Abstract
Very large genomes, that is, those above 20 Gb, are rare but widely distributed throughout the eukaryotes. They are found within the diatoms, dinoflagellates, metazoans and green plants, but so far have not been found in the excavates. There is a known positive correlation between genome size and the proportion of the genome composed of transposable elements (TEs). Very large genomes may therefore be expected to be almost entirely composed of TEs. Of the large genomes examined, in the angiosperms, gymnosperms and the dinoflagellates only a small portion of the genome was identified as TEs, most of these genomes were unidentified and may be novel or diverse TEs. In the salamanders and lungfish, 25 to 47% of the genome were identifiable retrotransposons, that is, TEs that copy themselves before insertion. However, the predominant class of TEs found in the lungfish was not the same as that found in the salamanders. The little data we have at the moment suggests therefore that the diversity and abundance of TEs is variable between taxa with large genomes, similar to patterns found in taxa with smaller genomes. Based on results from the human genome, we suggest that the ‘missing’ portion of the lungfish and salamander genomes are old, highly divergent, and therefore inactive copies of TEs. The data available indicate that, unlike plants with large genomes, neither the lungfish nor the salamanders show an increased risk of extinction. Based on a slow rate of DNA loss in salamanders it has been suggested that the large salamander genome is the result of run-away genome expansion involving genome size increases via TE proliferation associated with reduced recombination rate. We know of no studies on DNA loss or recombination rates in lungfish genomes, however a similar scenario could describe the process of genome expansion in the lungfish. A series of waves of TE transposition and sequence decay would describe the pattern of TE content seen in both the lungfish and the salamanders. The lungfish and salamanders, therefore, may accommodate their large load of TEs because these TEs have accumulated gradually over a long period of time and have been subject to inactivation and decay.
Keywords: genome size, transposable elements, eukaryotes, large genomes, evolution
Introduction
Haploid genome sizes in living organisms, excluding viruses, range by 6 orders of magnitude, from 1.39 × 10−4 Gb for the Tremblaya, a bacterial mealybug symbiont,1 to 148 Gb for the Japanese canopy plant, Paris japonica.2 The value of about 1,431 Gb (1,400 pg) estimated for amoeba in the 1960s is sometimes quoted as the largest known genome, however, there is some uncertainty in the accuracy of this value.3 While prokaryotic (eubacteria/archaea) genomes are much smaller overall than that of eukaryotes, there is a continuum in genome sizes between prokaryotes and eukaryotes. In particular, many smaller fungal and apicomplexan parasite genomes are within the same size range as that of many prokaryotes.2,4 In prokaryotes, coding DNA scales linearly with genome size. In eukaryotic genomes greater than 0.01 Gb, this changes, so that the expansion of coding DNA slows, and the expansion of non-coding DNA increases as genomes become larger.5 In large genomes much of this non-coding DNA consists of transposable elements.5 Non-coding DNA refers to DNA that is non-coding with reference to the host genome. Transposable elements code for proteins for their own replication but are considered non-coding in terms of the host genome.
Transposable elements fall into two major classes, Class I elements (retrotransposons) that copy themselves before insertion and Class II elements (DNA transposons) that leave the donor site before re-insertion.6 As genome size increases, the numerical contribution of Class II elements increases, but the fractional contribution does not; whereas the both the numerical and fractional contribution of Class I elements increases linearly.5 Class I elements can be divided into five orders on the basis of mechanistic features, organization and reverse transcriptase phylogeny: LTR retrotransposons, DIRS-like, Penelope-like, LINEs and SINES.6 In general, LINEs are more prevalent in metazoan than in land plant genomes, whereas LTR elements are more prevalent in land plant genomes.6
Why do genomes accumulate non-coding and potentially deleterious DNA, sometimes to the point that 85% of the genome is non-coding DNA, chiefly transposable elements? Lynch argues that the accumulation of non-coding DNA is a function of the ratio of the power of mutation to drift, the ‘mutational-hazard’ hypothesis, which is dependent on effective population size.5 In a 2003 seminal paper Lynch and Conery showed that there is a reduction by several orders of magnitude in effective population size when comparing prokaryotes to unicellular eukaryotes to multicellular eukaryotes.7 They suggested that this decrease in effective population size creates an environment with increased random genetic drift, allowing the accumulation of non-coding DNA that would otherwise be removed by purifying selection.8 Whitney and Garland (2010) re-analyzed a data set used by Lynch7 within a phylogenetic framework, found no relationship, and suggest genome size is unlikely to be explained by a single factor such as population size, but don’t present an alternative hypothesis. More recently, Jurka et al. have posited the ‘carrier subpopulation hypothesis’ (CASP),9 where they suggest that waves of TE propagation coincide with speciation because both phenomena are triggered by the division of large populations into small sub-populations in which drift is important, in effect an extension of the Lynch hypothesis.
