Abstract
DNA methylation contributes to tumor formation, development and metastasis. Epigenetic dysregulation of stem cells is thought to predispose to malignant development. The clinical significance of DNA methylation in ovarian tumor-initiating cells (OTICs) remains unexplored. We analyzed the methylomic profiles of OTICs (CP70sps) and their derived progeny using a human methylation array. qRT–PCR, quantitative methylation-specific PCR (qMSP) and pyrosequencing were used to verify gene expression and DNA methylation in cancer cell lines. The methylation status of genes was validated quantitatively in cancer tissues and correlated with clinicopathological factors. ATG4A and HIST1H2BN were hypomethylated in OTICs. Methylation analysis of ATG4A and HIST1H2BN by qMSP in 168 tissue samples from patients with ovarian cancer showed that HIST1H2BN methylation was a significant and independent predictor of progression-free survival (PFS) and overall survival (OS). Multivariate Cox regression analysis showed that patients with a low level of HIST1H2BN methylation had poor PFS (hazard ratio (HR), 4.5; 95% confidence interval (CI), 1.4–14.8) and OS (HR, 4.3; 95% CI, 1.3–14.0). Hypomethylation of both ATG4A and HIST1H2BN predicted a poor PFS (HR, 1.8; 95% CI, 1.0–3.6; median, 21 months) and OS (HR, 1.7; 95% CI, 1.0–3.0; median, 40 months). In an independent cohort of ovarian tumors, hypomethylation predicted early disease recurrence (HR, 1.7; 95% CI, 1.1–2.5) and death (HR, 1.4; 95% CI, 1.0–1.9). The demonstration that expression of ATG4A in cells increased their stem properties provided an indication of its biological function. Hypomethylation of ATG4A and HIST1H2BN in OTICs predicts a poor prognosis for ovarian cancer patients.
INTRODUCTION
Ovarian cancer is the most deadly malignancy of the female reproductive system (1). Epithelial ovarian cancer (EOC) represents one of the most common causes of death in women worldwide. The etiology is unclear, and the poor outcome has persisted for decades. Ovarian tumors, especially high-grade serous tumors, often produce vague symptoms, and the disease is often diagnosed in advanced stages (2,3). After cytoreductive surgery, although treatment with platinum or taxane produces a complete responses in 70% of patients, most will relapse and experience cisplatin-resistant disease within 18 months, especially in advanced-stage patients (4). Clinicopathological variables such as the patient's age and tumor histology, stage and grade cannot predict clinical outcomes accurately, nor do they provide biological insight into the clinical progression of tumors (3,5). Individualized cancer treatment is becoming more important in personalized medicine. Using molecular markers to stratify patients may help to identify subgroups of patients who will be responsive to current treatments and may help to avoid unnecessary side effects in patients with predicted poor responses.
Epigenetic changes play an important role in the development of cancer, which can be considered to be a developmental disorder. Of these epigenetic changes, DNA methylation has been studied intensively. Compared with the normal genome, cancer genomes are globally hypomethylated and hypermethylated or hypomethylated at specific sequences usually in enhancer or promoter regions. Aberrant DNA methylation is a common epigenetic event leading to the inactivation of tumor suppressor genes (TSG) in ovarian cancer (6,7). Several studies have indicated that the methylation of specific genes may serve as a biomarker for diagnostic screening and prognostic prediction for ovarian cancer (8,9). Epigenetic silencing by promoter hypermethylation of TSG that repress the expression in various cancer cells, such as ARLTS1 (10), DLEC1 (11), OPCML (12,13), RASSF1A (13,14) and TCEAL7 (15), is commonly observed in ovarian cancers. Silencing of the development-associated transcription factors HOXA10 and HOXA11 (16) is associated with ovarian cancer initiation and progression to chemotherapy resistance (17). In addition, hypermethylation of many genomic regions associated with transcriptional silencing has also been shown in different histological subtypes of ovarian cancer (18). DNA methylation might serve as a marker for the early diagnosis of cancer and as a means of assessing the prognosis of cancer patients.
