Abstract
The 2007 report “Toxicity Testing in the 21st Century: A Vision and A Strategy” argued for a change in toxicity testing for environmental agents and discussed federal funding mechanisms that could be used to support this transformation within the USA. The new approach would test for in vitro perturbations of toxicity pathways using human cells with high throughput testing platforms. The NRC report proposed a deliberate timeline, spanning about 20 years, to implement a wholesale replacement of current in-life toxicity test approaches focused on apical responses with in vitro assays. One approach to accelerating implementation is to focus on well-studied prototype compounds with known toxicity pathway targets. Through a series of carefully executed case studies with four or five pathway prototypes, the various steps required for implementation of an in vitro toxicity pathway approach to risk assessment could be developed and refined. In this article, we discuss alternative approaches for implementation and also outline advantages of a case study approach and the manner in which the cases studies could be pursued using current methodologies. A case study approach would be complementary to recently proposed efforts to map the human toxome, while representing a significant extension toward more formal risk assessment compared to the profiling and prioritization approaches offered by programs such as the EPA’s ToxCast effort.
I. Background
The U.S. National Research Council report on “Toxicity Testing in the 21st Century: A Vision and a Strategy” (NRC, 2007; Krewski et al., 2010) argued for a large-scale shift in toxicity testing. Testing protocols for 21st century toxicology should focus on mechanistic in vitro assays, typically using human cells in a high-throughput context. The in vitro tests would measure perturbations of “toxicity pathways”. Perturbations of these key biological signaling pathways are expected to be associated with specific adverse responses when a high-level of perturbation persists in vivo for sufficient durations. The NRC report has been extensively discussed in the toxicology and risk assessment communities (Andersen and Krewski, 2009; Krewski et al., 2009). The dialog has highlighted four key questions about a shift to in vitro tests for assessing risks of chemicals (Andersen and Krewski, 2010): (1) how will we define adversity from in vitro tests; (2) how will the in vitro test results be used to predict expected outcomes in animals and people who come in contact with the test compounds; (3) how will regulatory agencies set exposure standards for human populations based on in vitro test results; and (4) how will authorities accustomed to the current whole animal testing procedures develop the confidence required to move to another platform for testing and risk assessment. These questions squarely capture the core challenges that need attention in order to develop 21st century toxicology for both toxicity testing and for using these test results for human health risk assessment. The NRC report suggested a process in which incremental advances in key technologies would all have to be completed before making a wholesale transition to in vitro test-based risk assessment platforms. Another suggestion that has surfaced is to use case studies with prototype pathways and compounds to accelerate implementation of the Toxicity Testing in the 21st Century (TT21C) paradigm by developing working examples of the necessary methodologies. Case studies could show each component of this new paradigm in action and effectively probe Questions 1 through 3 noted above. This short article begins with considerations of approaches to implementing the NRC’s TT21C vision and then considers the process by which case studies might be pursued.
II. Developing the Knowledge Base for Testing
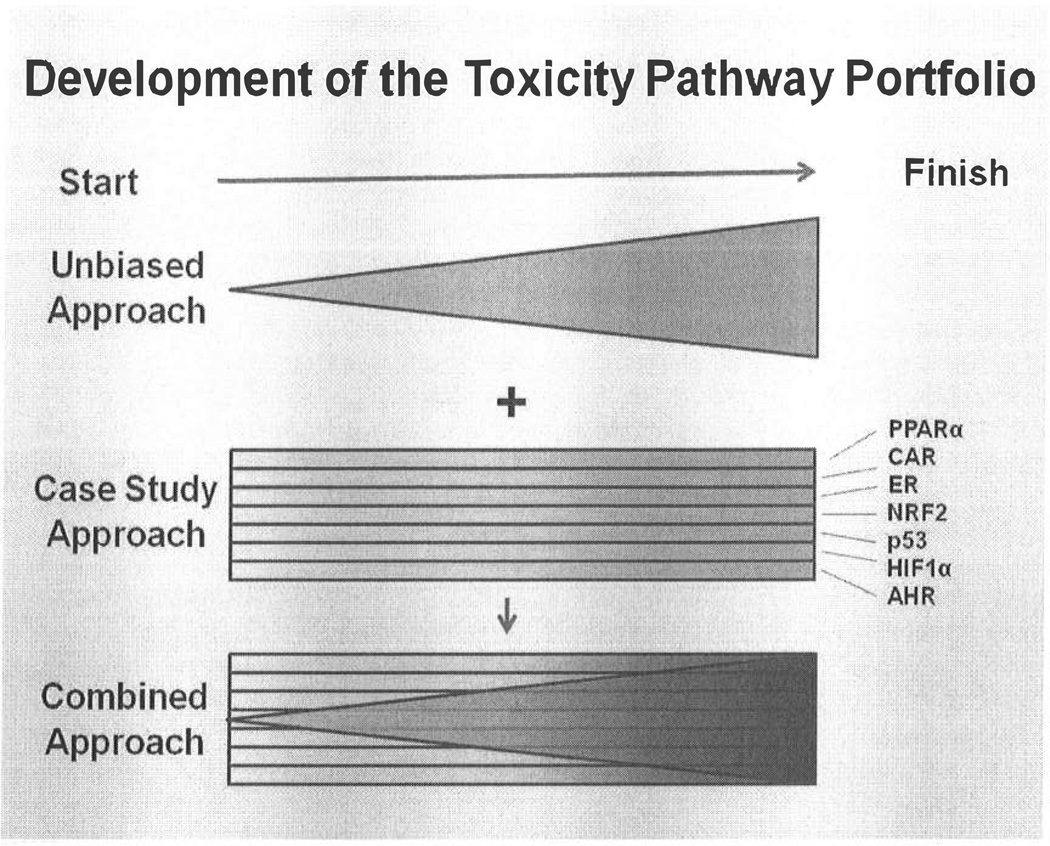
Implementing the new in vitro testing paradigm will require the development of new knowledge on toxicity pathways and how they interact and function. Conceptually, this new knowledge base can be accumulated either by using what has been called ‘an unbiased approach’, by using a case study approach, or by a combination of these two approaches (Figure 1). In the unbiased approach, a great deal of attention would be devoted to designing a convenient human cell based in vitro test system that would be expected to include as much of the known biological response landscape as possible. For example, the test system would be designed to include specialized functions of differentiated cell types and the pathways mediating paracrine interactions. This test system would likely include complex bioengineered multicellular test platforms and genetically modified cells with an array of differentiated expression patterns, displaying as complete a representation of known biology as possible. Once this test platform was in place, testing would be expected to gradually reveal the key toxicity pathways and their interactions (Figure 1). While toxicants with “known” effects would be included in the testing program, in this unbiased approach the inherent responses of the test platforms would be used to define points of departure (POD). The advantages of this unbiased approach include its lack of reliance on past animal-based test results, the open-ended nature of the inquiry, and the continual improvements in reliability and predictivity as more testing takes place. Disadvantages include the importance of the test platform itself as a unique model with unknown universal applicability, and the amount of time required to accumulate a comprehensive understanding of the toxicity pathways and their interactions before their use in human risk assessment.