How do TEs affect a genome? The relationship between TEs and the host genome has been described as a continuum from extreme parasitism to mutualism.10 TEs modify genomes from the small scale to the large scale, by inserting within or next to genes, by mediating chromosomal rearrangements, or by expanding/contracting genomes. TEs may be domesticated, that is all or part of the TE may be co-opted by the genome as a gene, regulatory or structural element.11 Insertion of a TE near or within a gene may result in changes in gene expression.11 At the phenotypic level, genome size affects cell size and consequently, in some cases, the rate of cell division and metabolic rate.12 Genomes have a variety of defenses against TEs, for example, transcriptional silencing via methylation,5 by small RNA repression,13 or by repeat-induced point mutation.5 Despite occasionally being of benefit, TEs are generally considered as having a negative effect on host fitness. Threatened plant species, that is, those close to extinction, have on average larger genomes than those that are not threatened.14 Higher numbers of copies of TEs in Drosophila is associated with reduced fitness.15 In humans, at least 48 LINE-1 mediated events associated with diseases have been identified.16 How then do genomes support the burden of TEs, in some cases very large burdens?
Here we review what is known about the transposable element content of genomes at the extreme of genome size, in very large genomes, defined here as larger than 20 Gb. We examine the number and distribution of the very large genomes within eukaryotes, the TE content of large genomes which have been examined, and whether a particular type of TE is found predominantly in very large genomes. Finally, we ask if species with very large genomes can survive over a very long time or are they condemned to quickly go extinct?
Very Large Genomes and Transposable Element Content
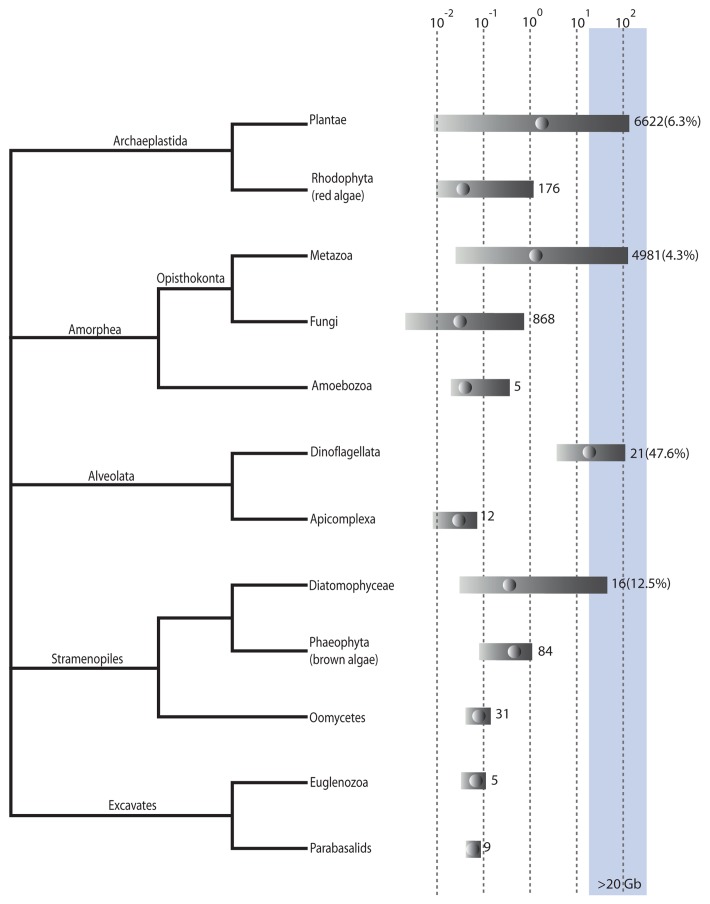
Genome sizes vary among eukaryotes by nearly five orders of magnitude with the largest genomes, that is, those greater than 20 Gb, found in the diatoms, dinoflagellates, metazoans and green plants (Fig. 1). The number of records available, however, is highly skewed, with the metazoans, fungi and green plants being much more highly represented than other groups (Fig. 1). Genome sizes shown in this figure, therefore, may reflect only a fraction of actual genome size diversity, particularly in the less well examined groups.
Figure 1. Genome sizes in eukaryotes. Relationships between groups based on Adl et al. (2012).19 Genome size is log(10) haploid genome size in Gb. Bars indicates min to max genome sizes for a particular group. Dots within the bars indicate median genome size. Figures to the right of the bar are the number of records for that group, the figure within brackets is the percentage of genomes within that group that are > 20 Gb. The blue shading indicates genomes larger than 20 Gb. Most of the data is taken from databases listed on DOGs database.2 Other references are too numerous to list but can be obtained from the authors on request. Duplicate records were removed so that the minimum genome size listed was kept.
