Abstract
Background
We explore the factor structure of DSM-5 cannabis use disorders, examine its prevalence across European- and African-American respondents as well as its genetic underpinnings, utilizing data from a genome-wide study of single nucleotide polymorphisms (SNPs). We also estimate the heritability of DSM-5 cannabis use disorders explained by these common SNPs.
Methods
Data on 3053 subjects reporting a lifetime history of cannabis use were utilized. Exploratory and confirmatory factor analyses were conducted to create a factor score, which was used in a genomewide association analysis. P-values from the single SNP analysis were examined for evidence of gene-based association. The aggregate effect of all SNPs was also estimated using Genome-Wide Complex Traits Analysis.
Results
The unidimensionality of DSM-5 cannabis use disorder criteria was demonstrated. Comparing DSM-IV to DSM-5, a decrease in prevalence of cannabis use disorders was only noted in European-American respondents and was exceedingly modest. For the DSM-5 cannabis use disorders factor score, no SNP surpassed the genome-wide significance testing threshold. However, in the European-American subsample, gene-based association testing resulted in significant associations in 3 genes (C17orf58, BPTF and PPM1D) on chromosome 17q24. In aggregate, 21% of the variance in DSM-5 cannabis use disorders was explained by the genomewide SNPs; however, this estimate was not statistically significant.
Conclusions
DSM-5 cannabis use disorder represents a unidimensional construct, the prevalence of which is only modestly elevated above the DSM-IV version. Considerably larger sample sizes will be required to identify individual SNPs associated with cannabis use disorders and unequivocally establish its polygenic underpinnings.
Keywords: Cannabis, DSM-5, GWAS, association, genetics, heritability
1. INTRODUCTION
Cannabis is the most commonly used illicit psychoactive substance in developed nations (Degenhardt and Hall, 2012). While a majority of cannabis users do not report problems, 10-30% of those who ever use cannabis meet criteria for a lifetime history of cannabis abuse or dependence as defined by the fourth edition of the Diagnostic and Statistical Manual (DSM-IV; American Psychiatric Association, 1994). Recently, changes to the diagnostic criteria for substance use disorder have been made in DSM-5 (American Psychiatric Association, 2013), including several for the diagnosis of cannabis use disorders (Hasin et al., 2013). Across the broad range of substance use disorders, (i) the distinction between abuse and dependence has been replaced by a unidimensional symptom count, with endorsement of 2 or more symptoms resulting in a DSM-5 diagnosis of substance use disorder (endorsement of specific numbers of symptoms define a mild, moderate or severe diagnosis); (ii) the DSM-IV criterion of legal problems has been eliminated from the diagnostic repertoire; and (iii) a new criterion for the DSM-5, craving (a long held substance dependence criterion in the International Classification of Disease, ICD) has been added. More specifically for cannabis, withdrawal is now a criterion. A wealth of psychometric evaluations in epidemiological (Agrawal et al., 2008; Compton et al., 2009; Gillespie et al., 2005; Hartman et al., 2008; Hasin et al., 2012, 2008; Langenbucher et al., 2004; Lynskey and Agrawal, 2007; Martin et al., 2006; Piontek et al., 2011; Wu et al., 2009, 2012) and clinical samples (Budney, 2006; Budney and Hughes, 2006) support these recommendations; however, the impact of these revisions on the prevalence of cannabis use disorders under the new DSM-5 classification remains largely unexplored. A recent study of Australian adults found a modest reduction in the rate of cannabis use disorder with the transition from DSM-IV to DSM-5 (Mewton et al., 2013), while another study of individuals with substance use disorders noted a modest increase of 4% (Peer et al., 2013).
Twin studies indicate that 50-60% of the variation in cannabis use disorders (abuse/dependence, variously defined using DSM-IIIR, DSM-IV and ICD) can be attributed to heritable influences (Verweij et al., 2010). Despite this robust heritability estimate, association studies for cannabis use disorders have largely failed to identify genetic variants of significant and replicable effect. A prior genome-wide association study (GWAS) of DSM-IV cannabis dependence, conducted in the sample used in this study, failed to identify genetic variants at a statistically significant level (Agrawal et al., 2011b). This has resulted in speculation regarding the biological underpinnings of cannabis use disorders; in particular, the question of whether common variation available in commercially available genome-wide arrays captures it (Sullivan et al., 2012).
Aggregating the effects of all single nucleotide polymorphisms (SNPs) on commercial arrays might quantify the overall role of common SNPs as well as causal variants in linkage disequilibrium (LD) with these SNPs on the trait of interest (Yang et al., 2010, 2011b). When significant, this would indicate that heritable variation in the trait is at least partially captured by these SNPs in a highly polygenic manner. Applying this methodology, investigators have successfully attributed 23-51% of the variation in current smoking, major depression, schizophrenia and human intelligence to genetic influences (Davies et al., 2011; Lee et al., 2012; Lubke et al., 2012).
The present study uses a multi-pronged phenotypic and genomic approach to evaluate, respectively, the architecture and genetic underpinnings of DSM-5 cannabis use disorders, defined as a quantitative phenotype. Instead of relying on a diagnostic measure, we first utilize item response models to construct a factor representing liability to DSM-5 cannabis use disorders, while accounting for sex and ethnic differences. Second, we use this psychometrically constructed factor score in a genome-wide association analysis. Finally, we evaluate whether genome-wide SNPs and putative causal variants in linkage disequilibrium with them explain a significant proportion of the heritable variation in DSM-5 cannabis use disorders.
2. METHODS
2.1 Sample
The Study of Addictions: Genes and Environment (SAGE) includes 3988 individuals ascertained from 3 study sources: the Collaborative Study of the Genetics of Alcoholism (N=1410; Begleiter et al., 1995; Reich et al., 1998), the Collaborative Study of the Genetics of Nicotine Dependence (N=1406; Bierut et al., 2007) and the Family Study of Cocaine Dependence (N=1172; Bierut et al., 2008). Further details regarding the study are available elsewhere (Bierut et al., 2010). The study includes substantial numbers of individuals who have used cannabis and experience problem use. For these analyses, data on 3053 (77% of the sample) individuals reporting a history of ever using cannabis were used.
2.2 Measures
2.2.1 DSM criteria
Twelve criteria from DSM-IV and DSM-5 (American Psychiatric Association, 1994) were utilized (Table 1). These included DSM-IV abuse criteria of (i) failure to fulfill major role obligations (role failure), (ii) recurrent use in hazardous situations (hazard), (iii) recurrent social/interpersonal problems because of use (social/interpersonal), and (iv) legal problems (legal), as well as the six DSM-IV dependence criteria of (v) tolerance, (vi) using in larger quantities or for longer than intended (larger/longer), (vii) persistent failed quit attempts (quit), (viii) spending a great deal of time using cannabis (time spent), (ix) giving up important activities to use cannabis (give up) and (x) experiencing physical or psychological problems because of cannabis use (problems). In addition, the two DSM-5 criteria of (xi) withdrawal and (xii) craving were also used.
Table 1.
