Abstract
Part of the substantial unexplained familial aggregation of breast cancer may be due to interactions between common variants, but few studies have had adequate statistical power to detect interactions of realistic magnitude. We aimed to assess all two-way interactions in breast cancer susceptibility between 70 917 single nucleotide polymorphisms (SNPs) selected primarily based on prior evidence of a marginal effect. Thirty-eight international studies contributed data for 46 450 breast cancer cases and 42 461 controls of European origin as part of a multi-consortium project (COGS). First, SNPs were preselected based on evidence (P < 0.01) of a per-allele main effect, and all two-way combinations of those were evaluated by a per-allele (1 d.f.) test for interaction using logistic regression. Second, all 2.5 billion possible two-SNP combinations were evaluated using Boolean operation-based screening and testing, and SNP pairs with the strongest evidence of interaction (P < 10−4) were selected for more careful assessment by logistic regression. Under the first approach, 3277 SNPs were preselected, but an evaluation of all possible two-SNP combinations (1 d.f.) identified no interactions at P < 10−8. Results from the second analytic approach were consistent with those from the first (P > 10−10). In summary, we observed little evidence of two-way SNP interactions in breast cancer susceptibility, despite the large number of SNPs with potential marginal effects considered and the very large sample size. This finding may have important implications for risk prediction, simplifying the modelling required. Further comprehensive, large-scale genome-wide interaction studies may identify novel interacting loci if the inherent logistic and computational challenges can be overcome.
INTRODUCTION
We recently identified 47 novel breast cancer susceptibility loci in a multi-centre case–control study of single nucleotide polymorphisms (SNPs) with prior evidence of association from a combined analysis of genome-wide association studies (GWAS) (1–3), raising the total number of susceptibility loci for breast cancer to >70 (4–20). Despite this and other successful efforts to identify breast cancer susceptibility variants by association, linkage and sequencing studies, a large portion of the observed familial aggregation of the disease remains unexplained (3,21–25). Part of this unexplained heritability may be due to variants that modify breast cancer risk through interaction effects (26), where (and throughout this paper) an interaction is defined in the statistical sense that the relative risks for the combined genotypes differ from those predicted by multiplying the marginal relative risks for each SNP. Few studies have evaluated genetic interactions with sufficient statistical power to reach meaningful conclusions and no SNP–SNP interactions in association with breast cancer risk have been convincingly replicated.
We aimed to assess, agnostically, two-SNP interactions for association with breast cancer risk between 70 917 common SNPs with potential marginal effects, which were selected for genotyping based primarily on a combined analysis of nine GWAS. Interactions were assessed using cases and controls of white European origin from the Breast Cancer Association Consortium (BCAC).
RESULTS
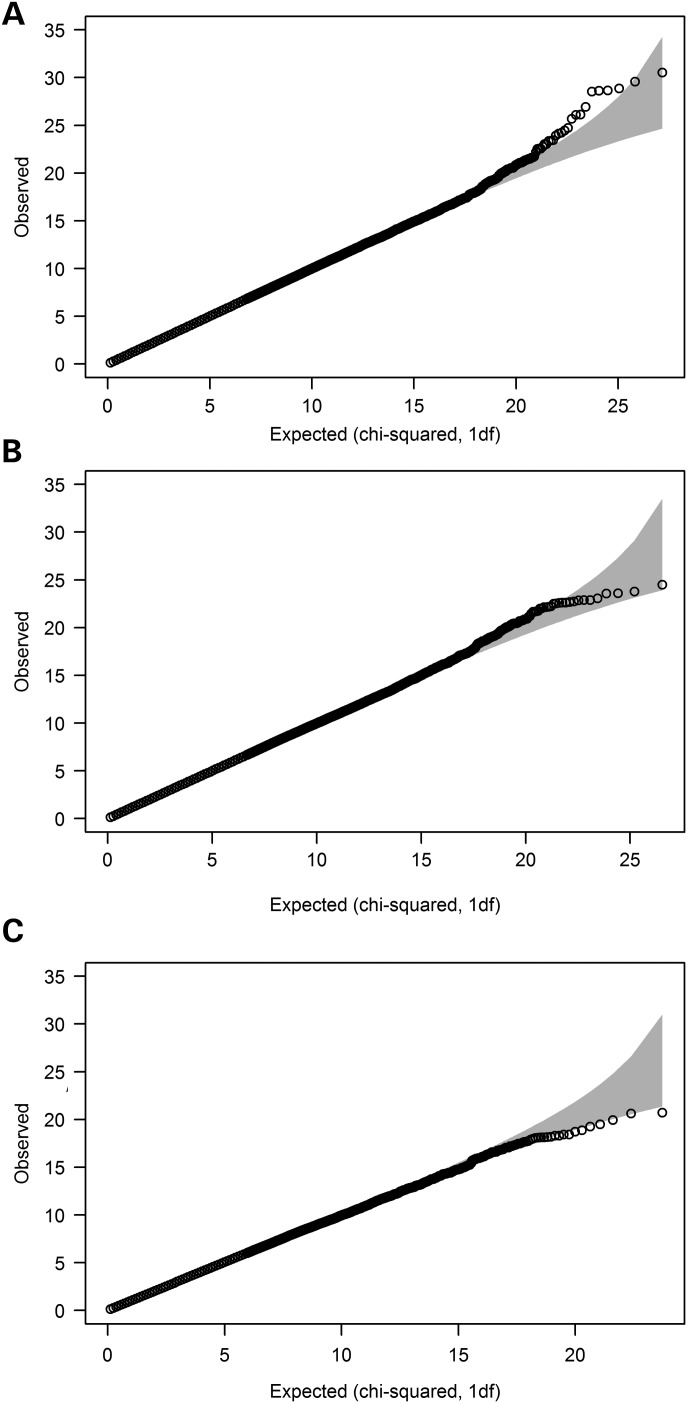
We first preselected 3277, 2788 and 1342 SNPs for the assessment of interactions in regard to risk for overall, oestrogen receptor (ER)-positive and ER-negative disease, respectively, based on statistical evidence of a marginal per-allele effect (P < 0.01). An evaluation of all two-SNP combinations (1 degree-of-freedom, d.f.) revealed no clear evidence of interaction in any of the three analyses (Fig. 1). For risk of breast cancer overall, of the 5.4 million interactions considered, 13 had a P-value of <10−6 (compared with 5.4 predicted), but none had a P-value of <10−8. The 13 interactions at P < 10−6 represented eight potential interaction signals after accounting for SNPs in high linkage disequilibrium (LD) (r2 > 0.85, Table 1), and three of these involved the same SNP (rs17117532) along with one of three others in modest LD (0.58 ≤ r2 ≤ 0.77). The lowest P-value was 3.3 × 10−8, which corresponded to a Bonferroni-corrected value of 0.16 considering all interactions evaluated. The corrected P-value was 0.058 based on the number of possible interactions between the estimated 1898 effective independent loci represented by the total 3277. In all 13 instances, the interaction effect was in the opposite direction to the main effects for the two SNPs involved, with very little LD between the two potentially interacting SNPs (r2 ≤ 0.073).
Figure 1.
Q–Q plots from the first set of analyses based on the χ2 statistics from the 1 d.f. LRT for (A) overall breast cancer, (B) oestrogen receptor (ER)-positive breast cancer and (C) ER-negative breast cancer.
Table 1.