Clearly, the best estimates of TE content of a genome will be from the analysis of a fully sequenced and assembled genome. However, no very large genome has been sequenced yet, due chiefly to technical and financial restraints,3 and search of the literature and databases suggests that there are no plans to fully sequence a very large genome. At least 100 metazoan genomes,17 mostly angiosperms (median genome size 3 Gb) and 25 green plant genomes,18 all anigosperms (median genome size 2.4 Gb), have been completely sequenced (Fig. 2). Apart from angiosperms and mammals, many of the genomes sequenced, or planned to be sequenced, are that of fungi, nematodes or drosophilidae, all of which have smaller genomes (median genome size 0.03 Gb, 0.08 Gb and 0.2 Gb respectively).2 However, some large genomes have been sampled for TE content, that of several salamanders, two angiosperm species (Fritillaria) and one each of a lungfish (Neoceratodus forsteri), dinoflagellate (Alexandrium ostenfeldii) and a gymnosperm (Pinus taeda). Below we summarize the results of large genome sampling, and also look at groups with large genomes that haven’t yet been examined.
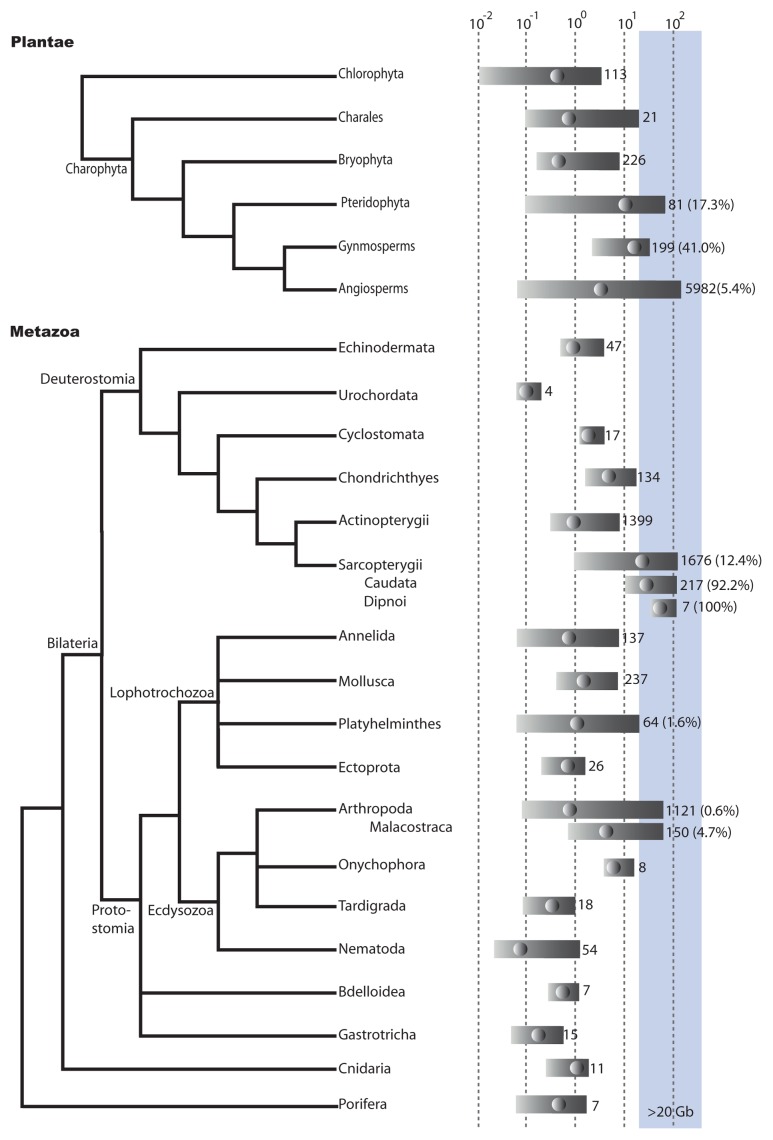
Figure 2. Genome sizes in metazoans and plants. Relationships between metazoans groups is based on a consensus of Philippe et al. (2011)55 and Rota-Stabelli et al. (2011).56 Relationships within the plants is based on Adl et al. (2012).17 Genome size is log(10) haploid genome size in Gb. Bars indicates min to max genome sizes for a particular group. Dots within the bars indicate median genome size. Figures to the right of the bar are the number of records for that group, the figure within brackets is the percentage of genomes within that group that are > 20 Gb.The blue shading indicates genomes larger than 20 Gb. Groups with less than 3 records were not included. Most of the data is taken from databases listed on DOGs database.2 Other references are too numerous to list but can be obtained from the authors on request. Duplicate records were removed so that the minimum genome size listed was kept.
There are two major lineages of green plants (Fig. 2). The first, the chlorophytes, is comprised of what is classically considered ‘green algae’. The second lineage, the charophyta, is comprised of land plants and several groups of ‘green algae’, the charales which are more closely related to land plants.19 All chlorophyta genomes examined are less than 20 Gb, the largest genome is that of Codium fragile at about 3.5 Gb.
Within the charophyta the largest genomes are found within the charales, ferns, angiosperms and gymnosperms (Fig. 2). The largest charale genomes are within the Chara genus, the largest genome is that of Chara contraria with a genome size of 19.6 pg (~19 Gb), just under our designated limit of a large genome of 20 Gb. No charale genomes have been sequenced, or sampled for TE content as far as we could determine.