Prevalence (%) of individual DSM-IV and proposed DSM-5 criteria for cannabis use disorder in 3053 lifetime cannabis users of European-American (EA) and African-American (AA) ancestry.
| Males | Females | |||
|---|---|---|---|---|
| EA | AA | EA | AA | |
| Role Obligations | 26.0a | 25.8a | 10.0b | 13.2b |
| Hazard | 49.2 | 37.3 | 21.8 | 16.8 |
| Legal | 4.8a | 3.2a | 1.4b | 1.4b |
| Social/Interpersonal | 31.3a | 26.9a | 14.2b | 13.8b |
| Tolerance | 34.0a | 32.5a | 13.2 | 17.2 |
| Withdrawal | 21.2a | 24.8a | 9.6 | 14.6 |
| Larger/Longer | 26.5 | 35.8 | 13.7 | 19.4 |
| Quit | 29.1 | 35.1 | 13.9 | 23.8 |
| Time Spent | 34.4a | 37.3a | 12.9 | 20.8 |
| Give up | 24.3a | 21.6a | 8.4b | 10.4b |
| Problems | 26.1a | 27.2a | 14.6b | 16.8b |
| Craving | 17.9a | 19.8a | 7.0 | 12.0 |
| DSM-IV abuse/dependence | 55.4a | 52.6a | 28.1b | 30.2b |
| DSM-5 use disorder | 51.1a | 52.6a | 25.1 | 31.8 |
Prevalence with the same superscripts could be statistically equated to each other at p> 0.05
Prevalence with the same superscripts could be statistically equated to each other at p> 0.05
2.2.2 DSM-5 factor score
Based on the factor analyses described below, a score representing liability to DSM-5 cannabis use disorders was used as a quantitative index.
2.3 Genotyping
The genotyping and quality control procedures applied to these data are explained in detail in earlier publications (Bierut et al., 2010; Laurie et al., 2010). In brief, DNA samples from 3988 individuals were genotyped on the Illumina Human 1M beadchip by the Center for Inherited Diseases Research (CIDR) at Johns Hopkins University. As described earlier, 948,658 SNPs passed data cleaning protocols. No imputed data were used for these analyses. HapMap genotyping controls, duplicates, related subjects, and outliers were removed. For the current analyses, data on 3,053 (77% of the sample) individuals reporting a lifetime history of cannabis use were used. Self-identified ethnicity (consistent with analysis of genetic data) was 2,018 European Americans and 1,035 African Americans.
2.4 Statistical Analysis
2.4.1 Phenotypic Factor Analysis
We used MPlus (v5; Muthen and Muthen, 2007) to conduct exploratory and confirmatory factor analyses of the 12 DSM-IV/DSM-5 criteria in the same sample. Exploratory analyses were conducted in the full sample, while subsequent confirmatory factor analyses were conducted in African-American (AA) and European-Americans (EA), separately by sex, using a multi-group framework. Initially, factor loadings and thresholds were constrained across the ethnic groups and across sexes. Individual submodels were tested to determine whether allowing the factor loading and threshold for each criterion to vary across the groups resulted in a significant improvement in model fit. The model that accommodated all statistically significant differences was used to generate factor scores that were subsequently used for genome-wide association analysis.
2.4.2 Genetic Analyses
GWAS: A linear regression model, in the PLINK (Purcell et al., 2007) software package, was used. Genotype was coded log-additively (i.e. increasing copies of the minor allele, selected from the full sample with ethnicities combined). Analyses were conducted separately in the EA (N=2018) and AA (N=1035) subsamples, adjusting for further ethnic differences via the inclusion of 2 principal components (generated via EIGENSTRAT; Price et al., 2006)). Other covariates included age at interview (dummy-coded to represent the lower three quartiles with the oldest age group used as a reference), sex and study source (whether the participants were drawn from COGA, COGEND or FSCD). The results from the EA and AA subsamples were meta-analyzed in METAL (Willer et al., 2010) using inverse-variance weighting.
2.4.3 Gene-based association analysis
We used the Versatile Gene-based Association Study (VEGAS) program to conduct gene-based tests of association (Liu et al., 2010). VEGAS assigns individual SNPs to each of the 17,787 autosomal genes (by physical position on the UCSC hg18 Genome Browser assembly). P-values from the GWAS are converted to upper-tailed chi-square statistics and then used to examine whether the chi-square distribution for each gene deviates from the null distribution. Due to differing linkage disequilibrium patterns across ethnicities, gene-based association was conducted on results when EA (using CEU) and AA (using YRI) subjects were analyzed separately.
2.4.4 Estimation of total genomic variation (heritability)
Genome-wide Complex Trait Analysis (GCTA; Yang et al., 2011a) was used to estimate the proportion of genomic variation explained by all SNPs available from the GWAS. Univariate analyses were conducted for the factor score, and covariates (sex, age, study site and ethnicity as indexed by principal components) were including in all computations. Analyses were restricted to those of self-reported EA ancestry. Analyses were not conducted in the AA subset because the modest sample size would likely have resulted in a large standard error.
3. RESULTS
3.1 Sample characteristics
The sample used for analyses was restricted to those who reported at least a one lifetime use of cannabis (N=3053; 49% male; 32.5% from COGA, 38.5% from COGEND, 29% from FSCD; 66% self-reported EA; mean age of 38.1 [18-68 years]). These individuals are characterized with respect to the 12 individual DSM-IV/DSM-5 criteria in Table 1. Prevalence of each criterion was higher in males than females for both ethnic groups, and males, regardless of ethnicity, were more likely than females to meet criteria for DSM-IV and DSM-5 diagnoses. However, several intriguing ethnic differences emerged. For both sexes, hazardous use, use of larger amounts or for a longer period of time and desire to quit or multiple failed quit attempts were differentially endorsed by EA and AA. EA men and women were more likely to endorse hazardous use and less likely to endorse using larger amounts or for longer than intended and failed quit attempts than their AA counterparts. In addition, tolerance, time spent using cannabis and the DSM-5 criteria of withdrawal and craving were more commonly reported by AA women than their EA counterparts – similar differences were not noted for men. The prevalence of DSM-IV cannabis abuse/dependence was higher in men compared with women, but no within-sex ethnic differences were noted. For DSM-5, cannabis use disorder was again more common in men than women, and there were no ethnic differences in men. Howeverr, AA women were more likely to meet criteria compared with their EA counterparts (31.8% versus 25.1%). Comparing the prevalence of DSM-IV versus DSM-5 cannabis use disorders - within each group, very modest changes were observed. Decrease in overall prevalence was noted for EA, while AA women showed a slight increase and AA men remained unchanged. Examining the [95%] confidence limits for the point estimates, only the decrease in prevalence in the EA was statistically significant (For men: 55.4% [47.9-54.1] vs. 51.1% [52.2-58.6]; for women: 28.1 [25.4-30.9] vs. 25.1% [22.6-27.9]) while the estimates in AA subjects could be equated across diagnostic classification scheme (For men: 52.6% [48.3-56.9] vs. 52.6 [47.2-55.8]; for women: 30.2% [26.2-34.4] vs. 31.8% [27.7-36.1].