SNP pairs with per-allele P-values for interaction <10−6
| Position | MAF (r2) | Single-SNP analysisa OR (95% CI), P-value | Interaction analysisb OR (95% CI), P-value | |
|---|---|---|---|---|
| Overall breast cancer | ||||
| rs10822036 | Chr10: 64044366 | 0.18 | 1.04 (1.01–1.06); 5.2 × 10−3 | 0.88 (0.82–0.94) |
| rs1379805 | Chr18: 18748550 | 0.36 | 1.04 (1.02–1.06); 1.4 × 10−4 | 0.92 (0.87–0.97) |
| Interaction | (<0.01) | 1.10 (1.06–1.14); 7.7 × 10−7 | ||
| Overall breast cancer | ||||
| rs132274 | Chr22: 27855126 | 0.35 | 1.03 (1.01–1.05); 9.4 × 10−3 | 1.19 (1.12–1.26) |
| rs17478824 | Chr5: 87821380 | 0.09 | 1.04 (1.01–1.08); 1.1 × 10−2 | 1.28 (1.18–1.41) |
| Interaction | (<0.01) | 0.88 (0.84–0.93); 6.6 × 10−7 | ||
| Overall breast cancer | ||||
| rs1573998 | Chr5: 56312250 | 0.20 | 1.06 (1.03–1.08); 2.6 × 10−6 | 0.87 (0.81–0.93) |
| rs16886496c | Chr5: 56253286 | 0.09 | 1.13 (1.09–1.17); 3.7 × 10−13 | 0.88 (0.80–0.96) |
| Interaction | (0.073) | 1.15 (1.09–1.21); 7.8 × 10−8 | ||
| Overall breast cancer | ||||
| rs17117532 | Chr14: 82392717 | 0.10 | 1.05 (1.01–1.08); 5.3 × 10−3 | 0.84 (0.77–0.92) |
| rs1936396d | Chr10: 44868074 | 0.23 | 1.03 (1.01–1.06); 4.3 × 10−3 | 0.87 (0.81–0.93) |
| Interaction | (<0.01) | 1.16 (1.10–1.23); 5.4 × 10−8 | ||
| Overall breast cancer | ||||
| rs17117532 | Chr14: 82392717 | 0.10 | 1.05 (1.01–1.08); 5.3 × 10−3 | 0.84 (0.77–0.92) |
| rs2018728e | Chr10: 44837353 | 0.34 | 1.03 (1.01–1.05); 5.6 × 10−3 | 0.88 (0.83–0.94) |
| Interaction | (<0.01) | 1.14 (1.09–1.20); 9.2 × 10−8 | ||
| Overall breast cancer | ||||
| rs17117532 | Chr14: 82392717 | 0.10 | 1.05 (1.01–1.08); 5.3 × 10−3 | 0.84 (0.77–0.91) |
| rs7905526f | Chr10: 44837353 | 0.27 | 1.03 (1.01–1.06); 2.1 × 10−4 | 0.87 (0.82–0.93) |
| Interaction | (<0.01) | 1.15 (1.10–1.21); 3.3 × 10−8 | ||
| Overall breast cancer | ||||
| rs17355209 | Chr4: 84672332 | 0.50 | 1.04 (1.02–1.06); 3.0 × 10−5 | 1.19 (1.13–1.26) |
| rs4980025 | Chr10: 80510787 | 0.11 | 0.94 (0.92–0.97); 3.0 × 10−4 | 1.17 (1.07–1.29) |
| Interaction | (<0.01) | 0.90 (0.86–0.94); 8.6 × 10−7 | ||
| Overall breast cancer | ||||
| rs7714708 | Chr5: 58330771 | 0.36 | 1.03 (1.01–1.05); 4.1 × 10−3 | 1.17 (1.11–1.23) |
| rs836808 | Chr5: 80010326 | 0.26 | 0.97 (0.95–0.99); 7.5 × 10−3 | 1.12 (1.06–1.19) |
| Interaction | (<0.01) | 0.92 (0.89–0.95); 4.0 × 10−7 | ||
| ER-positive disease | ||||
| rs11604821 | Chr11: 69061318 | 0.33 | 1.04 (1.02–1.07); 2.8 × 10−4 | 1.22 (1.14–1.30) |
| rs11600497 | Chr11: 68882499 | 0.14 | 0.95 (0.92–0.98); 3.0 × 10−3 | 1.17 (1.07–1.27) |
| Interaction | (<0.01) | 0.88 (0.84–0.93); 7.5 × 10−7 | ||
MAF, minor allele frequency; r, correlation coefficient; OR, odds ratio per copy of the minor allele(s); CI, confidence interval.
aThe single-SNP analysis modelled main effects, per copy of the minor allele, for each SNP separately (it was on this basis that SNPs were preselected for inclusion in the interaction analysis under the first analytical approach).
bThe interaction analysis included main effects for each of the two SNPs plus an interaction effect, in all cases per copy of each minor allele.
cSNP in LD with two SNPs, rs12655019 (r2 = 0.94) and rs16886525 (r2 = 0.99), for which P = 2.1 × 10−7 and 8.8 × 10−8, respectively, for interaction with rs1573998. r2 was 0.58 between rs1936396 and rs7905526, 0.77 between rs1936396 and rs7905526 and 0.74 between rs2018728 and rs7905526.
dSNP in LD with another SNP, rs11471 (r2 = 0.94), for which P = 3.3 × 10−7 for interaction with rs17117532. r2 was 0.58 between rs1936396 and rs7905526, 0.77 between rs1936396 and rs7905526 and 0.74 between rs2018728 and rs7905526.
eSNP in LD with another SNP, rs2018728 (r2 = 0.86), for which P = 9.0 × 10−7 for interaction with rs17117532. r2 was 0.58 between rs1936396 and rs7905526, 0.77 between rs1936396 and rs7905526 and 0.74 between rs2018728 and rs7905526.
fSNP in LD with another SNP, rs6593456 (r2 = 0.98), for which P = 8.7 × 10−8 for interaction with rs17117532. r2 was 0.58 between rs1936396 and rs7905526, 0.77 between rs1936396 and rs7905526 and 0.74 between rs2018728 and rs7905526.
For risk of ER-positive breast cancer, the strongest evidence of interaction was observed for rs7603983 and rs10490346 (P = 2.6 × 10−10). These two SNPs are 45 kb apart and in modest LD (r2 = 0.65). A re-evaluation of the cluster plot revealed poor cluster separation for rs7603983 and it was noted that data from one study in particular (Oulu Breast Cancer Study, OBCS) were overrepresented among borderline genotype determinations. When the 407 cases and 414 controls from the OBCS (1.1% of the sample) were excluded, there was no evidence of interaction (P = 0.58), suggesting that the original result was an artefact of the poor clustering. For none of the other 3.9 million SNP pairs considered for ER-positive disease risk was evidence of interaction observed after correction for multiple testing (VeffLi = 1647; P* ≥ 0.64) and only one pair had an uncorrected P-value of <10−6 (Table 1). Using this first approach, no evidence of two-SNP interactions was observed in regard to the risk of ER-negative breast cancer (VeffLi = 949; P* ≥ 0.91).
We evaluated the statistical power of our first approach to detect interactions between the preselected SNPs at a nominal significance level of 10−8, in the absence of main effects (Table 2). We estimated that for overall breast cancer, we had >90% power to detect a per-allele interaction odds ratio (ORint) as low as 1.16 between SNPs with minor allele frequency (MAF) >0.20. The corresponding minimum detectable ORint were 1.29 and 1.58 for SNPs with a MAF as low as 0.10 and 0.05, respectively. For pairs of SNPs with a equal MAF of 0.20, 0.10 and 0.05, an ORint of 1.16, 1.29 and 1.58, respectively, in the absence of main effects gives rise to a marginal OR of 1.08, 1.06 and 1.07, respectively, when the interactions are not accounted for. The corresponding statistical power estimates for the detection of these marginal OR in single-SNP analysis at P < 0.01 (i.e. to preselect SNPs for inclusion in the assessment of interactions) were 99, 88 and 72%. The power was similar for ER-positive disease, particularly for the more common SNPs (MAF ≥ 0.10). The power was lower for ER-negative disease, although for SNPs with MAF >0.20, the power was >90% for ORint as low as 1.30.