Figure 1. The toxicity pathway knowledge base can be developed using an unbiased approach, a case study approach, or a combined approach.
The knowledge needed to develop a functioning in vitro test system will be acquired over time (left-to-right). This knowledge can be acquired by having a comprehensive test platform that is challenged by numerous toxicants, and over time the interconnections and important response pathways emerge (unbiased approach). In this “unbiased approach”, knowledge expands over time as pathway interactions are understood (reflected by the width of the triangle increasing and the shading darkening from left to right). Alternatively, using a case study approach, a specific pathway can be evaluated in detail. While the density of knowledge increases (the shade darkens with time), the knowledge area covered does not change (the width of the knowledge area remains constant). Combining these approaches may be the best way to take advantage of the strengths of each approach; however, projects to demonstrate the applicability of the new toxicity testing paradigm may be most rapidly developed using a case study approach.
In the case study approach, existing knowledge of specific toxicity pathways would be used to design appropriate test systems that would then be exercised with model toxicants before exploring the responses of unknown toxicants (Figure 1). The advantages of this approach include the focus on well understood pathways and responses that build on our current understanding of cell and molecular biology, and the rapid applicability of the test system to a defined set of anticipated responses. The disadvantages include the narrow restriction of the inquiry to known biology which could lead to missing important unknown responses, and the need to develop different test platforms that are specific to each case study area. In this case study approach, the POD in the in vitro test system would be informed by the known in vivo responses to the model compounds, and would not rely solely on the platform responses. This characteristic would allow alignment of results from new in vitro approaches with past whole animal and mode-of-action research.
Ideally, implementation of the new toxicity testing paradigm will be achieved using a combination of unbiased and case study approaches (Figure 1). By combining approaches, the advantages of open-ended inquiry with the focused application of expert knowledge gives an opportunity to develop the most complete toxicity assessment capability. In the near term, however, what is most needed is a demonstration of the feasibility of these new approaches and their ability to be both reliable and predictive. These near-term goals, in our opinions, can most effectively be met with demonstration projects using a case study approach.
One important consideration in implementing the test platforms themselves is the difference between the platforms used to develop the deep biological knowledge base of pathway function and the platforms that will ultimately be used for testing. Because of the absence of the requisite knowledge, the assay platforms for studying specific pathways must be designed to yield integrated data on the interacting molecular systems that control cell function, including transcriptomics, transcriptional factor analysis, phosphoproteomics, metabolomics, etc. Time dependent changes in cellular function will also need to be assessed to develop a familiarity with the important temporal windows of response. The assay systems for routine testing, however, are likely to be much simpler and high throughput, assessing important integrative nodal responses that adequately assess and reflect the functionality of the cell. The more complex systems will be initially developed to assess assay function; the final applications will simply evaluate high-throughput dose-response. As described by Eric Berlow, an ecologist and network theorist, in a recent TED Talk (see www.ericlberlow.net), “simplicity often lies on the other side of complexity.” Berlow makes a distinction between the complex and the complicated, with complexity arising from lots of interactions and details while complication results from an obscuring of the details. During the development of our understanding of toxicity pathways, the details may at first seem overwhelming and unconnected, but ultimately it will be possible to identify those details that are both important and most representative of the overall responses. The case study approach then lays many larger issues aside (Question 4 in the list) in an effort to get started and show how in vitro toxicity testing information can be used in specific situations. Success with the case studies should refine the process and the nature of specific pathway assays, and help to provide context for the more unbiased approaches.