3.2 Factor analysis
An exploratory factor analysis of the full sample revealed that a single factor solution provided a reasonable fit to the data (Comparative Fit Index (CFI): 0.996, Root Mean Square Error of Approximation (RMSEA): 0.054). While a 2-factor exploratory solution modestly improved these fit indices (e.g., 2 factor solution: CFI: 0.999, RMSEA: 0.036), the inter-factor correlation was 0.90. Hence, we proceeded with the more parsimonious single factor confirmatory analysis, which readily approximates item response parameters. Confirmatory factor analysis of the 4 DSM-IV abuse, 6 DSM-IV dependence and the DSM-5 withdrawal and craving criteria revealed high factor loadings (0.75 – 0.90) for all criteria except legal problems (0.23), which was excluded (consistent with DSM-5) from further analyses comparing factor loadings and thresholds for each individual criterion across EA and AA males and females. The factor loadings and thresholds (all significant at p < .0001) from the model allowing for statistically significant differences across individual items are shown in Table 2. Factor loadings and thresholds could not be constrained across the groups for hazardous use, interpersonal problems, withdrawal, using more than intended, repeated/failed quit attempts, time spent and physical/psychological problems (please see Supplemental eTable 1 for fit indices1). Factor scores that accommodated these differing thresholds and factor loadings were created for each of the four subgroups and used for genomic analyses.
Table 2.
Standardized factor loadings [95% confidence intervals] from one factor confirmatory factor analysis in 3053 lifetime cannabis users of European-American (EA) and African-American (AA) ancestry.
| Males | Females | |||
|---|---|---|---|---|
| EA | AA | EA | AA | |
| Role Obligations | 0.90 [0.87-0.92] | 0.90 [0.87-0.92] | 0.90 [0.87-0.92] | 0.90 [0.87-0.92] |
| Hazard | 0.83 [0.78-0.88] | 0.73* [0.64-0.82] | 0.83 [0.78-0.88] | 0.71* [0.60-0.82] |
| Social/Interpersonal | 0.92 [0.88-0.95] | 0.90 [0.85-0.95] | 0.89 [0.85-0.94] | 0.89 [0.82-0.96] |
| Tolerance | 0.87 [0.85-0.90] | 0.87 [0.85-0.90] | 0.87 [0.85-0.90] | 0.87 [0.85-0.90] |
| Withdrawal | 0.88 [0.83-0.93] | 0.85 [0.78-0.92] | 0.92 [0.88-0.96] | 0.93 [0.89-0.98] |
| Larger/Longer | 0.84 [0.79-0.90] | 0.89 [0.84-0.94] | 0.91 [0.87-0.95] | 0.91 [0.86-0.97] |
| Quit | 0.75* [0.69-0.82] | 0.73* [0.63-0.83] | 0.85 [0.80-0.90] | 0.79* [0.70-0.88] |
| Time Spent | 0.87 [0.82-0.91] | 0.90 [0.84-0.94] | 0.92 [0.88-0.96] | 0.94 [0.89-0.93] |
| Give up | 0.92 [0.89-0.94] | 0.92 [0.89-0.92] | 0.92 [0.89-0.94] | 0.92 [0.89-0.94] |
| Problems | 0.91 [0.88-0.95] | 0.90 [0.84-0.95] | 0.92 [0.88-0.96] | 0.84 [0.75-0.92] |
| Craving | 0.90 [0.88-0.93] | 0.90 [0.88-0.93] | 0.90 [0.88-0.93] | 0.90 [0.88-0.93] |
| Mean Factor Score | 0.49 [0.31-0.67] | 0.50 [0.30-0.69] | −0.23 [−0.42- −0.05] | 0.00 [reference] |
Note: If factor loadings could be constrained, then they are shown as such. Differing factor loadings may not statistically differ from each other [i.e. as indicated by overlapping confidence limits] but may have been unconstrained in models because of differing thresholds, however those factor loadings with an * could be equated to each other but not to other estimates.
3.3 GWAS
Individual signals did not surpass the Bonferroni corrected genome-wide significance threshold of p < 5×10−8. The results for the top 20 SNPs are presented in Table 3 (the top 100 results for the EA and AA subsamples are available in eTable 2 and 3, respectively2). For the EA subsample, 11 SNPs on 17q23-24 appeared to be associated at nominal levels of significance although none surpassed the genomewide threshold of 5×10−8.The top SNP, rs6504555, was an intronic variant in the bromodomain PHD finger transcription factor (BPTF) gene – a regional association plot for this region of chromosome 17 is shown in Figure 1, indicating a high degree of linkage disequilibrium across the associated SNPs. With the exception of rs11870068, the remaining chromosome 17 SNPs were in moderate to high linkage disequilibrium (r2 ranging 0.66 to 1.0). In the AA subsample, results did not aggregate in any particular chromosomal region. The most significant SNP, rs4364205, on chromosome 3, was intergenic.
Table 3.
Top 20 association results from genome wide association study of the DSM-5 cannabis use disorders factor scores in 2018 European-American and 1,035 African-American lifetime cannabis users. Also shown are p-values from a meta-analysis of the results from both ethnic groups.