Table 2.
Minimum interaction odds ratio detectable with 90% power at P < 10−8 in the absence of main effects
| MAFSNP2 | MAFSNP1 |
|||
|---|---|---|---|---|
| 0.05 | 0.10 | 0.20 | 0.40 | |
| Overall breast cancer | ||||
| 0.05 | 1.58 (1.06, 56%) | 1.41 (1.04, 47%) | 1.30 (1.03, 48%) | 1.24 (1.03, 69%) |
| 0.10 | 1.41 (1.08, 85%) | 1.29 (1.06, 88%) | 1.21 (1.04, 78%) | 1.17 (1.04, 93%) |
| 0.20 | 1.30 (1.12, 99%) | 1.21 (1.08, 99%) | 1.16 (1.07, 99%) | 1.13 (1.05, 99%) |
| 0.40 | 1.24 (1.20, 99%) | 1.17 (1.14, 99%) | 1.13 (1.11, 99%) | 1.11 (1.09, 99%) |
| ER-positive breast cancer | ||||
| 0.05 | 1.69 (1.07, 55%) | 1.48 (1.05, 53%) | 1.35 (1.04, 60%) | 1.28 (1.03, 51%) |
| 0.10 | 1.48 (1.10, 89%) | 1.34 (1.07, 87%) | 1.25 (1.05, 83%) | 1.20 (1.04, 81%) |
| 0.20 | 1.35 (1.14, 99%) | 1.25 (1.10, 99%) | 1.19 (1.08, 99%) | 1.15 (1.06, 99%) |
| 0.40 | 1.28 (1.23, 99%) | 1.20 (1.17, 99%) | 1.15 (1.12, 99%) | 1.12 (1.10, 99%) |
| ER-negative breast cancer | ||||
| 0.05 | 2.17 (1.11, 52%) | 1.80 (1.08, 52%) | 1.57 (1.06, 53%) | 1.45 (1.05, 54%) |
| 0.10 | 1.80 (1.16, 88%) | 1.55 (1.11, 84%) | 1.40 (1.08, 82%) | 1.32 (1.07, 88%) |
| 0.20 | 1.57 (1.23, 99%) | 1.40 (1.16, 99%) | 1.30 (1.12, 99%) | 1.24 (1.10, 99%) |
| 0.40 | 1.45 (1.38, 99%) | 1.32 (1.27, 99%) | 1.24 (1.20, 99%) | 1.20 (1.17, 99%) |
MAF, minor allele frequency.
In parenthesis are the resulting marginal OR for SNP1 and power to detect it in the individual main-effect analysis (the latter is the probability of selecting the SNP for inclusion under the first approach at P < 0.01).
As an alternative approach, we exhaustively investigated all 2.5 billion possible two-SNP interactions using Boolean operation-based screening and testing (BOOST) and identified 278 387, 278 240 and 275 214 SNP pairs potentially associated (P < 10−4) with risk of overall, ER-positive and ER-negative disease, respectively. These interactions were then evaluated using logistic regression. For overall breast cancer risk, 18 SNP pairs had associated P-values of <10−10 from the genotype-based test for interaction (Supplementary Material, Table S1). Six of these included the SNP rs9625520, which on individual inspection of the cluster plot was determined to have failed genotyping due to merged clusters, despite having passed the previous quality-control filters based on the automatic genotype calls. All seven SNPs forming these six SNP pairs were in high LD (0.89 ≤ r2 ≤ 1.00) and all but rs9625520 were well-clustered; we re-checked the evidence of interaction between these and observed very little (all P[4 d.f.] ≥ 0.018) for any of 15 other possible two-SNP combinations. A further 11 pairs in the top 18 included the SNP rs7603983 (identified using the first analytical approach) and another correlated SNP within 220 kb (r2 ≥ 0.50); the evidence of interaction disappeared for each of these when data from the OBCS were excluded (P ≥ 0.82). For one of these SNP pairs, evidence of interaction was also observed in the per-allele model (P = 8.3 × 10−11), but not once data from the OBCS were excluded (P = 0.71). The remaining SNP-pair comprised two SNPs in high LD (r2 = 0.94); visual inspection of cluster plots showed that one of these (rs6989466) was poorly called (merged clusters). Since the only SNP in high LD with rs6989466 was the potentially interacting one (rs2013845), the interaction could not be assessed by proxy. No other SNP pairs had interaction-associated P-values of <10−10 from the per-allele test for interaction (1 d.f.).
No evidence of interaction in susceptibility to ER-positive or ER-negative breast cancer was observed for any additional SNP pairs using this second approach. The seven SNP pairs with an interaction P-value of <10−10 for ER-positive breast cancer risk were a subset of the 18 observed for overall breast cancer risk, all involved either rs7603983 (for which evidence disappeared after exclusion of data from the OBCS), rs9625520 or rs6989466 (both of which were deemed to have been poorly called after visual inspection of cluster plots) (Supplementary Material, Table S1).
DISCUSSION
We have assessed two-way combinations of >70 000 SNPs selected because they were associated with, or potentially associated with, breast cancer risk and found little evidence of interaction, defined as departure from multiplicativity between SNP main effects. This was the case for overall breast cancer and disease subtypes defined by ER status. We considered genotype-specific interactions (4 d.f.) for all 2.5 billion SNP pairs and per-allele departure (1 d.f.) for all pairwise combinations of SNPs selected based on evidence of a marginal effect.
Few SNP–SNP interactions have been identified (27) and none associated with breast cancer risk has been convincingly replicated. That said, no large-scale systematic evaluations have been published to date. Tao et al. (28) used BOOST to analyse data from two GWAS of prostate cancer, totalling 3140 cases and 4273 controls, and found no convincing evidence of interactions. To our knowledge, our study of 46 450 breast cancer cases and 42 461 controls is by far the largest of its kind. The statistical power of our first analysis of selected SNPs was very high (>90%) to detect ORint as low as 1.29 between common SNPs (MAF ≥ 0.10). Also, assuming an epistasis model in which an associated risk is restricted to individuals carrying variant alleles at both loci, which would imply a weak marginal effect for each SNP, the probability that SNPs involved in interactions of this magnitude were selected was also high (>80%). It was also highly likely that more common SNPs (MAF ≥ 0.20), involved in interactions with ORint as low as 1.16, were selected. Although no SNP pair achieved P < 10−8, we did observe an excess of pairs with an interaction P < 10−6 (13 versus 5.4 predicted). This suggests that some of these associations could be real and may be confirmed by even larger studies, if they could be carried out. In addition, it should be noted that the current analysis was based on a set of 70 917 SNPs and was, therefore, not genome-wide. On the other hand, 70% of the SNPs were included because evidence of per-allele association at P < 0.008 was observed in the combined GWAS (Stage 1) analysis of >10 000 cases and 10 000 controls; based on the scenarios considered in Table 2, the statistical power of this Stage 1 analysis was as low as 40% (although in most cases much higher) for more common SNPs. It should also be noted that the effect sizes for untyped causal variants may be greater than those associated with the tagging SNPs (tagSNPs) on genotyping arrays, and that the relative reduction in power to detect disease association based on tag-SNPs may be greater for interaction effects than for main effects. We cannot therefore rule out that more common SNPs involved in interactions, with ORint comparable to those reported above, were not selected for the iCOGS array and thus not included in our analysis. Nevertheless, our findings, based on a very large number of SNPs, are consistent with the assertion of Hill (2010) that interactions are likely to be small and very difficult to detect if the main effects are already small.