Although the prize for the largest genome ever identified goes to an angiosperm, the Japanese canopy plant, Paris japonica, at 152 pg (~148 Gb),20 prompting a rather memorable title of the blog, Byte Size Biology, ‘Now that’s a f***ing big genome!’,21 in fact most angiosperms have genomes on the smaller side (median 2.4 Gb) (Fig. 2). The only angiosperm genome larger than 20 Gb to be characterized for TE content are two Fritillaria (Liliaceae) species22 at 45 pg (~44 Gb) and 43pg (~42 Gb). Four fosmid clones selected for their repetitive nature were sequenced. Identifiable repetitive elements comprised 77.1 and 89.6% of the fosmid clones, chiefly LTR-retrotransposons. Only a small portion of the repetitive fraction of the genome was identified, using dot-blot hybridization they estimated that LTR-RTs are only 4.7 and 6.7% of the genomes. The authors therefore suggested that the Fritillaria genomes are composed of many diversified families of transposable elements.22
Gymnosperms have much larger genomes (median 17 Gb) than those of the angiosperms (Fig. 2). The TE content of the Loblolly pine (Pinus taeda, genome size ~21 Gb) has been estimated by two groups.23,24 The repetitive content of the Loblolly pine has been estimated at 75% by reassociation kinetics.25 Morse et al.,24 using BAC sequencing and massive parallel DNA sequencing of repetitive fractions of the genome, identified an LTR-RT, Gymny, as occupying a small fraction of the genome. Using sequencing of 10 BACs and whole genome shotgun sequencing, representing 7.5% of genome, Kovach et al.23 identified the 3 most common repetitive elements in the genomes as two different LTR-retrotransposons and a tandem centromeric repeat, but found that they represent less than 5% the genome. They suggested that the majority of elements in the pine are ‘novel’.
The ferns (Pteridophyta) also have larger genomes, from 0.7 Gb to 71 Gb, with a median of ~9 Gb (Fig. 2) and are known for their exceptionally high chromosome numbers.26 Despite this, most ferns appear to have neo-polyploidization levels similar to that of the angiosperms and diploid expression profiles.27 Barker et al. (2010)27 suggested that diploidization occurred in ferns but with physical loss of genetic material occurring at lower rates than in angiosperms. Very little is known about the genomes of the ferns.
Dinoflagellates (Dinoflagellata) are an important group in aquatic environments.28 As zooxanthellae they are found in symbiosis with corals, cnidarians, and clams and provide their hosts with essential metabolites. They can also produce a diverse array of toxins that have a significant impact on marine ecosystems and fisheries. Not only do they have some of the largest eukaryotic genomes (median 20 Gb) (Fig. 1), they have uncommon genomes, with unusual DNA bases and atypical histones.29 An analysis of the Alexandrium ostenfeldii (~112 Gb) genome by BAC end and fosmid clone sequencing, representing 0.0059% of the genome, suggests that the majority of the genome consists of large tandem arrays.30 These arrays fell into 5 categories and together comprise at least 58% of the genome.30 This accords with previous studies on the repetitive nature of the genome using different methods.30 However, the authors were only able to identify a tiny portion of the sequence (less than 1%) as transposable elements and for the tandem repeats were unable to find any homology to known repetitive elements nor to any domains in the Pfam database.30
Diatoms (Diatomophyceae) are one of the predominant contributors to global carbon fixation and account for 40% of the total primary production in the ocean.31 As far as we can tell, no large diatom genomes have been sampled for TE content but two smaller genomes have been sequenced. The major TE component of both the Phaeodactylum tricornutum and Thalassiosira pseudonana genomes is LTR-RTs, as in plants, but with a much higher abundance in P. tricornutum.32