III. Doing Risk Assessments from in vitro Test Results
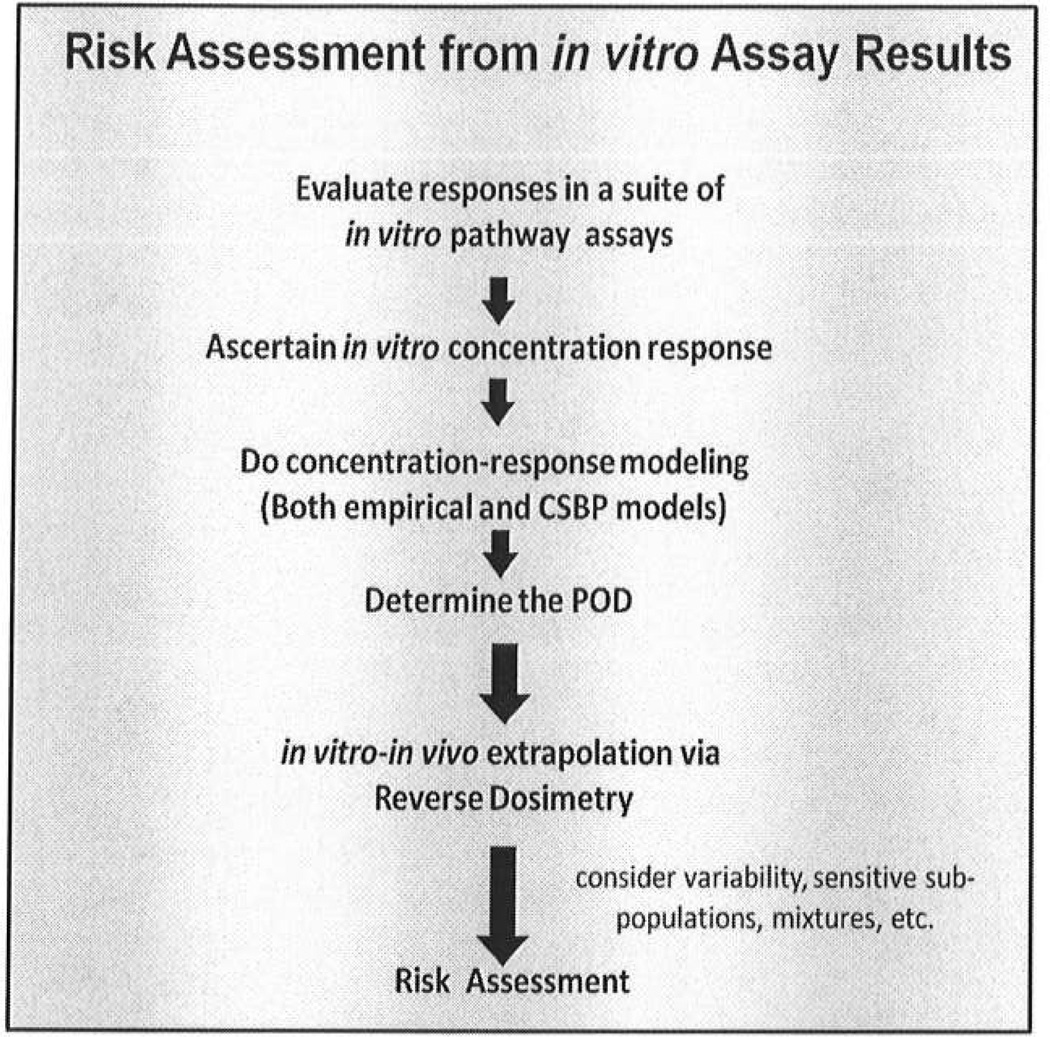
The manner in which in vitro testing results could be used for risk assessment was outlined in a generic fashion in the NRC report (NRC 2007) and in other thought pieces on new directions in toxicity testing (Krewski, Westphal et al. 2011). These contributions showed the correspondence of individual components of the proposed toxicity testing methods to the risk assessment paradigm from the 1983 NRC report, Risk Assessment in the Federal Government: Managing the Process (NRC, 1983). An essay on the manner in which in vitro test results will provide measures of adversity (Boekelheide and Campion 2010) sparked a broader dialog on designing a more explicit process to develop human exposure guidelines from an in vitro concentration deemed to cause an adverse cellular response (Boekelheide and Andersen 2010; Bhattacharya, Zhang et al. 2011). A simple, linear diagram of the likely process (Figure 2) shows steps involved in obtaining an in vitro a POD and then moving from the POD to a regulatory standard.
Figure 2. A Schematic showing steps in a toxicity pathway-based risk assessment.
Results from the panel of assays identify the pathway targets and generate a point of departure (POD) for the subsequent risk assessment as an in vitro concentration. Computational systems biology pathway (CSBP) modeling of circuitry and dynamics for the assay system indicates the expected shape of the dose response at lower doses, leading to a POD. The POD concentration is then converted to an exposure standard through techniques of reverse dosimetry implemented by pharmacokinetic modeling. This step takes advantages of in vitro-in vivo extrapolation (IVIVE).
Chemical risk assessment in the future would start by having a suite of ‘validated’ in vitro assays and determining the pathway assay or assays that had responses at the lowest test concentration. Most of the assays would be pathway specific with a few that represented a more global view of cellular responses. The pathway specific assays would have already been validated using positive controls known to affect pathway responses, such as 17-β estradiol for an estrogenic pathway assay or ionizing radiation for a DNA-damage pathway before use in general testing. The NRC report talked about using high-throughput assays capable of testing hundreds or thousands of compounds in very short times. The goal is not simply high-throughput, but having suitable in vitro assays that can be run quickly over a wide range of test concentrations. Testing over a wide range of concentrations is at the heart of quantitative high-throughput screening (q-HTS). With q-HTS, the empirical dose response curve for the assays would span multiple orders of concentration (Inglese, Auld et al. 2006).
From the results of a suite of toxicity pathway tests, the pathways affected at the lowest treatment concentrations would be identified. Computational systems biology pathway (CSBP) models, defined in the process of assay development and validation, would take pathway multi-point dose-response curves and predict transitions from sub-threshold doses, to doses causing adaptive changes in the pathway function, to doses expected to have adverse consequences. These changes in response patterns with increasing levels of perturbations are referred to as dose-dependent transitions (Slikker et al., 2004a; Slikker et al., 2004b). The in vitro pathway assay would provide the dose response information for assessing adversity. CSBP models for the assay would inform the low dose extrapolation to give the POD. Then, together with policy considerations, such as sensitive populations, response variability in a diverse population, etc., the in vitro adverse concentration would be adjusted to give an acceptable human plasma concentration. The last step in this process is estimating the in vivo human exposure expected to produce the in vitro concentration. This extrapolation relies on technologies referred to as in vitro-in vivo extrapolation - IVIVE (Shiran, Proctor et al. 2006; Gibson and Rostami-Hodjegan 2007) - and reverse dosimetry (Clewell, Tan et al. 2008) – and would have to include considerations of variability in expected response in different populations, co-exposures in mixtures, and other factors.
In vitro to in vivo extrapolation (IVIVE) estimates the environmental exposures to a chemical that could produce target tissue exposures in humans equivalent to those associated with effects in an in vitro toxicity test. Through a combination of quantitative structure property relationship (QSPR) modeling, physiologically based pharmacokinetic (PBPK) modeling, and collection of in vitro data on metabolism, transport, binding, etc., IVIVE provides estimates of the likelihood of harmful effects from expected environmental exposures. Generic IVIVE approaches have measured metabolic clearance in primary hepatocytes, plasma protein binding and estimates of renal filtration extrapolated from protein binding. These data were input to a population-based in vitro-to-in vivo extrapolation program (SIMCYP™) for estimating the human oral equivalent dose necessary to produce a steady-state in vivo concentration equivalent to in vitro AC50 (concentration at 50% of maximum activity) and LEC (lowest effective concentration) values from the ToxCast data (Rotroff et al., 2010).