Although our study possessed lower statistical power to detect two-way interactions between less common SNPs (MAF < 0.10), the study sample size required to detect any such interactions may not be achievable. Zuk et al. (2012) concur with this, suggesting that detecting genetic interactions may require a sample size in the range of 500 000. It is also possible that interactions could exist in the absence of marginal main effects, if each SNP had associated effects that were in opposite directions, depending on the genotype of the other SNP. Such SNPs would not have been systematically selected for the iCOGS array nor preselected for the assessment of interactions using our first analytical approach. However, while mathematically possible, such qualitative interactions seem less biologically plausible.
Had we adopted a more stringent P-value criterion (than P < 0.01) to select SNPs based on evidence of a marginal effect using the first approach, one result in particular would have stood out. This involved two SNPs (rs1573998 and rs16886496) in low LD (r2 = 0.073), both with marginal associations significant at P < 10−5 (Table 1), located within 60 kb of each other, at an established breast cancer susceptibility region on 5q11.2 (5). Such cis-interactions have a somewhat different interpretation to those between SNPs located in different regions, in that they may reflect haplotype-specific risks or, equivalently, the effects of untyped rarer variants. The iCOGS array included several hundred additional variants across this region in order to inform the search for the likely causal variant(s); these fine-mapping SNPs were not included in the present analysis since they are being analysed as part of a separate, ongoing project, but it may be informative to perform a separate analysis of all possible cis-interactions in the future.
The results from our exhaustive assessment of all two-way interactions using BOOST were consistent with the largely null result from our first set of analyses, although the power of this second analysis is difficult to ascertain. BOOST is generally more powerful than other comparable methods, except in the presence of per-allele departure from independent SNP effects (29). The 18 SNP pairs for which strongest evidence of interaction (P < 10−10) was observed all included one of three SNPs with poor or questionable genotype clustering and a second SNP relatively close to (≤178 kb) and in LD with (r2 ≥ 0.50) the first. All 18 were ruled out as possible interactions, either because the evidence disappeared in a sensitivity analysis excluding a very small proportion of the data from one study with particularly poor genotype clustering or because the SNP was considered to have failed genotyping on visual inspection of the intensity cluster plots. Where possible, interactions were reassessed with other SNPs in LD with the failed SNP and no consistent results were observed. This highlights the need to check manually the cluster plots of SNPs potentially involved in interactions, since poor clustering is not always picked up in standard quality-control checks for high-throughput genotyping data.
It is possible that other types of two-way SNP–SNP interaction (not tested for in the present study) exist, as may higher order interactions. These might be discovered by other analytical approaches. Our results, particularly those from the 4 d.f. genotype-based test, suggest that the genotyping quality of SNPs involved in potential interactions should be checked to rule out artefacts resulting from poor clustering. Even so, our results are consistent with those of other large studies that have assessed two-way interactions between established susceptibility SNPs and other risk factors for breast cancer (30–33). Together, these results suggest that established risk factors for breast cancer tend to be related to disease risk such that their associated effects, expressed as ORs, can be multiplied together. This has important implications for risk prediction, simplifying the modelling required. Related to this, Aschard et al. (27) have shown by simulation that even if gene–gene and gene–environment interactions exist in regard to breast cancer risk, they are unlikely to improve dramatically the discrimination ability of risk-prediction models.
That we found no strong evidence of two-way SNP interactions might be surprising, given the consistent evidence of genetic epistasis in model systems (34). However, the main effects in model systems also tend to be larger than those observed for SNPs. It may be that the influence on disease risk of the biological processes that are modified by SNP interactions is too small to be detected, even using combined studies with large sample size, at least for breast cancer. This may also be related to the much more complex genetic background present in humans, perhaps together with the influence of lifestyle factors that may dilute genetic interactions. More sophisticated analyses could possibly tease out SNP interactions in breast cancer susceptibility, but this will probably require a much better understanding of how to classify SNPs in terms of their functional basis. Two-way SNP interactions have been reported for other diseases such as Psoriasis, Ankylosing Spondylitis and Behcet's disease, suggesting that such interactions can be found for complex phenotypes in humans (35–37).
The key strengths of our study are, in addition to the extremely large sample size (and resulting high statistical power), the very large set of SNPs (potentially associated with breast cancer susceptibility) considered, the uniform genotyping procedures and quality-control measures adopted and the large-scale analyses conducted to evaluate two-way interactions in a comprehensive way. A non-trivial issue for analyses of this kind is the establishment of a statistical significance threshold that adequately controls the proportion of false-positive findings. Since permutation-testing was not feasible, we dealt with the issue of non-independence of the multiple tests for interaction under our first approach by estimating the effective number of independent SNPs and used this to compute an effective number of independent interactions. No SNP pairs were robust to correction for multiple testing on this basis. However, further work is required to determine whether this approach gives a reasonable estimate for the effective number of interaction tests. The appropriateness of the Bonferroni correction in this context could also be questioned. For the second approach, we applied a statistical significance threshold of 10−10, which is almost an order of magnitude greater than the Bonferroni-corrected value based on the 2.5 billion interactions tested and can, therefore, be considered liberal.
Association analyses, including analyses of interactions, may be subject to confounding due to population structure. We aimed to correct for potential population stratification by adjusting for study and the six leading principal components. Little evidence of inflation of test statistics was observed after these adjustments in the main-effects analysis of the iCOGS data (3), suggesting that correction for population structure was adequate, and that any bias in the interaction tests was likely to be small. In any event, such confounding would be more likely to lead to false–positive associations, which we did not observe.
In conclusion, we observed little evidence of two-way SNP interactions for breast cancer susceptibility, despite the large number of SNPs (with potential marginal effects) considered and the very large sample size. More comprehensive, large-scale genome-wide interaction studies may identify novel interacting loci if the inherent logistic and computational challenges can be overcome.
METHODS
Study subjects
A total of 38 case–control studies contributed data and DNA samples for 46 450 breast cancer cases and 42 461 controls of white European origin from 13 European countries, Canada, Australia and the USA. Details are provided in Table 3 and Supplementary Material, Table S2. The ER status of the tumour was known for 34 479 cases, 27 074 were ER-positive and 7405 were ER-negative. All study participants gave informed consent and all studies were approved by the corresponding local research ethics committees.
Table 3.