Within the metazoans very large genomes are found in the shrimps and amphipods (Malacostraca), flatworms (Platyhelminthes), lungfish (Dipnoi) and the salamanders and newts (Caudata). Many of the malacostracans with large genomes are shrimps and amphipods found in Arctic regions.33 No large genomes have been sampled for TE content but two smaller genomes have been examined. Interestingly, 21% of the black tiger shrimp genome (2.5 Gb) showed moderate similarity to a highly lethal and contagious virus, suggesting that much of the genome is composed of proviral remnants.34
The largest genome size estimated for the Platyhelminthes is that for Otomesostoma auditivum, just on 20 Gb. Like the crustaceans, no large genomes have been examined but three smaller genomes have been sequenced. The percentage of the genomes of Schistosoma mansoni (0.38 Gb), S. japonica (0.40 Gb), S. hematobium (0.38 Gb), estimated to be repetitive were similar, 40% -47%. However, the TE composition varied. For S. janonica and S. mansoni only 20% of the genome was identified as TEs, 13% nonLTR-RTs and 6% LTR-RTs in S. janonica,35 and in S. mansoni the inverse, 5% nonLTR-RTs, 15% LTR-RTs.36 For S. hematobium 32% of the genome was estimated to be nonLTR-RTs and 11% LTR-RTs.37
Genome sizes in the Caudata range from 10 to 120 Gb, with a median of ~30 Gb (Fig. 2) and in the lungfish from ~40 to 129 Gb with a median of 51 Gb (Fig. 2). The genomes of six salamanders, with genome sizes ranging from ~14 to 50 Gb, were examined using next generation sequencing, representing 0.07–1.9% of the genomes.38 A pipeline, including RepeatModeler to identify de novo repeats was used to identify repetitive elements. Twenty-five to 47% of the genome was identified as repetitive, the percentage depending on the species.38 The largest identified component was LTR-RTs, unlike other metazoan genomes, of which the largest TE component is usually LINEs. The TE component of the Australian lungfish (52 pg, ~50 Gb), was estimated using random sequencing, representing only a small fraction of the genome, and found to be about 40% TEs, 22% of that two closely related types of LINEs.39
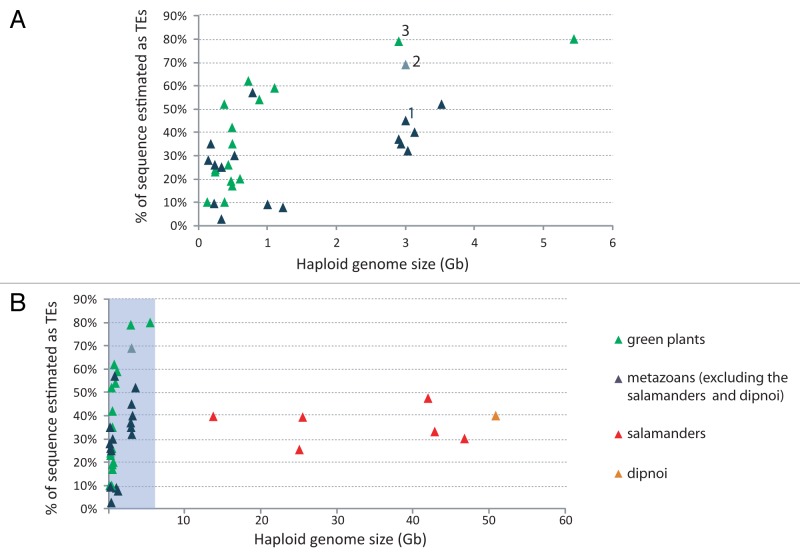
Among the large genomes sampled to date, the highest portion of the genome identified as TEs is for the salamanders (25–47%)38 and the lungfish (40%).39 An extrapolation of graphs showing the relationship between percentage of TE content against genome size would suggest that genomes this large would have a higher percentage of TE content.3 Intriguingly, these percentages are similar to that obtained for other well characterized, but much smaller, metazoan genomes (Fig. 3). In contrast, larger plant genomes can be up to 82% TEs (Fig. 3).40
Figure 3. The relationship between the percentage of sequences identified as transposable elements and genome size for plants and metazoans. (A) Genomes < 6 Gb (shaded in gray in B). (B) All genomes shown. Data from many sources and references are available from the authors on request. 1 = Homo sapiens;57 2 = Homo sapiens estimated using Pclouds;42 3 = Zea mays.40
The percentage of the lungfish and salamander genomes identified as TEs is almost certainly an underestimation because of difficulties in identifying TEs.41 A recent publication on the human genome,42 which had been estimated to be about 45% TEs, suggests that the human genome is at least 66–69% repetitive, chiefly TEs, which is much closer to estimates for larger plant genomes (Fig. 3B).42 The authors attributed the increase of the repetitive content detected to the ability of their approach to better detect short sequences and sequences from older and more diverse TE families.42 This finding suggests that the TE contents in the salamander and lungfish genomes could be underestimates due to the presence of undetected old and diverse TEs.