| Chromosome | SNP | Gene | Allele | Frequency | Beta | Lower 95% | Upper 95% | P-value | P-value other* | Dir. | Meta P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| European Americans | |||||||||||
| 17 | rs6504555 | BPTF | A | 0.22 | 0.15 | 0.09 | 0.20 | 1.73E-06 | 0.35 | ++ | 2.00E-05 |
| X | rs7884312 | DMD | A | 0.12 | 0.21 | 0.13 | 0.30 | 1.73E-06 | 0.87 | +- | 1.59E-03 |
| X | rs7880016 | DMD | A | 0.11 | 0.22 | 0.13 | 0.31 | 2.65E-06 | 0.53 | +- | 6.43E-03 |
| 17 | rs8071463 | BPTF | G | 0.22 | 0.14 | 0.08 | 0.20 | 3.25E-06 | 0.29 | -- | 2.16E-05 |
| 2 | rs3731808 | PDE11A | T | 0.01 | −0.58 | −0.83 | −0.33 | 4.82E-06 | 0.47 | -+ | 5.14E-04 |
| 12 | rs10082916 | RARG | T | 0.04 | −0.27 | −0.39 | −0.15 | 4.84E-06 | 0.60 | -- | 4.93E-04 |
| 12 | rs12307672 | RARG | A | 0.04 | −0.27 | −0.39 | −0.15 | 4.84E-06 | 0.71 | -- | 9.12E-04 |
| 17 | rs9897982 | BPTF | T | 0.22 | 0.14 | 0.08 | 0.20 | 5.10E-06 | 0.60 | ++ | 1.29E-04 |
| 17 | rs3935969 | BPTF | C | 0.22 | 0.14 | 0.08 | 0.20 | 5.13E-06 | 0.16 | -- | 1.35E-05 |
| 17 | rs2365468 | BPTF | T | 0.22 | 0.14 | 0.08 | 0.20 | 5.40E-06 | 0.55 | ++ | 1.12E-04 |
| 17 | rs9890629 | BPTF | A | 0.22 | 0.14 | 0.08 | 0.20 | 5.52E-06 | 0.15 | ++ | 1.26E-05 |
| 17 | rs8074078 | BPTF | G | 0.22 | 0.14 | 0.08 | 0.20 | 5.65E-06 | 0.15 | -- | 1.19E-05 |
| 17 | rs11870068 | BPTF | C | 0.22 | 0.14 | 0.08 | 0.20 | 5.80E-06 | 0.17 | -+ | 1.84E-03 |
| 12 | rs11065202 | C | 0.42 | 0.11 | 0.06 | 0.16 | 5.91E-06 | 0.95 | -+ | 2.39E-04 | |
| 17 | rs9891146 | C17orf58 | T | 0.28 | 0.13 | 0.07 | 0.18 | 6.42E-06 | 0.98 | ++ | 3.04E-04 |
| 17 | rs6504548 | BPTF | C | 0.22 | 0.14 | 0.08 | 0.20 | 6.49E-06 | 0.59 | -- | 1.41E-04 |
| 12 | rs2066938 | MGC5139 | G | 0.27 | 0.13 | 0.07 | 0.18 | 6.86E-06 | 0.09 | -- | 3.16E-06 |
| 17 | rs7208663 | BPTF | C | 0.22 | 0.14 | 0.08 | 0.20 | 7.13E-06 | 0.17 | -- | 1.85E-05 |
| 8 | rs12056774 | T | 0.23 | 0.13 | 0.07 | 0.19 | 7.71E-06 | 0.46 | ++ | 9.16E-05 | |
| 22 | rs165685 | PCQAP | T | 0.18 | 0.15 | 0.08 | 0.21 | 9.31E-06 | 0.65 | ++ | 2.16E-05 |
| African Americans | |||||||||||
| 3 | rs4364205 | T | 0.41 | −0.19 | −0.26 | −0.12 | 1.30E-07 | 0.06 | -+ | 1.10E-01 | |
| 1 | rs16853258 | G | 0.07 | −0.34 | −0.47 | −0.20 | 1.06E-06 | 0.16 | +- | 1.87E-06 | |
| 11 | rs1981990 | A | 0.30 | 0.19 | 0.11 | 0.26 | 2.62E-06 | 0.80 | +- | 8.70E-03 | |
| 2 | rs7601137 | AFF3 | C | 0.14 | 0.25 | 0.14 | 0.35 | 3.39E-06 | 0.62 | ++ | 2.61E-03 |
| 8 | rs2410545 | NAT1 | A | 0.12 | −0.27 | −0.39 | −0.16 | 4.08E-06 | 0.87 | -+ | 7.50E-02 |
| 2 | rs12479422 | A | 0.09 | 0.30 | 0.17 | 0.42 | 4.57E-06 | 0.29 | -- | 2.18E-04 | |
| 8 | rs11203943 | NAT1 | A | 0.05 | −0.38 | −0.54 | −0.22 | 4.67E-06 | 0.32 | ++ | 2.85E-03 |
| 10 | rs493965 | A | 0.48 | −0.17 | −0.24 | −0.10 | 5.83E-06 | 0.52 | -+ | 3.11E-02 | |
| 8 | rs6586712 | NAT1 | G | 0.12 | −0.26 | −0.37 | −0.15 | 8.95E-06 | 0.64 | +- | 1.56E-01 |
| 8 | rs16871627 | T | 0.17 | −0.21 | −0.31 | −0.12 | 9.41E-06 | 0.43 | -+ | 1.40E-05 | |
| 2 | rs865108 | STEAP3 | G | 0.47 | 0.17 | 0.09 | 0.24 | 9.56E-06 | 0.54 | -+ | 4.74E-02 |
| 22 | rs9627601 | TBC1D22A | C | 0.15 | −0.23 | −0.33 | −0.13 | 1.01E-05 | 0.23 | +- | 2.47E-05 |
| 14 | rs11850171 | G | 0.23 | 0.19 | 0.10 | 0.27 | 1.18E-05 | 0.92 | ++ | 1.31E-05 | |
| 3 | rs6774262 | A | 0.43 | 0.16 | 0.09 | 0.24 | 1.30E-05 | 0.19 | -- | 2.13E-04 | |
| 1 | rs950601 | A | 0.16 | −0.22 | −0.32 | −0.12 | 1.63E-05 | 0.20 | ++ | 1.35E-03 | |
| 1 | rs6677326 | G | 0.04 | 0.40 | 0.22 | 0.58 | 1.69E-05 | 0.87 | -+ | 2.30E-05 | |
| 10 | rs2815527 | G | 0.42 | 0.16 | 0.09 | 0.24 | 2.00E-05 | 0.67 | ++ | 6.75E-03 | |
| 22 | rs12170279 | TBC1D22A | G | 0.20 | −0.20 | −0.29 | −0.11 | 2.20E-05 | 0.30 | +- | 4.59E-05 |
| 7 | rs12668723 | A | 0.21 | 0.19 | 0.10 | 0.27 | 2.33E-05 | 0.12 | -- | 1.41E-04 | |
| 10 | rs2815523 | G | 0.42 | 0.16 | 0.09 | 0.24 | 2.54E-05 | 0.68 | ++ | 7.58E-03 | |
| 2 | rs7557254 | C | 0.37 | −0.16 | −0.24 | −0.09 | 2.60E-05 | 0.14 | -- | 2.50E-05 | |
P-value other indicates the p-value for that SNP in the other ethnicity (i.e. AA p-value for analyses conducted in European-Americans and vice versa).
Figure 1.
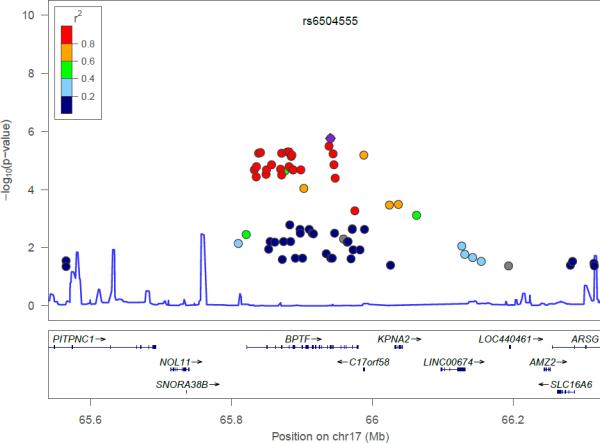
Regional association plot of chromosome 17 results from the European-American subsample. SNP with lowest p-value (diamond-shape, in purple).
Meta-analysis of the results from the EA and AA subsamples did not yield a boost in statistical significance (Table 3). This was evident from a comparison of results in the EA and AA subsamples. Of all SNPs with p-values < 0.05 in EA subsample, only 5% had corresponding p-values < 0.05 in AA subsample. However, particularly for the SNPs for the EA subsample shown in Table 2, the direction of effect in the AA subsample predominantly (with the exception of 5 of 20 SNPs) concurred with the EA subsample.