Studies contributing white European cases and controls
| Study | Country | Controls | Cases | ER+ | ER− |
|---|---|---|---|---|---|
| Australian Breast Cancer Family Study (ABCFS)a | Australia | 551 | 790 | 456 | 261 |
| Amsterdam Breast Cancer Study (ABCS) | Netherlands | 1429 | 1325 | 420 | 153 |
| Bavarian Breast Cancer Cases and Controls (BBCC) | Germany | 458 | 564 | 460 | 83 |
| British Breast Cancer Study (BBCS) | UK | 1397 | 1554 | 507 | 114 |
| Breast Cancer In Galway Genetic Study (BIGGS) | Ireland | 719 | 836 | 495 | 154 |
| Breast Cancer Study of the University Clinic Heidelberg (BSUCH) | Germany | 954 | 852 | 499 | 154 |
| CECILE Breast Cancer Study (CECILE) | France | 999 | 1019 | 797 | 144 |
| Copenhagen General Population Study (CGPS) | Denmark | 4086 | 2901 | 1919 | 357 |
| Spanish National Cancer Centre Breast Cancer Study (CNIO-BCS) | Spain | 876 | 902 | 242 | 88 |
| California Teachers Study (CTS) | USA | 71 | 68 | 0 | 17 |
| ESTHER Breast Cancer Study (ESTHER) | Germany | 502 | 478 | 304 | 98 |
| Gene Environment Interaction and Breast Cancer in Germany (GENICA) | Germany | 427 | 465 | 328 | 119 |
| Helsinki Breast Cancer Study (HEBCS) | Finland | 1234 | 1664 | 1295 | 237 |
| Hannover-Minsk Breast Cancer Study (HMBCS) | Belarus | 130 | 690 | 37 | 0 |
| Karolinska Breast Cancer Study (KARBAC) | Sweden | 662 | 722 | 338 | 63 |
| Kuopio Breast Cancer Project (KBCP) | Finland | 251 | 445 | 304 | 97 |
| kConFab/Australian Ovarian Cancer Study (kConFab/AOCS) | Australia | 897 | 613 | 162 | 59 |
| Leuven Multidisciplinary Breast Centre (LMBC) | Belgium | 1388 | 2671 | 2071 | 379 |
| Mammary Carcinoma Risk Factor Investigation (MARIE) | Germany | 1778 | 1818 | 1349 | 399 |
| Milan Breast Cancer Study Group (MBCSG) | Italy | 400 | 488 | 149 | 42 |
| Mayo Clinic Breast Cancer Study (MCBCS) | USA | 1931 | 1862 | 1486 | 295 |
| Melbourne Collaborative Cohort Study (MCCS) | Australia | 511 | 614 | 352 | 119 |
| Multi-ethnic Cohort (MEC) | USA | 741 | 731 | 415 | 87 |
| Montreal Gene-Environment Breast Cancer Study (MTLGEBCS) | Canada | 436 | 489 | 421 | 64 |
| Norwegian Breast Cancer Study (NBCS) | Norway | 70 | 22 | 0 | 22 |
| Oulu Breast Cancer Study (OBCS) | Finland | 414 | 507 | 407 | 100 |
| Ontario Familial Breast Cancer Registry (OFBCR)b | Canada | 511 | 1175 | 630 | 268 |
| Leiden University Medical Center Breast Cancer Study (ORIGO) | Netherlands | 327 | 357 | 211 | 70 |
| NCI Polish Breast Cancer Study (PBCS) | Poland | 424 | 519 | 519 | 0 |
| Karolinska Mammography Project for Risk Prediction of Breast Cancer (pKARMA) | Sweden | 5537 | 5434 | 3672 | 702 |
| Rotterdam Breast Cancer Study (RBCS) | Netherlands | 699 | 664 | 368 | 131 |
| Singapore and Sweden Breast Cancer Study (SASBAC) | Sweden | 1378 | 1163 | 663 | 144 |
| Sheffield Breast Cancer Study (SBCS) | UK | 848 | 843 | 377 | 105 |
| Study of Epidemiology and Risk factors in Cancer Heredity (SEARCH) | UK | 8069 | 9347 | 5160 | 1181 |
| Städtisches Klinikum Karlsruhe Deutsches Krebsforschungszentrum Study (SKKDKFZS) | Germany | 29 | 136 | 0 | 136 |
| IHCC-Szczecin Breast Cancer Study (SZBCS) | Poland | 315 | 365 | 165 | 60 |
| Triple Negative Breast Cancer Consortium Study (TNBCC) | Various | 542 | 881 | 0 | 881 |
| UK Breakthrough Generations Study (UKBGS) | UK | 470 | 476 | 96 | 22 |
| Total | 42 461 | 46 450 | 27 074 | 7405 |
ER+, oestrogen-receptor-positive cases; ER−, oestrogen-receptor-negative cases.
aAustralian site of the Breast Cancer Family Registry.
bOntario site of the Breast Cancer Family Registry.
SNP selection and genotyping
SNP selection and genotyping methods have been described previously (3). The 75 380 SNPs eligible for inclusion in the present study were selected primarily based on statistical evidence of association with breast cancer risk from a combined analysis of nine GWAS involving 10 052 cases and 12 575 controls (46% SNPs) or from analyses of disease-subtype- or population-specific GWAS (24% SNPs). Additional SNPs were selected based on other evidence of association with breast (2% SNPs) or other cancers (4% SNPs), with overall survival following breast cancer (9% SNPs) or with risk factors for breast or other hormone-related cancers (12% SNPs) or because they tagged genes in the DNA repair pathway (2% SNPs).
Genotyping was carried out at four centres using a custom-built Illumina iSelect array (iCOGS) as part of a multi-consortium project (COGS), as described previously (3). Genotypes were called using Illumina's proprietary GenCall algorithm. Two percent of samples were provided in duplicate by all studies, and 270 HapMap2 samples were genotyped at all four centres. Subjects with an overall call-rate <95% were excluded. We excluded SNPs with call-rates <95% and those for which statistical evidence of deviation from Hardy–Weinberg equilibrium was observed for controls at a significance threshold of 10−7, based on a stratified 1 d.f. test in which the deviations were summed across study-based strata (38). We also excluded SNPs for which genotypes were discrepant for more than 2% of duplicate samples. A total of 72 611 of the eligible SNPs were successfully genotyped according to these criteria; 70 917 of those had a MAF of at least 0.01 in controls and were considered for the present analysis. The cluster plots of SNPs for which evidence of interaction was observed were individually re-evaluated and, where appropriate, manually recalled.
Statistical methods
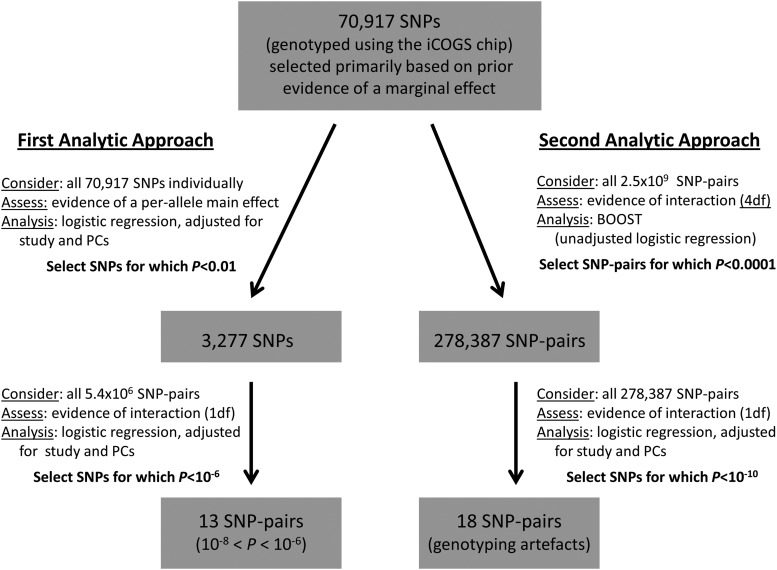
Evidence of interaction between SNP pairs in susceptibility to breast cancer was assessed using logistic regression, primarily based on a 1 d.f. likelihood ratio test (LRT) comparing two models: one model included main effects for study (categorical variable) and seven principal components (each continuous), a per-allele main effect for each of the two SNPs under consideration and an interaction parameter for the product of the latter two; the other model included main-effect parameters only. Thus, interaction was primarily considered to be per-allele departure from multiplicativity between ORs corresponding to SNP main effects. This was done for breast cancer risk overall, for risk of ER-positive breast cancer and for ER-negative breast cancer. Based on a series of pilot analyses, we estimated that, even with access to super-computing facilities with 1000 processors, an exhaustive analysis of all 2.5 billion (×109) two-way SNP interactions, while desirable (39), would take more than one year to complete. This was due to the number of SNP pairs to be tested, the large sample size and the inclusion of 37 dummy variables in the adjustment for study, the latter two factors increasing substantially the run-time per interaction.