IV. Developing Assays
Human cells in culture
The NRC report favored assays using human cells, cell-lines or organotypic cell cultures in order for the toxicity pathway testing to be more representative of expected effects of test compounds on human biology. In designing pathway assays, there will be compromises about available cells, the nature of pathway assay read-outs, and the presence of pathway components in specific cells or cell lines. Some assay development could be done in cell lines, but the goal would be to have the final suite of pathway assays testing conducted in human cells. This vision looks more accessible in 2011 than in 2007 due to the advances made in using tissue specific stem cells rather than having to harvest human cells from donors. For instance, hepatoblasts can be differentiated from liver stem cell precursors, maintained, then grown and differentiated into primary hepatocytes for testing in cultures (Wang et al., 2010). As noted in the discussion of unbiased and case study approaches, the development of these assays would require developing a deep biological knowledge of the networks, nodes, and dynamics of pathway responses to test compounds.
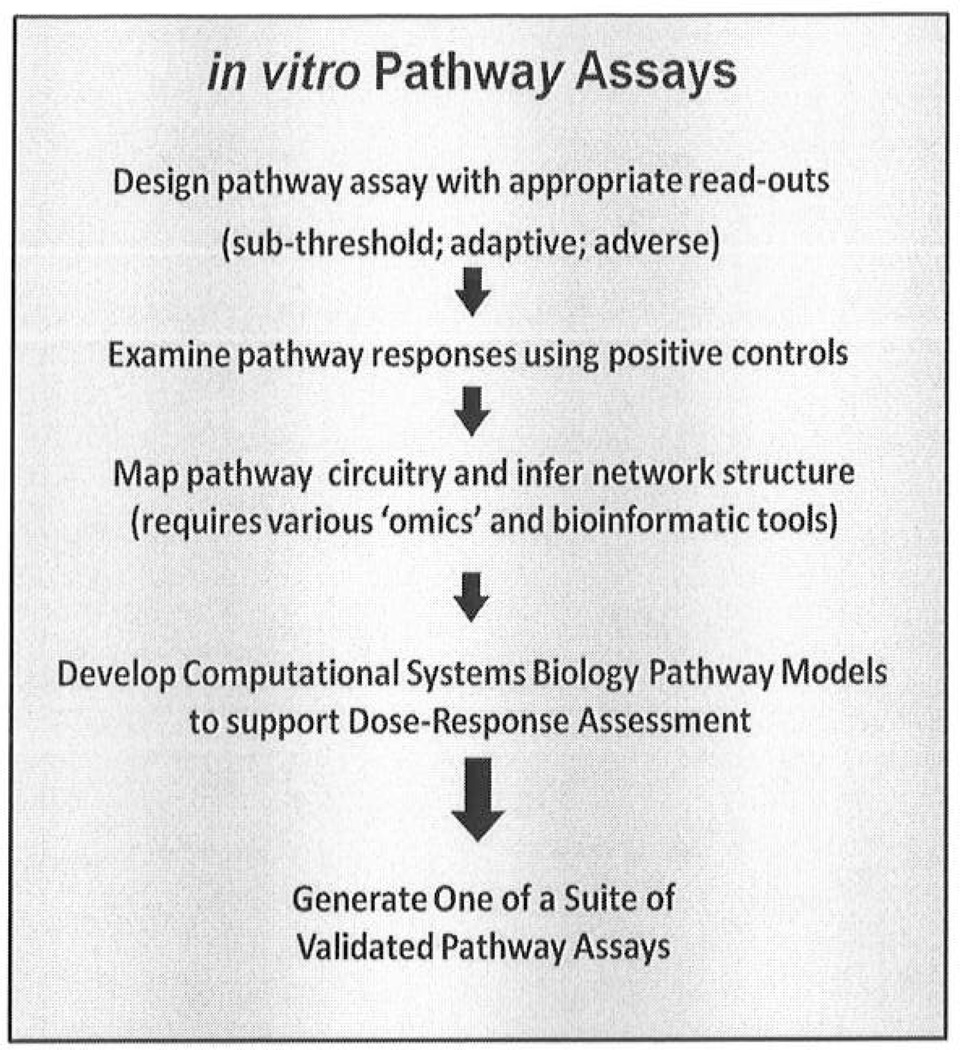
Assays then would be developed first with positive control test compounds to illuminate the range of responses associated with different degrees of severity of effect. To illustrate the case study process, we describe approaches to assay design with the DNA-damage, p53 pathway, and then use the PPAR-α pathway to discuss aspects of network inference and CSBP modeling; these are two toxicity pathways currently being developed as case studies. A schematic for design of assays for specific toxicity pathways (Figure 3) needs to capture aspects of design for purpose, validation against pathway structure and pathway dynamics, and ability to use the results for mechanistic dose response assessment through computational pathway models. Assays used for routine testing would have to first be ‘validated’ through these steps.
Figure 3. Developing Pathway Assays.
The steps shown here constitute the process of assuring that pathway assay are fit for purpose, i.e., for identifying activity in a specific pathway, assessing the structure and dynamics of the overall signaling network, and providing computational systems biology pathway models to assist dose-response modeling of assay results and low-dose extrapolations. The process of pathway development is fundamental to their use as part of a testing suite for risk assessment, as shown in Figure 2.
DNA-damage Assays
Our studies with the DNA-damage related pathways represent an on-going, productive collaboration between Unilever and The Hamner. For this pathway, we are using high content imaging and flow cytometry methods to visualize and quantify markers of DNA-damage (e.g., H2AX and TP53 binding to DNA and micronucleus formation) and activation of the p53 response pathway, including p-53, phosphorylated-p-53, p21 expression and cell cycle arrest (Lahav et al., 2004; Bryce et al., 2008; van Attikum and Gasser, 2009). Currently, a research collaboration between The Hamner Institutes and Unilever uses a human osteosarcoma cell line, HT-1080 (Sun et al., 2011). Although this is a cell line rather than a primary human cell, it has functioning native p53 (Benchimol et al., 1982). The goal in this project is to develop a suite of assays at the cellular level that link DNA-damage, pathway responses to DNA-damage, and ultimately mutation. Initially, the responses are modeled using empirical dose response models, including models that test for thresholds (Lutz and Lutz, 2009). These empirical dose-response models differentiate the doses causing specific responses, provide evidence for thresholds, show relative potencies for the multiple responses, and guide CSBP models for DNA-damage that provide a mechanistic basis for these dose-dependent transitions.