3.4 Gene-based association results
Three genes surpassed the conservative gene-based Bonferroni threshold of 2.8 × 10−6 in the EA, but not the AA subsample (Table 4). In the EA subsample, association was noted on chromosome 17q23-24 for C17orf158 (chromosome 17 open reading frame 58), and the adjacent genes BPTF and PPM1D (protein phosphatase Mg2+/Mn2+ dependent, 1D). Multiple other neighboring genes also showed aggregation of association signals although none surpassed gene-based correction. As VEGAS allows for SNPs to be assigned to the 50 kb region flanking the gene (footprint), this clustering of genes might be attributed to SNPs being assigned to the footprints of multiple neighboring genes. The association appeared to be specific to the EA subsample with corresponding p-values > 0.05 in the AA subsample. For results from the AA subsample, the lowest p-value for the gene-based association test was .00013 for Patched domain containing 3 (PTCHD3), which had a corresponding p-value of 0.61 in the EA results.
Table 4.
Top 10 genes showing association via gene-based association analysis in European- and African-American subsamples analyzed separately.
| Gene | Chromosome | Start basepair | End basepair | P-value (EA) | P-value (AA) |
|---|---|---|---|---|---|
| European-American subsample (N=2,018) | |||||
| C17orf58 | 17 | 63417678 | 63420227 | <1E-6 | 0.422 |
| PPM1D | 17 | 56032335 | 56096818 | <1E-6 | 0.149 |
| BPTF | 17 | 63252241 | 63410956 | 0.000001 | 0.351 |
| UNC119B | 12 | 11963221 | 11964582 | 0.000016 | 0.11 |
| LRRC37A3 | 17 | 60280949 | 60345365 | 0.000026 | 0.618 |
| ACADS | 12 | 11964795 | 11966219 | 0.000027 | 0.119 |
| KIAA0152 | 12 | 11960933 | 11962405 | 0.000033 | 0.136 |
| MYEF2 | 15 | 46218920 | 46257850 | 0.000137 | 0.944 |
| CABP1 | 12 | 11956280 | 11958951 | 0.000161 | 0.288 |
| ZNF681 | 19 | 23713836 | 23733533 | 0.000164 | 0.24 |
| African-American subsample (N=1,035) | |||||
| PTCHD3 | 10 | 27727122 | 27743303 | 0.000125 | 0.61 |
| AFF3 | 2 | 99530147 | 10012546 | 0.000381 | 0.466 |
| FILIP1L | 3 | 10103467 | 10131603 | 0.000415 | 0.224 |
| C3orf26 | 3 | 10101937 | 10138013 | 0.000527 | 0.215 |
| CNOT10 | 3 | 32701701 | 32790358 | 0.000556 | 0.171 |
| FANCM | 14 | 44674885 | 44739843 | 0.000565 | 0.407 |
| IAPP | 12 | 21417084 | 21423683 | 0.000588 | 0.493 |
| NMUR2 | 5 | 15175129 | 15176503 | 0.000603 | 0.674 |
| FKBP3 | 14 | 44654858 | 44674272 | 0.000668 | 0.311 |
| CCDC91 | 12 | 28301399 | 28594366 | 0.000686 | 0.257 |
3.5 Total genomic variation (heritability)
Genomic variation was responsible for 21% (SE=17.7) of the phenotypic variance in the factor score. However, the estimate was not statistically significant (p=0.13).
4. DISCUSSION
We sought to examine the phenotypic and genomic architecture of a continuously distributed cannabis use disorders factor, psychometrically derived from DSM-5 criteria, in samples ascertained for alcohol, nicotine and cocaine dependence. Our analyses revealed a high degree of support for the unidimensionality of cannabis use disorders. Analysis of ethnic differences indicated a modest reduction in the prevalence of DSM-5 cannabis use disorders, relative to DSM-IV, in EA. Genomic analyses, using a genome-wide scan, failed to identify SNPs that satisfied statistical thresholds for significance; however, gene-based association implicated genes on the q-arm of chromosome 17. A genome wide variance calculation revealed that 21% of the phenotypic variance in cannabis use disorders was captured by the available common variation on the genome-wide array, but this estimate had a large standard error and was not significant.
We used the factor score as our phenotype for genomic analyses. Incorporating withdrawal and craving, excluding legal problems and combining across DSM-IV abuse and dependence criteria, this factor embodies the ‘spirit’ of the new DSM-5 diagnostic scheme while not being encumbered by concerns that the threshold of 2 or more criteria for diagnosis of disorder is too lax (Martin et al., 2011b). From a psychometric perspective, our results are consistent with the extant literature (see Hasin et al. 2013 for a comprehensive overview). For instance, despite our sample being ascertained for alcohol, nicotine and cocaine dependence, which inflated endorsement rates of individual criteria (i.e., due to the high comorbidity between alcoholism and cannabis use disorder), our high rates of hazardous use were comparable with those reported for lifetime cannabis users from the general population as reflected in data from the National Epidemiological Survey of Alcohol and Related Conditions (Agrawal and Lynskey, 2007; Compton et al., 2009). Likewise, broadly consistent with numerous other studies, the DSM-IV abuse criterion of legal problems was infrequently endorsed and had a weak factor loading, affirming its proposed exclusion from DSM-5. The overall prevalence of the remaining criteria, although much higher than in general population cohorts, supports the presence of a unidimensional construct across sexes and ethnicities. Craving and withdrawal, both of which have been added to DSM-5, performed well, with high factor loadings supporting their inclusion.
Overall, rates of diagnostic DSM-5 cannabis use disorders appear to be modestly lower than those for DSM-IV abuse/dependence, but only in EA, particularly men. This finding is highly comparable with epidemiological analyses of alcohol symptomatology in U.S. (Agrawal et al., 2010; Martin et al., 2011a; Verges et al., 2011) and with results from the 2007 Australian National Survey of Mental Health and Wellbeing, which reported a decrease in the lifetime rate of cannabis use disorder from 6.2% to 5.4% when transitioning from DSM-IV to DSM-5 (Mewton et al., 2013). In our sample, this decrease was uniformly attributable to individuals who endorsed hazardous use alone, which results in a DSM-IV diagnosis of cannabis abuse but not a DSM-5 diagnosis of cannabis use disorder, because it falls below the latter's minimum two-symptom threshold. No differences were noted in AA men (or women), and this is also not surprising. Individuals endorsing this criterion alone tend to be of higher socio-economic standing (Keyes and Hasin, 2008) and tend to, overwhelmingly, endorse this criterion due to a history of drinking and driving (Agrawal et al., 2011a). That socio-economic status may correlate with ethnicity is expected – in our data, 45.9% of AA participants reported a gross annual income of less than $20,000, vs. 15.4% of their EA counterparts.