We used two strategies to reduce the number of SNP pairs considered (Fig. 2). Under the first, we selected individual SNPs based on evidence of a marginal per-allele effect at P < 0.01. For each outcome (overall, ER-positive and ER-negative), SNPs were selected and then all possible two-way combinations of this reduced set of SNPs were considered in testing for evidence of interaction for that outcome. This strategy was founded on the idea that even a purely epistatic interaction (i.e. where the risk associated with one SNP is only manifest in individuals carrying a particular allele of another SNP) will give rise to apparent marginal SNP effects when the interaction is not modelled (40). Since the screening test is independent of the subsequent interaction test, substantial gain in power is obtained after correction for multiple (interaction) testing (41). Since permutation testing was not possible, given the sample size and number of interactions considered, we estimated instead the effective number of independent SNPs (VeffLi) using the method described by Li and Ji (42). This method was applied via the web-interface matSpDlite (http://gump.qimr.edu.au/general/daleN/matSpDlite/, last accessed on 20 November 2013), based on the observed correlations between SNPs, which was input as a matrix of correlation coefficients (43). The number of two-way interactions between VeffLi SNPs (T) was then used as a basis on which to calculate Bonferroni-corrected P-values (P*).
Figure 2.
Schematic representation of the two strategies applies to assess pairwise interactions in susceptibility to overall breast cancer risk between the 70 917 SNPs considered.
Under the second strategy, we applied BOOST (29) to screen all possible SNP pairs and select a reduced set to test formally using the LRT defined above. BOOST runs the equivalent of a simplified logistic-regression-based LRT in a highly efficient way, thereby permitting all 2.5 billion possible interactions to be screened in a reasonable time-frame. However, it is based on a co-dominant, genotype-based interaction model (4 d.f.) and does not allow adjustment for covariates. We used this strategy to select SNP pairs with the strongest evidence of interaction (P < 10−4) for more careful assessment using the less analytically efficient, but more adequate, adjusted logistic-regression models. The study- and principal-component-adjusted 4 d.f. interaction LRT test was then applied to the selected SNP pairs, as was the 1 d.f. test described above. Since BOOST requires complete data for all variants for all individuals, missing genotypes were imputed as the most common of the three genotypes for each SNP, across all subjects with available data. Imputed genotypes were not used in the final logistic-regression analysis.
Statistical power calculations were performed based on a range of MAF and OR using Quanto (http://hydra.usc.edu/gxe/, last accessed on 20 November 2013), and all other statistical analyses were carried out using R version 2.13.2. Q–Q plots were drawn for the first set of analyses based on the χ2 statistics from the 1 d.f. LRT using the qq.chisq function (44).
SUPPLEMENTARY MATERIAL
FUNDING
Funding for the iCOGS infrastructure came from the European Community's Seventh Framework Programme under grant agreement number 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692), the National Institutes of Health (CA128978) and Post-cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112—the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation and the Ovarian Cancer Research Fund. The ABCFS and OFBCR work was supported by grant UM1 CA164920 from the National Cancer Institute (USA). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government or the BCFR. The ABCFR was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow and M.C.S. is a NHMRC Senior Research Fellow. The OFBCR work was also supported by the Canadian Institutes of Health Research ‘CIHR Team in Familial Risks of Breast Cancer’ program. The ABCS was funded by Dutch Cancer Society Grant no.NKI2007-3839; M.K.S. was funded by Dutch Cancer Society Grant no. NKI2009-4363. The work of the BBCC was partly funded by ELAN-Programme of the University Hospital of Erlangen. The BBCS is funded by Cancer Research UK and Breakthrough Breast Cancer and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). The BCAC is funded by CR-UK (C1287/A10118 and C1287/A12014). Meetings of the BCAC have been funded by the European Union COST programme (BM0606). D.F.E. is a Principal Research Fellow of CR-UK. E.S. is supported by NIHR Comprehensive Biomedical Research Centre, Guy's & St. Thomas' NHS Foundation Trust in partnership with King's College London, UK. I.T. is supported by the Oxford Biomedical Research Centre. The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ). The CECILE study was funded by the Fondation de France, the French National Institute of Cancer (INCa), the National League against Cancer, the National Agency for Environmental and Occupational Health and Food Safety (ANSES), the National Agency for Research (ANR) and the Association for Research against Cancer (ARC). The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council and Herlev Hospital. The CNIO-BCS was supported by the Genome Spain Foundation, the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI081120). The Human Genotyping-CEGEN Unit, CNIO is supported by the Instituto de Salud Carlos III. The CTS was initially supported by the California Breast Cancer Act of 1993 and the California Breast Cancer Research Fund (contract 97-10500) and is currently funded through the National Institutes of Health (R01 CA77398). Collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885. H.A.C. receives support from the Lon V. Smith Foundation (LVS39420). The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany (grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114), the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, the Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, the Institute of the Ruhr University Bochum (IPA) and the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. The HEBCS was supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (132473), the Finnish Cancer Society, The Nordic Cancer Union and the Sigrid Juselius Foundation. The HMBCS was supported by short-term fellowships from the German Academic Exchange Program [to N.B], and the Friends of Hannover Medical School [to N.B.]. Financial support for KARBAC was provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, the Swedish Cancer Society, The Gustav V Jubilee foundatin and Bert von Kantzows foundation. The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, the Academy of Finland and by the strategic funding of the University of Eastern Finland. kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC, the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia and the Cancer Foundation of Western Australia. The kConFab Clinical Follow Up Study was funded by the NHMRC [145684, 288704, 454508]. Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command [DAMD17-01-1-0729], the Cancer Council of Tasmania and Cancer Foundation of Western Australia and the NHMRC [199600]. G.C.T. and P.W. are supported by the NHMRC. LMBC is supported by the ‘Stichting tegen Kanker’ (232-2008 and 196-2010). The MARIE study was supported by the Deutsche Krebshilfe e.V. [70-2892-BR I], the Hamburg Cancer Society, the German Cancer Research Center and the genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. MBCSG is supported by grants from the Italian Association for Cancer Research (AIRC) and by funds from the Italian citizens who allocated a 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects ‘5 × 1000’). The MCBCS was supported by the NIH grants [CA122340, CA128978] and a Specialized Program of Research Excellence (SPORE) in Breast Cancer [CA116201], the Breast Cancer Research Foundation and a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation. MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. The MEC was supported by NIH grants CA63464, CA54281, CA098758 and CA132839. The work of MTLGEBCS was supported by the Quebec Breast Cancer Foundation and the Ministry of Economic Development, Innovation and Export Trade (grant PSR-SIIRI-701). The NBCS was supported by grants from the Norwegian Research Council (155218/V40, 175240/S10 to A.L.B.D., FUGE-NFR 181600/V11 to V.N.K. and a Swizz Bridge Award to A.L.B.D.). The OBCS was supported by research grants from the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the Academy of Finland, the