Concluding Remarks
The mean of published genome sizes is 1.39 Gb. Or, to put it another way, 80% of eukaryotic genomes are less than 5 Gb, and 95% are less than 20 Gb. Very large genomes are therefore rare but they are not confined to any particular group, they are found throughout the tree of life; within the plants, metazoans, dinoflagellates and diatoms (Fig. 1). The lack of the very large genomes within the excavates should be taken with caution because of the extremely small sample size (Fig. 1). Are very large genomes large because of massive amplifications of a single type of TE? The little data available would suggest not; that like taxa with smaller genomes,11 the diversity and abundance of TEs is variable between taxa. In the Australian lungfish, the predominant type of TE is non-LTRs,39 while in the salamanders it is LTRs that predominate.38 For the other large genomes examined, the dinoflagellate (Alexandrium ostenfeldii), two angiosperm species (Fritillaria)22 and one a gymnosperm (Pinus taeda),23,24 either no TEs were identified, or TEs were identified but found to be only a small proportion of the genome. The authors of the papers examining the plant genomes suggest they are composed of diverse or novel TEs.22-24
Up to this point, we have assumed that genomes become very large due to the presence of TEs. In eukaryotes expansion in genomes between 0.01 and 5 Gb is correlated with TE abundance.5 However, could the difficulty in identifying TEs in very large genomes, such as the lillies, be because genomes on this scale are due to large segmental duplications or whole genome duplications (WGD) instead? Current evidence suggests that WGDs are not associated with long-term increases in genome size. In angiosperms, mean DNA amount per basic genome tends to actually decrease with increasing ploidy.43,44 In metazoans, lineages with additional WGD events compared with sister lineages, for example the teleosts, do not have larger genomes (Fig. 2).45 Most ferns appear to have neo-polyploidization levels similar to that of the angiosperms and diploid expression profiles27 and the lungfish karyotype and a phylogeny of several Hox genes suggest that the genome has not undergone a recent WGD event.39 In a recent analysis of the transcriptome of the salamander Ambystoma tigrinum the authors note that the size of the transcriptome is in line with those of other vertebrates, suggesting that the large genome (~30 Gb) is not the result of extensive segmental duplications.46
The fate of TEs within a genome are generally thought to be governed by a balance between transposition and selection, but other factors may also influence TE evolution and dynamics.11 Most copies of TEs within a genome are inactive; copies may acquire stop codons or frameshifts or another TE may insert within the copy. LINE elements are often 5′ truncated and therefore inactive upon insertion due to their transposition machinery. In addition, TE activity is often repressed by host genome control mechanisms.11 New, active TEs may be introduced into a genome by two means, by horizontal transfer or by the re-emergence of autonomous sequences as a result of recombination between inactive copies, defunct TEs may also use the transposition machinery of an active TE. Modeling of TE dynamics suggest that after transposition a TE can suffer a number of fates. These fates depend on various factors, such as how deleterious the TE insertion is on host fitness, interactions between different families of TEs, the rate of transposition of the TE, the strength of selection vs. drift, the effectiveness of the host control machinery, and the overall rate of DNA loss in the host genome.11
The well known positive correlation between genome size and TE abundance would appear not hold for very large genomes (Fig. 3). We have suggested that in fact it does, that the ‘missing’ TEs are old, and therefore highly divergent and difficult to detect. How may the dynamics of TEs and the host result in a very large genome? Here we use what is known about the salamanders and lungfish to posit that they have had large genomes for a long period of time and so are ‘accommodating’ the load. An estimation of the evolution of salamander genome sizes within a phylogenetic framework suggests that most of the increase in genome size occurred between the late Carboniferous, ~300 Ma (million years ago), and the Cenozoic, ~65 Ma.47 A non-phylogenetic estimation of the evolution of lungfish genome sizes showed that there was a rapid increase in genome size ~350–200 Ma, followed by little change in the lineage leading to the Australian lungfish but further increases in genome size in the South American lungfish lineage until ~100 Ma.48 Unlike plants, salamanders with large genomes are not more likely to be at increased risk for extinction.49 Similar statistical analyses have not been done for the lungfish, however, of the six species of lungfish, three are listed as being of ‘least concern’ on the IUCN Red List Threatened Species while the other three have not be assessed.50 The salamanders and the lungfish have similar TE profiles, in terms of the percentage of the genome identified as TEs.38,39
Genome size is correlated with TE content, but also with the recombination rates and rates of DNA loss.51-53 Sun et al. (2012) have shown that in the salamanders there is a slow rate of DNA loss.54 They suggest a scenario of run-away genome expansion in the salamanders whereby genome size increases via TE proliferation with the number of chromosome arms remaining constant, the recombination rate therefore decreases, reducing the rate of DNA loss, this reduction in DNA loss facilitates further genome expansion because insertions and deletions are less likely to be deleterious as coding density decreases.54 We know of no studies on DNA loss or recombination rates in lungfish genomes, however a similar scenario could describe the process of genome expansion in the lungfish. A series of waves of TE transposition and sequence decay would describe the pattern of TE content seen in both the lungfish and the salamanders. The lungfish and salamanders, therefore, accommodate their large load of TEs because these TEs have accumulated gradually over a long period of time and have been subject to inactivation and decay.
Acknowledgments
This work was supported by Centre National de la Recherche Scientifique under the program ‘Action Thématique Incitative sur Programme’ awarded to Didier Casane from 2006–2009. The authors would also like to thank the two reviewers for their comments and improvements to the manuscript.