Upon examining gender and ethnic differences within classification version (e.g. DSM-5 diagnoses across males and females), the only significant variation was noted for DSM-5 diagnoses in AA women who were more likely to receive a diagnosis of DSM-5, but not DSM-IV cannabis use disorder, relative to their EA female counterparts. Intriguingly, also relative to their EA counterparts, they were less likely to endorse hazardous use but more likely to endorse numerous other criteria, with the exception of giving up important activities and use despite physical/psychological problems. This finding may be attributable to the larger number of AA women that were ascertained from the cocaine dependence study (46% AA versus 18.5% EA women are drawn from FSCD) versus other studies. Although this observation holds true for the men as well, and the prevalence (or mean number of symptoms endorsed) did not vary across AA and EA women, it is possible that AA women (but not men or EA women) from the FSCD study represent a high-risk group. For instance, when compared to the alcohol and nicotine dependence studies, AA women from the cocaine (FSCD) study were more likely to report lower household income (59.6 vs. 34.4%) and a greater likelihood of less than a high school education (32.6 vs 17.4%). Thus, this vulnerability might reflect environmental adversity rather than increased genetic susceptibility, and in any case, is accounted for in the genomic analyses by incorporating study sample and gender as covariates.
From a genetic perspective, the single SNP analyses did not reveal any genome wide significant signals. This is likely because our sample is underpowered, even with a quantitative trait, to detect single variants of modest effect size. Using GWAPower (Feng et al., 2011), we estimated power available in our dataset to identify SNPs of varying effect size. Power was 80% when an effect size of 0.01 (1%) was anticipated (with covariates explaining about 20% of the variance, and Type 1 error set at 5 × 10−8). Increasing efforts to amass larger samples with comparable cannabis-related data would afford greater power to detect variants of more modest effect size via meta- and mega-analyses. However, few current studies have DSM-5 criteria data. In this regard, factor scores (or symptom counts) such as ours may prove to be useful phenotypes as they can accommodate DSM-IV and DSM-5 based assessments of vulnerability to cannabis use disorders.
In contrast, the gene-based analyses conducted with the European-American subsample identified a cluster of genes, of varied function, on the q-arm of chromosome 17 that appeared to contain an aggregation of variants associated with DSM-5 cannabis use disorders. The genes that surpassed gene-based correction were C17orf58, BPTF and PPM1D. PPM1D is in a region of chromosome 17 that is well documented to be amplified in breast cancer (Bernards, 2004), and the gene itself belongs to a family of serine/threonine phosphatases that are involved in stress signaling (Lowe et al., 2012). On the other hand, BPTF was originally identified in brain homogenates from deceased Alzheimer's patients (Jordan-Sciutto et al., 2000). It is putatively involved in chromatin remodeling (Landry et al., 2008). We hesitate to speculate about the potential role of these genes in the etiology of cannabis use disorders.
Contradictory to the extant twin literature positing 50% heritable variation in cannabis use disorders, the aggregate effects of SNPs on the array captured 21% of genetic variation; however, this estimate was not statistically significant. The lack of significance is primarily due to our sample size. For instance, with cigarette smoking, a sample size of 4181 yielded a heritability estimate of 19% at p=0.024. It is, however, worth noting that similar to other major psychiatric disorders (Lee et al., 2013), common variation on commercial arrays does not capture all the postulated heritability in complex traits. This may be attributable to imperfect linkage disequilibrium between these SNPs and rarer causal variants or due to other factors, such as epistasis, gene-environment interplay and other variation (e.g., copy number variants).
Some other limitations of the present study are worth noting. First and foremost, the present sample was ascertained from three family studies of substance use disorders for the express purpose of identifying genetic variants for alcoholism, nicotine and cocaine dependence and related psychopathology. Hence, the psychometric analyses may not generalize to other cohorts with different ascertainment criteria. Second, while we were able to include a measure of cannabis withdrawal in the analysis, the symptoms and diagnostic scheme (i.e., 3 or more of 7 withdrawal symptoms) used to assess withdrawal do not conform to those in DSM-5. This was unavoidable since all studies predated the DSM-5 by a considerable number of years. However, analyses using the DSM-5 criteria in an independent twin sample do not indicate any evidence for genetic influences on DSM-5 withdrawal that do not overlap with DSM-IV cannabis abuse/dependence (Verweij et al., 2012).
From a clinical and public health standpoint, it is also reassuring to note that a transition from DSM-IV to DSM-5 will likely involve only a modest alteration in prevalence of diagnosed individuals. However, future studies, particularly those aggregating individual-level genotypic and phenotypic data across multiple samples should explore the extent to which individual DSM-IV, and in particularly, the new DSM-5 criteria contribute to specificity of genetic signals identified.
Supplementary Material
Acknowledgements
None
Role of Funding Source: Funding sources were not involved in the conceptualization or execution of this study nor in the preparation of this manuscript. Dr. Agrawal is supported by K02DA032573 and R01DA23668. She has previously received peer-reviewed grant funding and support from the Foundation for Alcohol Research/ABMRF. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423, R01 DA019963). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary material can be found by accessing the online version of this paper at http://dx.doi.org and by entering doi:...
Contributors: AA, MTL, KKB and LJB conceived of analyses. AA conducted all analyses with statistical support from LA, DMD, HJE, TF, AG, DBH and SH. Data were collected via funding to LA, DMD, HJE, TF, AG, EOJ, VH, JRK, SK, JIN, MS and LJB. Phenotypic expertise was provided by MTL, KKB, DMD, EOJ, VH, JRK, SK, MS. Support with genetic analyses was provided by LA, DMD, HJE, TF, DBH, SH, AG and JIN. All authors read and critically reviewed drafts of the manuscript.
Supplementary material can be found by accessing the online version of this paper at http://dx.doi.org and by entering doi:...
Supplementary material can be found by accessing the online version of this paper at http://dx.doi.org and by entering doi:...
Conflict of Interest: Laura J. Bierut is listed as an inventor on Issued U.S. Patent 8,080,371,“Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction.