University of Oulu and the Oulu University Hospital. The ORIGO study was supported by the Dutch Cancer Society (RUL 1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. pKARMA is a combination of the KARMA and LIBRO-1 studies. KARMA was supported by Märit and Hans Rausings Initiative Against Breast Cancer. KARMA and LIBRO-1 were supported the Cancer Risk Prediction Center (CRisP; www.crispcenter.org), a Linnaeus Centre (Contract ID 70867902) financed by the Swedish Research Council. The RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124, DDHK 2009-4318). SASBAC was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH) and the Susan G. Komen Breast Cancer Foundation. K.C. was financed by the Swedish Cancer Society (5128-B07-01PAF). The SBCS was supported by Yorkshire Cancer Research S305PA. SEARCH is funded by a programme grant from Cancer Research UK [C490/A10124] and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. SKKDKFZS is supported by the DKFZ. Katarzyna Jaworska is a fellow of International PhD program, Postgraduate School of Molecular Medicine, Warsaw Medical University, supported by the Polish Foundation of Science. The TNBCC was supported by the NIH grant CA128978, the Breast Cancer Research Foundation, Komen Foundation for the Cure, the Ohio State University Comprehensive Cancer Center, the Stefanie Spielman fund for Breast Cancer Research and a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation. Part of the TNBCC (DEMOKRITOS) has been co-financed by the European Union (European Social Fund—ESF) and Greek national funds through the Operational Program ‘Education and Lifelong Learning’ of the National Strategic Reference Framework (NSRF)—Research Funding Program of the General Secretariat for Research & Technology: ARISTEIA. The UKBGS is funded by Breakthrough Breast Cancer and the Institute of Cancer Research (ICR). ICR acknowledges NHS funding to the NIHR Biomedical Research Centre.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the individuals who took part in these studies and all the researchers, study staff, clinicians and other healthcare providers, technicians and administrative staff who have enabled this work to be carried out. In particular, we thank Michael Lush, Maggie Angelakos, Judi Maskiell, Richard van Hien, Sten Cornelissen, Ellen van der Schoot, Alexander Hein, Michael Schrauder, Matthias Rübner, Sonja Oeser, Silke Landrith, Eileen Williams, Elaine Ryder-Mills, Kara Sargus, Niall McInerney, Gabrielle Colleran, Andrew Rowan, Angela Jones, Christof Sohn, Andeas Schneeweiß, Peter Bugert, the Danish Breast Cancer Group, Nuria Álvarez, Daniel Herrero, Primitiva Menendez, the CTS Steering Committee (including Leslie Bernstein, James Lacey, Sophia Wang, Huiyan Ma, Yani Lu and Jessica Clague DeHart at the Beckman Research Institute of the City of Hope; Dennis Deapen, Rich Pinder, Eunjung Lee and Fred Schumacher at the University of Southern California; Pam Horn-Ross, Peggy Reynolds and David Nelson at the Cancer Prevention Institute of California; and Hannah Park at the University of California Irvine), Hartwig Ziegler, Sonja Wolf, Volker Hermann, The GENICA network: Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany [H.B., Wing-Yee Lo, Christina Justenhoven], Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [Y.D.K., Christian Baisch], Institute of Pathology, University of Bonn, Bonn, Germany [Hans-Peter Fischer], Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum [DKFZ] Heidelberg, Germany [U.H.], Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Germany [T.B., Beate Pesch, Sylvia Rabstein, Anne Lotz], Institute for Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany [Volker Harth], Tuomas Heikkinen, Irja Erkkilä, Kirsimari Aaltonen, Karl von Smitten, Natalia Antonenkova, Peter Hillemanns, Hans Christiansen, Eija Myöhänen, Helena Kemiläinen, Heather Thorne, Eveline Niedermayr, the AOCS Management Group (D. Bowtell, G. Chenevix-Trench, A. deFazio, D. Gertig, A. Green and P. Webb), the ACS Management Group (A. Green, P. Parsons, N. Hayward, P. Webb and D. Whiteman), Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel, Kathleen Corthouts, Tracy Slanger, Elke Mutschelknauss, Ramona Salazar, S. Behrens, R. Birr, W. Busch, U. Eilber, B. Kaspereit, N. Knese, K. Smit, Siranoush Manoukian, Daniela Zaffaroni, Monica Barile, Irene Feroce, Bernardo Bonanni, the Cogentech Cancer Genetic Test Laboratory, The Mayo Clinic Breast Cancer Patient Registry, Mark Goldberg, France Labrèche, Martine Tranchant, Marie-France Valois, Annie Turgeon, Lea Heguy, Meeri Otsukka, Kari Mononen, Teresa Selander, Nayana Weerasooriya, OFBCR staff, E. Krol-Warmerdam, J. Molenaar, J. Blom, Louise Brinton, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner, Petra Bos, Jannet Blom, Ellen Crepin, Anja Nieuwlaat, Annette Heemskerk, the Erasmus MC Family Cancer Clinic, Sue Higham, Simon Cross, Helen Cramp, Dan Connley, the Ohio State University Human Genetics Sample Bank, Robert Pilarski and the SEARCH and EPIC teams.
This study would not have been possible without the contributions of Andrew Berchuck (OCAC); Rosalind A. Eeles, Ali Amin Al Olama, Zsofia Kote-Jarai and Sara Benlloch (PRACTICAL); Antonis Antoniou, Lesley McGuffog and Ken Offit (CIMBA); Andrew Lee, Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, Cambridge; the staff of the CNIO genotyping unit; Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière, Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre; Sune F. Nielsen and the staff of the Copenhagen DNA laboratory; Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility.
Conflict of Interest statement. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
REFERENCES
- 1.Bojesen S.E., Pooley K.A., Johnatty S.E., Beesley J., Michailidou K., Tyrer J.P., Edwards S.L., Pickett H.A., Shen H.C., Smart C.E., et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 2013;45:371–384. doi: 10.1038/ng.2566. doi:10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Closas M., Couch F.J., Lindstrom S., Michailidou K., Schmidt M.K., Brook M.N., Orr N., Rhie S.K., Riboli E., Feigelson H.S., et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat. Genet. 2013;45:392–398. doi: 10.1038/ng.2561. doi:10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L., Schmidt M.K., Chang-Claude J., Bojesen S.E., Bolla M.K., et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–361. doi: 10.1038/ng.2563. doi:10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox A., Dunning A.M., Garcia-Closas M., Balasubramanian S., Reed M.W., Pooley K.A., Scollen S., Baynes C., Ponder B.A., Chanock S., et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 2007;39:352–358. doi: 10.1038/ng1981. doi:10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 5.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. doi:10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas G., Jacobs K.B., Kraft P., Yeager M., Wacholder S., Cox D.G., Hankinson S.E., Hutchinson A., Wang Z., Yu K., et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. doi:10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed S., Thomas G., Ghoussaini M., Healey C.S., Humphreys M.K., Platte R., Morrison J., Maranian M., Pooley K.A., Luben R., et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. doi:10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milne R.L., Benitez J., Nevanlinna H., Heikkinen T., Aittomaki K., Blomqvist C., Arias J.I., Zamora M.P., Burwinkel B., Bartram C.R., et al. Risk of estrogen receptor-positive and -negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. J. Natl. Cancer Inst. 2009;101:1012–1018. doi: 10.1093/jnci/djp167. doi:10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stacey S.N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S.A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A., et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. doi:10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 11.Stacey S.N., Manolescu A., Sulem P., Thorlacius S., Gudjonsson S.A., Jonsson G.F., Jakobsdottir M., Bergthorsson J.T., Gudmundsson J., Aben K.K., et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. doi:10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 12.Milne R.L., Goode E.L., Garcia-Closas M., Couch F.J., Severi G., Hein R., Fredericksen Z., Malats N., Zamora M.P., Arias Perez J.I., et al. Confirmation of 5p12 as a susceptibility locus for progesterone-receptor-positive, lower grade breast cancer. Cancer Epidemiol. Biomarkers Prev. 2011;20:2222–2231. doi: 10.1158/1055-9965.EPI-11-0569. doi:10.1158/1055-9965.EPI-11-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L., et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. doi:10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antoniou A.C., Wang X., Fredericksen Z.S., McGuffog L., Tarrell R., Sinilnikova O.M., Healey S., Morrison J., Kartsonaki C., Lesnick T., et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet. 2010;42:885–892. doi: 10.1038/ng.669. doi:10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Q., Long J., Lu W., Qu S., Wen W., Kang D., Lee J.Y., Chen K., Shen H., Shen C.Y., et al. Genome-wide association study identifies breast cancer risk variant at 10q21.2: results from the Asia Breast Cancer Consortium. Hum. Mol. Genet. 