, Taracila M, Wallace CJ, Hujer KM, Bethel CR, Hornick JM, Bonomo R. Elucidating the role of Trp105 in the KPC-2 β-lactamase. Protein Sci. 2010;19:1714–1727. doi: 10.1002/pro.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papp-Wallace KM, Taracila M, Hornick JM, Hujer AM, Hujer KM, Distler AM, Endimiani A, Bonomo RA. Substrate Selectivity and a Novel Role in Inhibitor Discrimination by Residue-237 in the KPC-2 β-Lactamase. Antimicrob. Agents Chemother. 2010;54:2867–2877. doi: 10.1128/AAC.00197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagarajan R, Pratt RF. Thermodynamic Evaluation of a Covalently Bonded Transition State Analogue Inhibitor: Inhibition of β-Lactamases by Phosphonates. Biochemistry. 2004;43:9664–9673. doi: 10.1021/bi049309b. [DOI] [PubMed] [Google Scholar]
- 41.Becktel W, Schellman J. Protein Stability Curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 42.Beadle BM, McGovern SL, Patera A, Shoichet BK. Functional Analyses of AmpC β-Lactamase through Differential Stability. Protein Sci. 1999;8:1816–1824. doi: 10.1110/ps.8.9.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papp-Wallace KM, Bethel CR, Distler AM, Kasuboski C, Taracila M, Bonomo RA. Inhibitor Resistance in the KPC-2 β-Lactamase, a Preeminent Property of This Class A β-Lactamase. Antimicrob. Agents Chemother. 2010;54:890–897. doi: 10.1128/AAC.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu G, Robertson DH, Brooks CLR, Vieth M. Detailed Analysis of Grid-Based Molecular Docking: A Case Study of CDOCKER-A CHARMm-Based MD Docking Algorithm. J. Comput. Chem. 2003;24:1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 45.Drawz SM, Taracila M, Caselli E, Prati F, Bonomo RA. Exploring Sequence Requirements for C3/C4 Carboxylate Recognition in the Pseudomonas aeruginosa Cephalosporinase: Insights into Plasticity of the AmpC β-Lactamase. Protein Sci. 2011;20:941–958. doi: 10.1002/pro.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verlet L. Computer “Experiments” on Classical Fluids. I. Thermodynamical Properties of Lennard-Jones Molecules. Phys. Rev. 1967;159:98–103. [Google Scholar]
- 47.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford, UK: Oxford University Press; 1987. Periodic Boundary Conditions and Potential Truncation. [Google Scholar]
- 48.Haile JM. Molecular Dynamic Simulations: Elementary Methods. New York: John Wiley & Sons; 1992. [Google Scholar]
- 49.Eidam O, Romagnoli C, Caselli E, Babaoglu K, Teotico Pohlhaus D, Karpiak J, Bonnet R, Shoichet BK, Prati F. Design, Synthesis, Crystal Structures, and Antimicrobial Activity of Sulfonamide Boronic Acids as β-Lactamase Inhibitors. J. Med. Chem. 2010;53:7852–7863. doi: 10.1021/jm101015z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. [20] Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Shuttelkopf AW, van Aalten DM. PRODRG: A Tool for High-Throughput Crystallography of Protein–Ligand Complexes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.