Finding the Components of the Pathways and Networks
A recent food for thought article (Hartung and McBride, 2011) discussed mapping a human toxome and distinguished among definitions of nodes, pathways and networks. These case studies are designed to evaluate specific toxicity pathways (in this case specific targets with known biological modes-of-action), but application in principle requires assessing the structure of the signaling network in which the pathway is embedded. Our second case study with the PPAR-α receptor focuses on network inference and computational modeling of the dynamic network. Experimental and bioinformatic tools for inferring structures of biological networks are now widely available (Sachs et al., 2002; di Bernardo et al., 2005; Woolf et al., 2005; Shen et al., 2011), but these tools have not yet been used to examine dose-response or dose-dependent transitions across dose regions within the networks. Dose-response information, both in vivo and in vitro, is already available that would allow network inference for some pathways, e.g., estrogen receptor(ER) or aryl hydrocarbon receptor (AhR) function (Fertuck et al., 2003; Kwekel et al., 2005; Naciff et al., 2009).
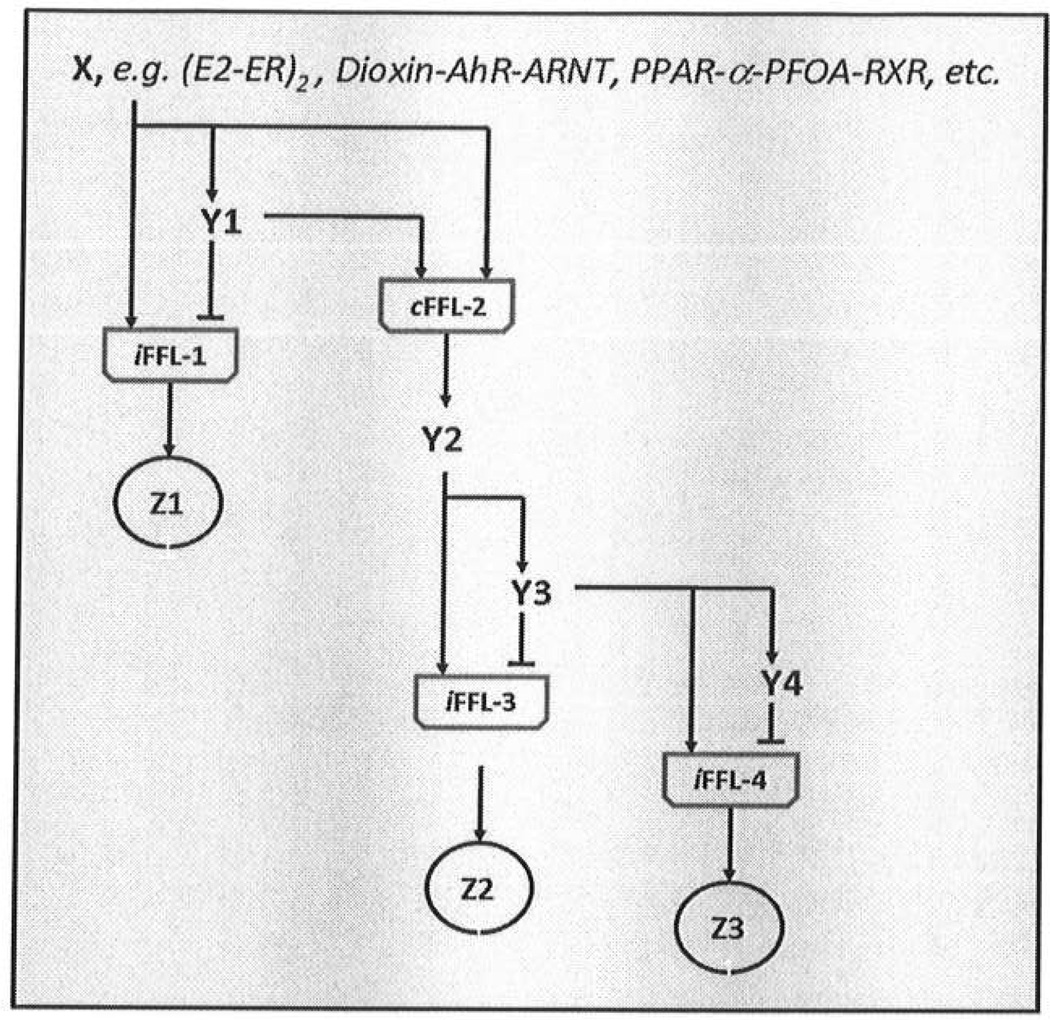
Using GW 7647, a specific PPAR-α agonist, these studies in primary hepatocytes from humans and rats have focused on gene expression and transcription factor binding after treatment (Woods et al., 2011). The study design follows that used to establish a cannabinoid receptor network (Bromberg et al., 2008b). Other inference methods (Shen et al., 2011) apply Bayesian approaches and combine gene expression, CHIP-Chip or CHIP-seq methods for transcription factor binding, and DNA-sequence analysis for locating DNA-response elements throughout the genome. The expected output for each pathway would be a grouping of nodes connected in a logical pattern leading to a sequential dose-dependent network that accounts for the consequences of pathway activation. The resulting circuit structure is likely to contain sequential nodes controlling gene expression, similar in structure to what has been called a “developmental network” (Alon, 2007). The schematic of a developmental network (Figure 4) would contain multiple nodes and feed-forward loops thereby creating a dynamic cascade of sequential activation of sub-components of the network. The cascade behavior of these networks (Landers and Spelsberg, 1992) is expected in turn to show both time and dose-dependencies in network function. In addition, high throughput- phosphoproteomic data now provide opportunities to construct extended pathways that link receptor activation with cellular phenotypes (Samaga et al., 2009; Melas et al., 2011); more limited evaluation of contributions of mitogen-activated protein kinases (MAPKs) to specific phenotypes after pathway activation have shown the differential control of key signaling nodes within overall networks (Bromberg et al., 2008a; Zorina et al., 2010). Integrated systems biology data – gene expression, phosphoprotein alterations, transcription factor analysis – will be needed to provide the dynamic and functional characterization of these assays and their associated toxicity pathways.