2011;20:4991–4999. doi: 10.1093/hmg/ddr405. doi:10.1093/hmg/ddr405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnbull C., Ahmed S., Morrison J., Pernet D., Renwick A., Maranian M., Seal S., Ghoussaini M., Hines S., Healey C.S., et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. doi:10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher O., Johnson N., Orr N., Hosking F.J., Gibson L.J., Walker K., Zelenika D., Gut I., Heath S., Palles C., et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J. Natl. Cancer Inst. 2011;103:425–435. doi: 10.1093/jnci/djq563. doi:10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 18.Haiman C.A., Chen G.K., Vachon C.M., Canzian F., Dunning A., Millikan R.C., Wang X., Ademuyiwa F., Ahmed S., Ambrosone C.B., et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat. Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. doi:10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghoussaini M., Fletcher O., Michailidou K., Turnbull C., Schmidt M.K., Dicks E., Dennis J., Wang Q., Humphreys M.K., Luccarini C., et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat. Genet. 2012;44:312–318. doi: 10.1038/ng.1049. doi:10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddiq A., Couch F.J., Chen G.K., Lindstrom S., Eccles D., Millikan R.C., Michailidou K., Stram D.O., Beckmann L., Rhie S.K., et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet. 2012;21:5373–5384. doi: 10.1093/hmg/dds381. doi:10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gracia-Aznarez F.J., Fernandez V., Pita G., Peterlongo P., Dominguez O., de la Hoya M., Duran M., Osorio A., Moreno L., Gonzalez-Neira A., et al. Whole exome sequencing suggests much of non-BRCA1/BRCA2 familial breast cancer is due to moderate and low penetrance susceptibility alleles. PLoS ONE. 2013;8:e55681. doi: 10.1371/journal.pone.0055681. doi:10.1371/journal.pone.0055681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mavaddat N., Antoniou A.C., Easton D.F., Garcia-Closas M. Genetic susceptibility to breast cancer. Mol. Oncol. 2010;4:174–191. doi: 10.1016/j.molonc.2010.04.011. doi:10.1016/j.molonc.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park D.J., Lesueur F., Nguyen-Dumont T., Pertesi M., Odefrey F., Hammet F., Neuhausen S.L., John E.M., Andrulis I.L., Terry M.B., et al. Rare mutations in XRCC2 increase the risk of breast cancer. Am. J. Hum. Genet. 2012;90:734–739. doi: 10.1016/j.ajhg.2012.02.027. doi:10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith P., McGuffog L., Easton D.F., Mann G.J., Pupo G.M., Newman B., Chenevix-Trench G., Szabo C., Southey M., Renard H., et al. A genome wide linkage search for breast cancer susceptibility genes. Genes Chromosomes Cancer. 2006;45:646–655. doi: 10.1002/gcc.20330. doi:10.1002/gcc.20354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson E.R., Doyle M.A., Ryland G.L., Rowley S.M., Choong D.Y., Tothill R.W., Thorne H., Barnes D.R., Li J., Ellul J., et al. Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genet. 2012;8:e1002894. doi: 10.1371/journal.pgen.1002894. doi:10.1371/journal.pgen.1002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuk O., Hechter E., Sunyaev S.R., Lander E.S. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. doi:10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aschard H., Chen J., Cornelis M.C., Chibnik L.B., Karlson E.W., Kraft P. Inclusion of gene-gene and gene-environment interactions unlikely to dramatically improve risk prediction for complex diseases. Am. J. Hum. Genet. 2012;90:962–972. doi: 10.1016/j.ajhg.2012.04.017. doi:10.1016/j.ajhg.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao S., Feng J., Webster T., Jin G., Hsu F.C., Chen S.H., Kim S.T., Wang Z., Zhang Z., Zheng S.L., et al. Genome-wide two-locus epistasis scans in prostate cancer using two European populations. Hum. Genet. 2012;131:1225–1234. doi: 10.1007/s00439-012-1148-4. doi:10.1007/s00439-012-1148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan X., Yang C., Yang Q., Xue H., Fan X., Tang N.L., Yu W. BOOST: a fast approach to detecting gene-gene interactions in genome-wide case-control studies. Am. J. Hum. Genet. 2010;87:325–340. doi: 10.1016/j.ajhg.2010.07.021. doi:10.1016/j.ajhg.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milne R.L., Gaudet M.M., Spurdle A.B., Fasching P.A., Couch F.J., Benitez J., Arias Perez J.I., Zamora M.P., Malats N., Dos Santos Silva I., et al. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case-control study. Breast Cancer Res. 2010;12:R110. doi: 10.1186/bcr2797. doi:10.1186/bcr2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickels S., Truong T., Hein R., Stevens K., Buck K., Behrens S., Eilber U., Schmidt M., Haberle L., Vrieling A., et al. Evidence of gene-environment interactions between common breast cancer susceptibility loci and established environmental risk factors. PLoS Genet. 2013;9:e1003284. doi: 10.1371/journal.pgen.1003284. doi:10.1371/journal.pgen.1003284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campa D., Kaaks R., Le Marchand L., Haiman C.A., Travis R.C., Berg C.D., Buring J.E., Chanock S.J., Diver W.R., Dostal L., et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J. Natl. Cancer Inst. 2011;103:1252–1263. doi: 10.1093/jnci/djr265. doi:10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Travis R.C., Reeves G.K., Green J., Bull D., Tipper S.J., Baker K., Beral V., Peto R., Bell J., Zelenika D., et al. Gene-environment interactions in 7610 women with breast cancer: prospective evidence from the Million Women Study. Lancet. 2010;375:2143–2151. doi: 10.1016/S0140-6736(10)60636-8. doi:10.1016/S0140-6736(10)60636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat. Rev. Genet. 2013;14:168–178. doi: 10.1038/nrg3404. doi:10.1038/nrg3404. [DOI] [PubMed] [Google Scholar]
- 35.Kirino Y., Bertsias G., Ishigatsubo Y., Mizuki N., Tugal-Tutkun I., Seyahi E., Ozyazgan Y., Sacli F.S., Erer B., Inoko H., et al. Genome-wide association analysis identifies new susceptibility loci for Behcet's disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 2013;45:202–207. doi: 10.1038/ng.2520. doi:10.1038/ng.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans D.M., Spencer C.C., Pointon J.J., Su Z., Harvey D., Kochan G., Oppermann U., Dilthey A., Pirinen M., Stone M.A., et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 2011;43:761–767. doi: 10.1038/ng.873. doi:10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strange A., Capon F., Spencer C.C., Knight J., Weale M.E., Allen M.H., Barton A., Band G., Bellenguez C., Bergboer J.G., et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 2010;42:985–990. doi: 10.1038/ng.694. doi:10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haldane J.B.S. An exact test for randomness of mating. J. Genet. 1954;52:631–635. [Google Scholar]
- 39.Evans D.M., Marchini J., Morris A.P., Cardon L.R. Two-stage two-locus models in genome-wide association. PLoS Genet. 2006;2:e157. doi: 10.1371/journal.pgen.0020157. doi:10.1371/journal.pgen.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Millstein J., Conti D.V., Gilliland F.D., Gauderman W.J. A testing framework for identifying susceptibility genes in the presence of epistasis. Am. J. Hum. Genet. 2006;78:15–27. doi: 10.1086/498850. doi:10.1086/498850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kooperberg C., Leblanc M. Increasing the power of identifying gene x gene interactions in genome-wide association studies. Genet. Epidemiol. 2008;32:255–263. doi: 10.1002/gepi.20300. doi:10.1002/gepi.20300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J., Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. doi:10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 43.Nyholt D.R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 2004;74:765–769. doi: 10.1086/383251. doi:10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. doi:10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.