REFERENCES
- Agrawal A, Bucholz KK, Lynskey MT. DSM-IV alcohol abuse due to hazardous use: a less severe form of abuse? J. Stud. Alcohol Drugs. 2010;71:857–863. doi: 10.15288/jsad.2010.71.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Heath AC, Lynskey MT. DSM-IV to DSM-5: the impact of proposed revisions on diagnosis of alcohol use disorders. Addiction. 2011a;106:1935–1943. doi: 10.1111/j.1360-0443.2011.03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT. Does gender contribute to heterogeneity in criteria for cannabis abuse and dependence? Results from the National Epidemiological Survey on Alcohol and Related Conditions. Drug Alcohol Depend. 2007;88:300–307. doi: 10.1016/j.drugalcdep.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT, Hinrichs A, Grucza R, Saccone SF, Krueger R, Neuman R, Howells W, Fisher S, Fox L, Cloninger R, Dick DM, Doheny KF, Edenberg HJ, Goate AM, Hesselbrock V, Johnson E, Kramer J, Kuperman S, Nurnberger JI, Jr., Pugh E, Schuckit M, Tischfield J, Rice JP, Bucholz KK, Bierut LJ. A genome-wide association study of DSM-IV cannabis dependence. Addict. Biol. 2011b;16:514–518. doi: 10.1111/j.1369-1600.2010.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Pergadia ML, Lynskey MT. Is there evidence for symptoms of cannabis withdrawal in the national epidemiologic survey of alcohol and related conditions? Am. J. Addict. 2008;17:199–208. doi: 10.1080/10550490802019519. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. American Psychiatric Association; Arlington, VA: 2013. [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li TK, Schuckit M, Edenberg H, Rice JP. The collaborative study on the genetics of alcoholism. Alcohol Health Res. World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Bernards R. Wip-ing out cancer. Nat. Genet. 2004;36:319–320. doi: 10.1038/ng0404-319. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI, Jr., Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, Fugman D, Goate AM, Hinrichs AL, Konvicka K, Martin NG, Montgomery GW, Saccone NL, Saccone SF, Wang JC, Chase GA, Rice JP, Ballinger DG. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Strickland JR, Thompson JR, Afful SE, Cottler LB. Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend. 2008;95:14–22. doi: 10.1016/j.drugalcdep.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budney AJ. Are specific dependence criteria necessary for different substances: how can research on cannabis inform this issue? Addiction 101 Suppl. 2006;1:125–33. doi: 10.1111/j.1360-0443.2006.01582.x. [DOI] [PubMed] [Google Scholar]
- Budney AJ, Hughes JR. The cannabis withdrawal syndrome. Curr. Opin. Psychiatry. 2006;19:233–238. doi: 10.1097/01.yco.0000218592.00689.e5. [DOI] [PubMed] [Google Scholar]
- Compton WM, Saha TD, Conway KP, Grant BF. The role of cannabis use within a dimensional approach to cannabis use disorders. Drug Alcohol Depend. 2009;100:221–227. doi: 10.1016/j.drugalcdep.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Ke X, Le HS, Christoforou A, Luciano M, McGhee K, Lopez L, Gow AJ, Corley J, Redmond P, Fox HC, Haggarty P, Whalley LJ, McNeill G, Goddard ME, Espeseth T, Lundervold AJ, Reinvang I, Pickles A, Steen VM, Ollier W, Porteous DJ, Horan M, Starr JM, Pendleton N, Visscher PM, Deary IJ. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol. Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt L, Hall W. Extent of illicit drug use and dependence, and their contribution to the global burden of disease. Lancet. 2012;379:55–70. doi: 10.1016/S0140-6736(11)61138-0. [DOI] [PubMed] [Google Scholar]
- Feng S, Wang S, Chen CC, Lan L. GWAPower: a statistical power calculation software for genome-wide association studies with quantitative traits. BMC Genet. 2011;12:12. doi: 10.1186/1471-2156-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie NA, Neale MC, Prescott CA, Aggen SH, Kendler KS. Factor and item-response analysis of DSM-IV criteria for abuse and dependence on cannabis, cocaine, hallucinogens, sedatives, stimulants and opioids. Addiction. 2005;102:920–30. doi: 10.1111/j.1360-0443.2007.01804.x. [DOI] [PubMed] [Google Scholar]
- Hartman CA, Gelhorn H, Crowley TJ, Sakai JT, Stallings M, Young SE, Rhee SH, Corley R, Hewitt JK, Hopfer CJ. Item response theory analysis of DSMIV cannabis abuse and dependence criteria in adolescents. J. Am. Acad. Child Adolesc. Psychiatry. 2008;47:165–173. doi: 10.1097/chi.0b013e31815cd9f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, Fenton MC, Beseler C, Park JY, Wall MM. Analyses related to the development of DSM-5 criteria for substance use related disorders: proposed DSM-5 criteria for alcohol, cannabis, cocaine and heroin disorders in 663 substance abuse patients. Drug Alcohol Depend. 2012;122:28–37. doi: 10.1016/j.drugalcdep.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, Keyes KM, Alderson D, Wang S, Aharonovich E, Grant BF. Cannabis withdrawal in the United States: results from NESARC. J. Clin. Psychiatry. 2008;69:1354–1363. doi: 10.4088/jcp.v69n0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, O'Brien CP, Auriacombe M, Borges G, Bucholz K, Budney A, Compton WM, Crowley T, Ling W, Petry NM, Schuckit M, Grant BF. DSM-5 criteria for substance use disorders: recommendations and rationale. Am. J. Psychiatry. 2013;170:834–851. doi: 10.1176/appi.ajp.2013.12060782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan-Sciutto KL, Dragich JM, Caltagarone J, Hall DJ, Bowser R. Fetal Alz-50 clone 1 (FAC1) protein interacts with the Myc-associated zinc finger protein (ZF87/MAZ) and alters its transcriptional activity. Biochemistry. 2000;39:3206–3215. doi: 10.1021/bi992211q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes KM, Hasin DS. Socio-economic status and problem alcohol use: the positive relationship between income and the DSM-IV alcohol abuse diagnosis. Addiction. 2008;103:1120–1130. doi: 10.1111/j.1360-0443.2008.02218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MS, Kuehn MR, Yamaguchi TP, Wu C. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenbucher J, Labouvie EW, Martin C, Sanjuan PM, Bavly L, Kirisci L, Chung T. An application of item response theory analysis to alcohol, cannabis and cocaine criteria in DSM-IV. J. Abnorm. Psychol. 2004;113:72–80. doi: 10.1037/0021-843X.113.1.72. [DOI] [PubMed] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, Gabriel SB, Harris EL, Hu FB, Jacobs KB, Kraft P, Landi MT, Lumley T, Manolio TA, McHugh C, Painter I, Paschall J, Rice JP, Rice KM, Zheng X, Weir BS. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, DeCandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, Keller MC, Visscher PM, Wray NR. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, Mowry BJ, Thapar A, Goddard ME, Witte JS, Absher D, Agartz I, Akil H, Amin F, Andreassen OA, Anjorin A, Anney R, Anttila V, Arking DE, Asherson P, Azevedo MH, Backlund L, Badner JA, Bailey AJ, Banaschewski T, Barchas JD, Barnes MR, Barrett TB, Bass N, Battaglia A, Bauer M, Bayes M, Bellivier F, Bergen SE, Berrettini W, Betancur C, Bettecken T, Biederman J, Binder EB, Black DW, Blackwood DH, Bloss CS, Boehnke M, Boomsma DI, Breen G, Breuer R, Bruggeman R, Cormican P, Buccola NG, Buitelaar JK, Bunney WE, Buxbaum JD, Byerley WF, Byrne EM, Caesar S, Cahn W, Cantor RM, Casas M, Chakravarti A, Chambert K, Choudhury K, Cichon S, Cloninger CR, Collier DA, Cook EH, Coon H, Cormand B, Corvin A, Coryell WH, Craig DW, Craig IW, Crosbie J, Cuccaro ML, Curtis D, Czamara D, Datta S, Dawson G, Day R, De Geus EJ, Degenhardt F, Djurovic S, Donohoe GJ, Doyle AE, Duan J, Dudbridge F, Duketis E, Ebstein RP, Edenberg HJ, Elia J, Ennis S, Etain B, Fanous A, Farmer AE, Ferrier IN, Flickinger M, Fombonne E, Foroud T, Frank J, Franke B, Fraser C, Freedman R, Freimer NB, Freitag CM, Friedl M, Frisen L, Gallagher L, Gejman PV, Georgieva L, Gershon ES, Geschwind DH, Giegling I, Gill M, Gordon SD, Gordon-Smith K, Green EK, Greenwood TA, Grice DE, Gross M, Grozeva D, Guan W, Gurling H, De HL, Haines JL, Hakonarson H, Hallmayer J, Hamilton SP, Hamshere ML, Hansen TF, Hartmann AM, Hautzinger M, Heath AC, Henders AK, Herms S, Hickie IB, Hipolito M, Hoefels S, Holmans PA, Holsboer F, Hoogendijk WJ, Hottenga JJ, Hultman CM, Hus V, Ingason A, Ising M, Jamain S, Jones EG, Jones I, Jones L, Tzeng JY, Kahler AK, Kahn RS, Kandaswamy R, Keller MC, Kennedy JL, Kenny E, Kent L, Kim Y, Kirov GK, Klauck SM, Klei L, Knowles JA, Kohli MA, Koller DL, Konte B, Korszun A, Krabbendam L, Krasucki R, Kuntsi J, Kwan P, Landen M, Langstrom N, Lathrop M, Lawrence J, Lawson WB, Leboyer M, Ledbetter DH, Lee PH, Lencz T, Lesch KP, Levinson DF, Lewis CM, Li J, Lichtenstein P, Lieberman JA, Lin DY, Linszen DH, Liu C, Lohoff FW, Loo SK, Lord C, Lowe JK, Lucae S, MacIntyre DJ, Madden PA, Maestrini E, Magnusson PK, Mahon PB, Maier W, Malhotra AK, Mane SM, Martin CL, Martin NG, Mattheisen M, Matthews K, Mattingsdal M, McCarroll SA, McGhee KA, McGough JJ, McGrath PJ, McGuffin P, McInnis MG, McIntosh A, McKinney R, McLean AW, McMahon FJ, McMahon WM, McQuillin A, Medeiros H, Medland SE, Meier S, Melle I, Meng F, Meyer J, Middeldorp CM, Middleton L, Milanova V, Miranda A, Monaco AP, Montgomery GW, Moran JL, Moreno-De-Luca D, Morken G, Morris DW, Morrow EM, Moskvina V, Muglia P, Muhleisen TW, Muir WJ, Muller-Myhsok B, Murtha M, Myers RM, Myin-Germeys I, Neale MC, Nelson SF, Nievergelt CM, Nikolov I, Nimgaonkar V, Nolen WA, Nothen MM, Nurnberger JI, Nwulia EA, Nyholt DR, O'Dushlaine C, Oades RD, Olincy A. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S. A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Cha H, Lee MO, Mazur SJ, Appella E, Fornace AJ., Jr. Landmark, editor. Regulation of the Wip1 phosphatase and its effects on the stress response. Front. Biosci. 2012;17:1480–1498. doi: 10.2741/3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubke GH, Hottenga JJ, Walters R, Laurin C, de Geus EJ, Willemsen G, Smit JH, Middeldorp CM, Penninx BW, Vink JM, Boomsma DI. Estimating the genetic variance of major depressive disorder due to all single nucleotide polymorphisms. Biol. Psychiatry. 2012;72:707–709. doi: 10.1016/j.biopsych.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynskey MT, Agrawal A. Psychometric properties of DSM assessments of illicit drug abuse and dependence: results from the National Epidemiological Survey on Alcohol and Related Conditions. Psychol. Med. 2007;37:1345–1355. doi: 10.1017/S0033291707000396. [DOI] [PubMed] [Google Scholar]
- Martin CS, Chung T, Kirisci L, Langenbucher JW. Item response theory analysis of diagnostic criteria for alcohol and cannabis use disorders in adolescents: implications for DSM-V. J. Abnorm. Psychol. 2006;115:807–814. doi: 10.1037/0021-843X.115.4.807. [DOI] [PubMed] [Google Scholar]
- Martin CS, Sher KJ, Chung T. Hazardous use should not be a diagnostic criterion for substance use disorders in DSM-5. J. Stud. Alcohol Drugs. 2011a;72:685–686. doi: 10.15288/jsad.2011.72.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CS, Steinley DL, Verges A, Sher KJ. The proposed 2/11 symptom algorithm for DSM-5 substance-use disorders is too lenient. Psychol. Med. 2011b;41:2008–2010. doi: 10.1017/S0033291711000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewton L, Slade T, Teesson M. An Evaluation of the proposed DSM-5 cannabis use disorder criteria using Australian national survey data. J. Stud. Alcohol Drugs. 2013;74:614–621. doi: 10.15288/jsad.2013.74.614. [DOI] [PubMed] [Google Scholar]
- Muthen LK, Muthen BO. Mplus User's Guide. Muthen & Muthen; Los Angeles, CA: 2007. [Google Scholar]
- Peer K, Rennert L, Lynch KG, Farrer L, Gelernter J, Kranzler HR. Prevalence of DSM-IV and DSM-5 alcohol, cocaine, opioid, and cannabis use disorders in a largely substance dependent sample. Drug Alcohol Depend. 2013;127:215–9. doi: 10.1016/j.drugalcdep.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontek D, Kraus L, Legleye S, Buhringer G. The validity of DSM-IV cannabis abuse and dependence criteria in adolescents and the value of additional cannabis use indicators. Addiction. 2011;106:1137–1145. doi: 10.1111/j.1360-0443.2010.03359.x. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van EP, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr., Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am. J. Med. Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verges A, Littlefield AK, Sher KJ. Did lifetime rates of alcohol use disorders increase by 67% in 10 years? A comparison of NLAES and NESARC. J. Abnorm. Psychol. 2011;120:868–877. doi: 10.1037/a0022126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij KJ, Agrawal A, Nat NO, Creemers HE, Huizink AC, Martin NG, Lynskey MT. A genetic perspective on the proposed inclusion of cannabis withdrawal in DSM-5. Psychol. Med. 2012:1–10. doi: 10.1017/S0033291712002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij KJ, Zietsch BP, Lynskey MT, Medland SE, Neale MC, Martin NG, Boomsma DI, Vink JM. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction. 2010;105:417–430. doi: 10.1111/j.1360-0443.2009.02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LT, Pan JJ, Blazer DG, Tai B, Stitzer ML, Brooner RK, Woody GE, Patkar AA, Blaine JD. An item response theory modeling of alcohol and marijuana dependences: a National Drug Abuse Treatment Clinical Trials Network study. J. Stud. Alcohol Drugs. 2009;70:414–425. doi: 10.15288/jsad.2009.70.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LT, Woody GE, Yang C, Pan JJ, Reeve BB, Blazer DG. A dimensional approach to understanding severity estimates and risk correlates of marijuana abuse and dependence in adults. Int. J. Methods Psychiatr. Res. 2012;21:117–133. doi: 10.1002/mpr.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011a;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de AM, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 2011b;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