Figure 4. A schematic for a “developmental network” that controls receptor-mediated signaling.
The network is controlled by several incoherent (IFFL) and coherent feed forward loops (cFFL) with nodes to either activate (arrows) or repress (blunt lines) gene expression through critical signaling nodes – the y and Z factors. The goal in network inference is to understand the circuitry of various toxicity pathways at this level of detail in order to describe exposures that are without appreciable effects, doses with activation of early portions of the pathway, and full activation. These dose dependencies are expected to coincide with areas of sub-threshold, adaptive and adverse perturbations as outlined in the NRC report (NRC, 2007). The proteins designated Z1, Z2, and Z3 would have non-monotonic dose response curves; Y1, Y2, Y3 and Y4 would have monotonically increasing, but sequentially delayed, responses. Based on similar networks previously described (Alon, 2007); the code can be obtained by contacting MEA (MAndersen@thehamner.ore).
CSBP Models
For each toxicity pathway, the goal is to have a CSBP model that accounts for the variable response dynamics, depending on the level of activation of the network. A variety of modeling tools are now available and have become widely used in the biomedical engineering community (Aldridge et al., 2006). Our primary efforts with CSBP models are currently focused on p53-mediated DNA-damage and repair networks. High-dose responses of p53-mediated DNA-damage pathways have been examined through iterative experimentation and pathway modeling from the biomedical community for the past 10 years (Lahav et al., 2004; Batchelor et al., 2008; Batchelor et al., 2009; Loewer et al., 2010). In application of the assays on a routine basis for testing (Figure 2), network inference and CSBP models would not be determined for each compound studied. These tools are part of the process of assay development, determination of fitness of the assays for purpose, and use a deep understanding of network biology to consider dose-dependent transitions in network activation.
V. Running Case Studies
A case study approach takes well-studied compounds that are known to affect specific pathways and runs them through a process to assess dose-response and convert the POD to a human exposure standard (Figure 2). Candidates for these case studies are compounds that have been studied with conventional in vivo testing methods and whose mode-of-action has been well-characterized from both the in vivo test results and secondary mechanistic research. The mode-of-action framework activities over the past decade could serve as the basis for selecting one or more candidates (Sonich-Mullin et al., 2001; Boobis et al., 2006; Boobis et al., 2008; Julien et al., 2009). Case study approaches with prototype compounds requires the availability of appropriate assays for specific cellular toxicity pathways and genomic/bioinformatic tools to infer network structure and to create computational systems biology pathway (CSBP) models. For specific test compounds, in vitro in vivo extrapolation (IVIVE) methods require either chemical or chemical class specific approaches for kinetic modeling to describe expected pharmacokinetics and to support reverse dosimetry. Some initial work has focused on both receptor-mediated toxicity pathways (PAPR-α, ER, AhR, etc.) and pathways associated with chemical reactivity, such as DNA-damage, oxidative stress, and metal stress pathways (Simmons et al., 2009). Receptor-mediated pathways, especially estrogen, androgen and thyroid hormone networks, are of great interest because of contemporary focus on endocrine –active compounds, aka endocrine disruptors, and the regulatory requirement for testing these compounds in a tiered approach from limited in vitro assays to more complete multi-generational in vivo studies. To gain confidence in their applicability, the case studies will require optimization and characterization of the assay systems (Figure 3), and then exercising of the overall process with model chemicals and other pathway active compounds (Figure 2).
VI. Conclusions
At least three different approaches to implementation of TT21C have now been articulated: a toxome approach (Hartung and McBride ,2011), the US EPA ToxCast program (http://www.epa.govlncct/toxcast/) together with the associated multi-agency Tox21 initiative (http://www.epa.sov/ncct/Tox21/), and the case study approach described here. Each of these approaches has strengths and contributes to the overall goal of modernizing toxicity testing. The toxome project would map the entirety of pathways of toxicity in humans; ToxCast/Tox21 seeks to develop ways to predict potential toxicity and to develop a cost-effective approach for prioritizing the thousands of chemicals that need toxicity testing (Collins et al., 2008). The case studies are intended to show the application of new understanding of toxicity pathways directly for human health risk assessment and do it quickly. Progress in developing case studies will require careful selection of prototype compounds and prototype pathways. In addition, contributions are necessary across disciplines – assay design, genomics/bioinformatics, computational modeling, pharmacokinetics and human health risk assessment. The case study approach offers some advantages. First, it focuses on developing the generic tools and the processes by which in vitro toxicity information will be used for setting regulatory standards in specific instances. Second, it proposes learning by doing. Many key issues relevant to the use of this new information will become apparent by moving ahead with examples rather than worrying over how to make wholesale changes. Two of the authors of this perspective (MEA and HJC, lll) can look to the advances in PBPK modeling over the past 30 years. It is no exaggeration to say that the majority of challenges required to implement PBPK modeling to a diverse set of compounds were clearly defined with results from the first two compounds – styrene and methylene chloride (Ramsey and Andersen, 1984; Andersen et al., 1987). History is likely to repeat itself with TT21C. After completing the first two or three pathway case studies, most of the issues will become clear and expansion of the testing to other pathways will be greatly accelerated. We are also very much aware from our experiences that in the process of developing case studies, the schematic of the application of toxicity pathway information for risk assessment (Figure 2) will likely change. In addition, we expect that development of the specific case studies will also help illuminate the steps that will be needed to go forward with the unbiased approaches (Figure 1).
Acknowledgments
MEA and HJC, lll thank the ACC-LRI (Long Range Research Initiative of the American Chemistry Council), Dow Chemical, Unilever, the Exxon Mobil Foundation and Dow Corning for support of method development and data generation for pursuing case study approaches to bringing the TT21C vision to life more quickly. Support for KB was provided in part by the Superfund Research Program of the National Institute of Environmental Health Sciences at the National Institutes of Health (P42 ES013660).