Glossary
Abbreviations:
- DIRS
dictyostelium intermediate repeat sequence
- Ginger-1
Gypsy-integrase-related type 1
- LINE
long interspersed nuclear element
- LTR
long terminal repeat
- Ma
million years ago
- RT
retrotransposon
- SINE
short interspersed nuclear element
- TE
transposable element
- WGD
whole genome duplication
Disclosure of Potential Conflicts of Interest
The authors state that they have no conflict of interest or financial disclosure statements to declare.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/24775
References
- 1.McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21:1366–72. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DOGS - Database of Genome Sizes www.cbs.dtu.dk/databases/DOGS/index.php
- 3.Gregory TR. Synergy between sequence and size in large-scale genomics. Nat Rev Genet. 2005;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- 4.Fungal Genome Size Database www.zbi.ee/fungal-genomesize
- 5.Lynch M. The Origins of Genome Architecture. 1st Ed. Sunderland, Massachussetts, USA: Sinauer Associates Inc., 2007. [Google Scholar]
- 6.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–82. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 7.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–4. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 8.Whitney KD, Garland T., Jr. Did genetic drift drive increases in genome complexity? PLoS Genet. 2010;6:e1001080. doi: 10.1371/journal.pgen.1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jurka J, Bao W, Kojima KK. Families of transposable elements, population structure and the origin of species. Biol Direct. 2011;6:44. doi: 10.1186/1745-6150-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 11.Hua-Van A, Le Rouzic A, Boutin TS, Filée J, Capy P. The struggle for life of the genome’s selfish architects. Biol Direct. 2011;6:19. doi: 10.1186/1745-6150-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory R. Genome Size Evolution in Animals. In: Gregory RT, ed. The Evolution of the Genome San Diego, USA: Elsevier Academic Press, 2005:3–87. [Google Scholar]
- 13.Lu J, Clark AG. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome Res. 2010;20:212–27. doi: 10.1101/gr.095406.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinogradov AE. Selfish DNA is maladaptive: evidence from the plant Red List. Trends Genet. 2003;19:609–14. doi: 10.1016/j.tig.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Pasyukova EG, Nuzhdin SV, Morozova TV, Mackay TFC. Accumulation of transposable elements in the genome of Drosophila melanogaster is associated with a decrease in fitness. J Hered. 2004;95:284–90. doi: 10.1093/jhered/esh050. [DOI] [PubMed] [Google Scholar]
- 16.Chen J-M, Stenson PD, Cooper DN, Férec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet. 2005;117:411–27. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 17.National Human Genome Research Institue www.genome.gov/10002154
- 18.CoGePedia http://genomevolution.org/wiki/index.php/Plant_Genome_Statistics
- 19.Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–93. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pellicer J, Fay M, Leitch I. The largest eukaryotic genome of them all? Bot J Linn Soc. 2010;164:10–5. doi: 10.1111/j.1095-8339.2010.01072.x. [DOI] [Google Scholar]
- 21.Byte Size Biology http://bytesizebio.net/index.php/2010/10/26/now-thats-a-fing-big-genome
- 22.Ambrozová K, Mandáková T, Bures P, Neumann P, Leitch IJ, Koblízková A, et al. Diverse retrotransposon families and an AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies. Ann Bot. 2011;107:255–68. doi: 10.1093/aob/mcq235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovach A, Wegrzyn JL, Parra G, Holt C, Bruening GE, Loopstra CA, et al. The Pinus taeda genome is characterized by diverse and highly diverged repetitive sequences. BMC Genomics. 2010;11:420. doi: 10.1186/1471-2164-11-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morse AM, Peterson DG, Islam-Faridi MN, Smith KE, Magbanua Z, Garcia SA, et al. Evolution of genome size and complexity in Pinus. PLoS One. 2009;4:e4332. doi: 10.1371/journal.pone.0004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plomion C, Chagne D, Pot D, Kumar S, Wilcox P, Burdon R, et al. Pines. In: Kole C, ed. Genome Mapping and Molecular Breeding in Plants, Volume 7 Forest Trees Berlin: Springer-Verlag, 2007; 7:29–92. [Google Scholar]
- 26.Nakazato T, Baker M, Rieseberg L, Gastony G. Evolution of the nuclear genome of ferns and lycophytes. In: edited by Rankder T, Haufler C, ed. Biology and evolution of ferns and lycophytes Cambridge, UK: Cambridge University Press, 2008. [Google Scholar]
- 27.Barker MS, Wolf PG. Unfurling Fern Biology in the Genomics Age. Bioscience. 2010;60:177–85. doi: 10.1525/bio.2010.60.3.4. [DOI] [Google Scholar]
- 28.Hackett JD, Anderson DM, Erdner DL, Bhattacharya D. Dinoflagellates: a remarkable evolutionary experiment. Am J Bot. 2004;91:1523–34. doi: 10.3732/ajb.91.10.1523. [DOI] [PubMed] [Google Scholar]
- 29.Wisecaver JH, Hackett JD. Dinoflagellate genome evolution. Annu Rev Microbiol. 2011;65:369–87. doi: 10.1146/annurev-micro-090110-102841. [DOI] [PubMed] [Google Scholar]