References
- Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8:1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nature Reviews. Genetics. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Andersen ME, Clewell HJ, 3rd, Gargas ML, Smith FA, Reitz RH. Physiologically based pharmacokinetics and the risk assessment process for methylene chloride. Toxicology and applied pharmacology. 1987;87:185–205. doi: 10.1016/0041-008x(87)90281-x. [DOI] [PubMed] [Google Scholar]
- Andersen ME, Krewski D. Toxicity testing in the 21st century: bringing the vision to life. Toxicological Sciences. 2009;107:324–330. doi: 10.1093/toxsci/kfn255. [DOI] [PubMed] [Google Scholar]
- Andersen ME, Krewski D. The Vision of Toxicity Testing in the 21st Century: Moving from Discussion to Action. Toxicological Sciences. 2010;117:17–24. doi: 10.1093/toxsci/kfq188. [DOI] [PubMed] [Google Scholar]
- Batchelor E, Loewer A, Lahav G. The ups and downs of p53: understanding protein dynamics in single cells. Nature Reviews Cancer. 2009;9:371–377. doi: 10.1038/nrc2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: A mechanism for triggering p53 pulses in response to DNA damage. Molecular Cell. 2008;30:277–289. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchimol S, Pim D, Crawford L. Radioimmunoassay of the cellular protein p53 in mouse and human cell lines. Embo J. 1982;1:1055–1062. doi: 10.1002/j.1460-2075.1982.tb01296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boobis AR, Cohen SM, Dellarco V, McGregor D, Meek ME, Vickers C, Willcocks D, Farland W. IPCS framework for analyzing the relevance of a cancer mode of action for humans. Crit Rev Toxicol. 2006;36:781–792. doi: 10.1080/10408440600977677. [DOI] [PubMed] [Google Scholar]
- Boobis AR, Doe JE, Heinrich-Hirsch B, Meek ME, Munn S, Ruchirawat M, Schlatter J, Seed J, Vickers C. IPCS framework for analyzing the relevance of a noncancer mode of action for humans. Crit Rev Toxicol. 2008;38:87–96. doi: 10.1080/10408440701749421. [DOI] [PubMed] [Google Scholar]
- Bromberg KD, lyengar R, He JC. Regulation of neurite outgrowth by G(i/o) signaling pathways. Frontiers in Bioscience. 2008a;13:4544–4557. doi: 10.2741/3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg KD, Ma'ayan A, Neves SR, lyengar R. Design logic of a cannabinoid receptor signaling network that triggers neurite outgrowth. Science. 2008b;320:903–909. doi: 10.1126/science.1152662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce SM, Avlasevich SL, Bemis JC, Lukamowicz M, Elhajouji A, Van Goethem F, De Boeck M, Beerens D, Aerts H, Van Gompel J, Collins JE, Ellis PC, White AT, Lynch AM, Dertinger SD. Interlaboratory evaluation of a flow cytometric, high content in vitro micronucleus assay. Mutat Res. 2008;650:181–195. doi: 10.1016/j.mrgentox.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Gray GM, Bucher JR. Toxicology. Transforming environmental health protection. Science. 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Bernardo D, Thompson MJ, Gardner TS, Chobot SE, Eastwood EL, Wojtovich AP, Elliott SJ, Schaus SE, Collins JJ. Chemogenomic profiling on a genome-wide scale using reverse-engineered gene networks. Nat Biotechnol. 2005;23:377–383. doi: 10.1038/nbt1075. [DOI] [PubMed] [Google Scholar]
- Fertuck KC, Eckel JE, Gennings C, Zacharewski TR. Identification of temporal patterns of gene expression in the uteri of immature, ovariectomized mice following exposure to ethynylestradiol. Physiol Genomics. 2003;15:127–141. doi: 10.1152/physiolgenomics.00058.2003. [DOI] [PubMed] [Google Scholar]
- Hartung T, McBride M. Food for Thought … on mapping the human toxome. Altex. 2011;28:83–93. doi: 10.14573/altex.2011.2.083. [DOI] [PubMed] [Google Scholar]
- Julien E, Boobis AR, Olin SS. The Key Events Dose-Response Framework: a cross-disciplinary mode-of-action based approach to examining dose-response and thresholds. Crit Rev Food Sci Nutr. 2009;49:682–689. doi: 10.1080/10408390903110692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewski D, Acosta D, Andersen M, Anderson H, Bailar JC, Boekelheide K, Brent R, Charnley G, Cheung VG, Green S, Kelsey KT, Kerkvliet NI, Li AA, McCray L, Meyer O, Patterson RD, Pennie W, Scala RA, Solomon GM, Stephens M, Yager J, Zeise L Staff of Committee on Toxicity, T., and Assessment of Environmental, A. Toxicity Testing in the 21st Century: A Vision and a Strategy. Journal of Toxicology and Environmental Health. Part B, Critical Reviews. 2010;13:51–138. doi: 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewski D, Andersen ME, Mantus E, Zeise L. Toxicity Testing in the 21st Century: Implications for Human Health Risk Assessment. Risk Analysis. 2009;29:474–479. doi: 10.1111/j.1539-6924.2008.01150.x. [DOI] [PubMed] [Google Scholar]
- Kwekel JC, Burgoon LD, Burt JW, Harkema JR, Zacharewski TR. A cross-species analysis of the rodent uterotrophic program: elucidation of conserved responses and targets of estrogen signaling. Physiol Genomics. 2005;23:327–342. doi: 10.1152/physiolgenomics.00175.2005. [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- Landers JP, Spelsberg TC. New concepts in steroid hormone action: transcription factors, proto-oncogenes, and the cascade model for steroid regulation of gene expression. Crit Rev Eukaryot Gene Expr. 1992;2:19–63. [PubMed] [Google Scholar]
- Loewer A, Batchelor E, Gaglia G, Lahav G. Basal Dynamics of p53 Reveal Transcriptionally Attenuated Pulses in Cycling Cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz WK, Lutz RW. Statistical model to estimate a threshold dose and its confidence limits for the analysis of sublinear dose-response relationships, exemplified for mutagenicity data. Mutat Res. 2009;678:118–122. doi: 10.1016/j.mrgentox.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Melas IN, Mitsos A, Messinis DE, Weiss TS, Alexopoulos LG. Combined logical and data-driven models for linking signalling pathways to cellular response. BMC Syst Biol. 2011;5:107. doi: 10.1186/1752-0509-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naciff JM, Khambatta ZS, Thomason RG, Carr GJ, Tiesman JP, Singleton DW, Khan SA, Daston GP. The genomic response of a human uterine endometrial adenocarcinoma cell line to 17alpha-ethynyl estradiol. Toxicol Sci. 2009;107:40–55. doi: 10.1093/toxsci/kfn219. [DOI] [PubMed] [Google Scholar]
- NRC. Risk assessment in the federal government : managing the process. Washington, D.C.: National Academy Press; 1983. [PubMed] [Google Scholar]
- NRC. Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: The National Academies Press; 2007. [Google Scholar]
- Ramsey JC, Andersen ME. A physiologically based description of the inhalation pharmacokinetics of styrene in rats and humans. Toxicology and applied pharmacology. 1984;73:159–175. doi: 10.1016/0041-008x(84)90064-4. [DOI] [PubMed] [Google Scholar]
- Rotroff DM, Beam AL, Dix DJ, Farmer A, Freeman KM, Houck KA, Judson RS, LeCluyse EL, Martin MT, Reif DM, Ferguson SS. Xenobiotic-Metabolizing Enzyme and Transporter Gene Expression in Primary Cultures of Human Hepatocytes Modulated by Toxcast Chemicals. Journal of Toxicology and Environmental Health. Part B, Critical Reviews. 2010;13:329–346. doi: 10.1080/10937404.2010.483949. [DOI] [PubMed] [Google Scholar]
- Sachs K, Gifford D, Jaakkola T, Sorger P, Lauffenburger DA. Bayesian network approach to cell signaling pathway modeling. Sci STKE. 2002;2002:pe38. doi: 10.1126/stke.2002.148.pe38. [DOI] [PubMed] [Google Scholar]
- Samaga R, Saez-Rodriguez J, Alexopoulos LG, Sorger PK, Klamt S. The logic of EGFR/ErbB signaling: theoretical properties and analysis of high-throughput data. PLoS computational biology. 2009;5:e1000438. doi: 10.1371/journal.pcbi.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Huang Y, Liu Y, Wang G, Zhao Y, Wang Z, Teng M, Wang Y, Flockhart DA, Skaar TC, Yan P, Nephew KP, Huang TH, Li L. A modulated empirical Bayes model for identifying topological and temporal estrogen receptor alpha regulatory networks in breast cancer. BMC Syst Biol. 2011;5:67. doi: 10.1186/1752-0509-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons SO, Fan CY, Ramabhadran R. Cellular Stress Response Pathway System as a sentinel Ensemble in Toxicological Screening. ToxicologicaI Sciences. 2009;111:202–225. doi: 10.1093/toxsci/kfp140. [DOI] [PubMed] [Google Scholar]
- Slikker W, Jr, Andersen ME, Bogdanffy MS, Bus JS, Cohen SD, Conolly RB, David RM, Doerrer NG, Dorman DC, Gaylor DW, Hattis D, Rogers JM, Setzer RW, Swenberg JA, Wallace K. Dose-dependent transitions in mechanisms of toxicity: case studies. Toxicology and applied pharmacology. 2004a;201:226–294. doi: 10.1016/j.taap.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Slikker W, Jr, Andersen ME, Bogdanffy MS, Bus JS, Cohen SD, Conolly RB, David RM, Doerrer NG, Dorman DC, Gaylor DW, Hattis D, Rogers JM, Woodrow Setzer R, Swenberg JA, Wallace K. Dose-dependent transitions in mechanisms of toxicity. Toxicology and applied pharmacology. 2004b;201:203–225. doi: 10.1016/j.taap.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Sonich-Mullin C, Fielder R, Wiltse J, Baetcke K, Dempsey J, Fenner-Crisp P, Grant D, Hartley M, Knaap A, Kroese D, Mangelsdorf I, Meek E, Rice JM, Younes M. IPCS conceptual framework for evaluating a mode of action for chemical carcinogenesis. Regul Toxicol pharmacol. 2001;34:146–152. doi: 10.1006/rtph.2001.1493. [DOI] [PubMed] [Google Scholar]
- Sun B, Daniels K, Ross S, Zhang Q, Andersen M, Carmichael PL, Clewell R. Dose-Response Evaluation of p53 and H2AX DNA Damage Response by Methyl Methanesulfonate, Etoposide, and Quercetin. The Toxicologist. 2011:24. [Google Scholar]
- van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–277. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yao HL, Cui CB, Wauthier E, Barbier C, Costello MJ, Moss N, Yamauchi M, Sricholpech M, Gerber D, Loboa EG, Reid LM. Paracrine signals from mesenchymal cell populations govern the expansion and differentiation of human hepatic stem cells to adult liver fates. Hepatology. 2010;52:1443–1454. doi: 10.1002/hep.23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG, Yarborough KM, Pluta LJ, Thomas RS, Andersen ME. Constructing a PPARa-mediated transcriptional network in primary human and rat heaptocytes. The Toxicologist. 2011:78. [Google Scholar]
- Woolf PJ, Prudhomme W, Daheron L, Daley GQ, Lauffenburger DA. Bayesian analysis of signaling networks governing embryonic stem cell fate decisions. Bioinformatics. 2005;21:741–753. doi: 10.1093/bioinformatics/bti056. [DOI] [PubMed] [Google Scholar]
- Zorina Y, Iyengar R, Bromberg KD. Cannabinoid 1 Receptor and Interleukin-6 Receptor Together induce Integration of Protein Kinase and Transcription Factor Signaling to Trigger Neurite Outgrowth. Journal of Biologicol Chemistry. 2010;285:1358–1370. doi: 10.1074/jbc.M109.049841. [DOI] [PMC free article] [PubMed] [Google Scholar]