- 30.Jaeckisch N, Yang I, Wohlrab S, Glöckner G, Kroymann J, Vogel H, et al. Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii. PLoS One. 2011;6:e28012. doi: 10.1371/journal.pone.0028012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarthou G, Timmermans KR, Blain S, Tréguer P. Growth physiology and fate of diatoms in the ocean: a review. J Sea Res. 2005;53:25–42. doi: 10.1016/j.seares.2004.01.007. [DOI] [Google Scholar]
- 32.Maumus F, Allen AE, Mhiri C, Hu H, Jabbari K, Vardi A, et al. Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genomics. 2009;10:624. doi: 10.1186/1471-2164-10-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rees DJ, Belzile C, Glémet H, Dufresne F. Large genomes among caridean shrimp. Genome. 2008;51:159–63. doi: 10.1139/G07-108. [DOI] [PubMed] [Google Scholar]
- 34.Huang S-W, Lin Y-Y, You E-M, Liu T-T, Shu H-Y, Wu K-M, et al. Fosmid library end sequencing reveals a rarely known genome structure of marine shrimp Penaeus monodon. BMC Genomics. 2011;12:242. doi: 10.1186/1471-2164-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–51. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–8. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young ND, Jex AR, Li B, Liu S, Yang L, Xiong Z, et al. Whole-genome sequence of Schistosoma haematobium. Nat Genet. 2012;44:221–5. doi: 10.1038/ng.1065. [DOI] [PubMed] [Google Scholar]
- 38.Sun C, Shepard DB, Chong RA, López Arriaza J, Hall K, Castoe TA, et al. LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders. Genome Biol Evol. 2012;4:168–83. doi: 10.1093/gbe/evr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metcalfe CJ, Filée J, Germon I, Joss J, Casane D. Evolution of the Australian lungfish (Neoceratodus forsteri) genome: a major role for CR1 and L2 LINE elements. Mol Biol Evol. 2012;29:3529–39. doi: 10.1093/molbev/mss159. [DOI] [PubMed] [Google Scholar]
- 40.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–6. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 41.Flutre T, Permal E, Quesneville H. In search of lost trajectories: Recovering the diversification of transposable elements. Mob Genet Elements. 2011;1:151–4. doi: 10.4161/mge.1.2.17094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leitch I, Bennett M. Genome downsizing in polyploid plants. Biol J Linn Soc Lond. 2004;82:651–63. doi: 10.1111/j.1095-8312.2004.00349.x. [DOI] [Google Scholar]
- 44.Ozkan H, Tuna M, Arumuganathan K. Nonadditive changes in genome size during allopolyploidization in the wheat (aegilops-triticum) group. J Hered. 2003;94:260–4. doi: 10.1093/jhered/esg053. [DOI] [PubMed] [Google Scholar]
- 45.Santini F, Harmon LJ, Carnevale G, Alfaro ME. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol Biol. 2009;9:194. doi: 10.1186/1471-2148-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eo SH, Doyle JM, Hale MC, Marra NJ, Ruhl JD, DeWoody JA. Comparative transcriptomics and gene expression in larval tiger salamander (Ambystoma tigrinum) gill and lung tissues as revealed by pyrosequencing. Gene. 2012;492:329–38. doi: 10.1016/j.gene.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 47.Organ CL, Canoville A, Reisz RR, Laurin M. Paleogenomic data suggest mammal-like genome size in the ancestral amniote and derived large genome size in amphibians. J Evol Biol. 2011;24:372–80. doi: 10.1111/j.1420-9101.2010.02176.x. [DOI] [PubMed] [Google Scholar]
- 48.Thomson K. An attempt to reconstruct evolutionary changes in the cellular DNA content of lungfish. J Exp Zool. 1972;180:363–72. doi: 10.1002/jez.1401800307. [DOI] [Google Scholar]
- 49.Vinogradov AE. Genome size and extinction risk in vertebrates. Proc Biol Sci. 2004;271:1701–5. doi: 10.1098/rspb.2004.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.IUCN. 2012. The IUCN Red List of Threatened Species. Version 2012.2. www.iucnredlist.org
- 51.Petrov DA, Sangster TA, Johnston JS, Hartl DL, Shaw KL. Evidence for DNA loss as a determinant of genome size. Science. 2000;287:1060–2. doi: 10.1126/science.287.5455.1060. [DOI] [PubMed] [Google Scholar]
- 52.Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006;23:450–68. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- 53.Nam K, Ellegren H. Recombination drives vertebrate genome contraction. PLoS Genet. 2012;8:e1002680. doi: 10.1371/journal.pgen.1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun C, López Arriaza JR, Mueller RL. Slow DNA loss in the gigantic genomes of salamanders. Genome Biol Evol. 2012;4:1340–8. doi: 10.1093/gbe/evs103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Philippe H, Brinkmann H, Copley RR, Moroz LL, Nakano H, Poustka AJ, et al. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature. 2011;470:255–8. doi: 10.1038/nature09676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rota-Stabelli O, Campbell L, Brinkmann H, Edgecombe GD, Longhorn SJ, Peterson KJ, et al. A congruent solution to arthropod phylogeny: phylogenomics, microRNAs and morphology support monophyletic Mandibulata. Proc Biol Sci. 2011;278:298–306. doi: 10.1098/rspb.2010.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]