One emerging model for the development of drug-resistant tumors is a pool of self-renewing malignant progenitors known as tumor-initiating cells (TICs) or cancer stem cells. TICs have the ability to resist chemoradiotherapy and give rise to cancer metastasis and recurrence (19). In theory, TICs are at the top of the hierarchy of cell types in distinct stages of differentiation (20). Epigenetic mechanisms are key components of the dynamic regulation of embryonic stem cell differentiation. Methylation changes of embryonic stem cell loci have been reported to hold prognostic potential in various cancers (21). However, which epigenetic stem cell features are retained or changed in human cancers during carcinogenesis is unclear, and there are few reports on epigenetic dynamics in cancer stem cells and their clinical relevance.
In this study, we hypothesized that epigenetic modulation of ovarian tumor-initiating cells (OTICs) occurs during the repopulation of cancer cells. We aimed to identify changes in DNA methylation during this process and to test their potential as prognostic biomarkers for clinical applications.
RESULTS
DNA methylation-based marker discovery pipeline
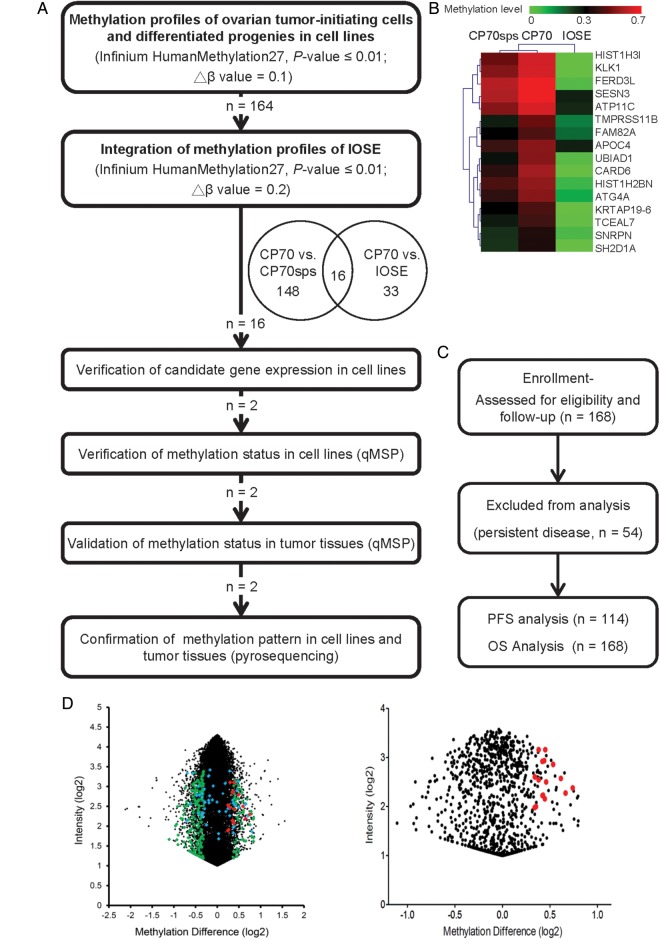
Because TICs are associated with a poor prognosis, we hypothesized that some genes related to stemness should be unmethylated in these cells and would become methylated during repopulation. Our aim was to identify some genes with low methylation and high mRNA expression in TICs and with high methylation and low expression levels in repopulated cancer cells. We devised a comprehensive and systematic strategy to identify DNA methylation markers for EOC that are present in cancer tissues but not in TICs. The logistics of the study are summarized in Figure 1A. The genes that exhibited global differences between OTICs (CP70sps) and differentiated cancer cells (CP70) are shown in Figure 1D (left). The probes of the methylation array that failed (detection P-value > 0.01) in any sample were omitted from the analysis. There were 164 genes that were hypermethylated in the differentiated cancer cells (CP70) compared with the OTICs (CP70sps). To narrow the candidate gene list, we integrated our results with the methylomic profiling of an immortalized ovarian surface epithelial (IOSE) cell line that mimics the normal ovarian epithelium. The distribution of differential methylation probes is shown in Figure 1D. Forty-nine genes were hypermethylated in the cancer cell line compared with IOSE cells. Sixteen genes were commonly hypermethylated in the CP70 cell line but not in the CP70sps and IOSE cell lines. The methylomic profiles of these 16 genes were clustered hierarchically, as shown in Figure 1B.
Figure 1.
Flow chart of the present study. (A) The HumanMethylation27 BeadChip used to interrogate simultaneously the DNA methylation status of 27 578 probes spanning 14 489 unique genetic loci, which were used for methylomic profiling of OTICs (CP70sps), cells with differentiated progenies (CP70) and IOSE cells. qMSP was performed in a TaqMan probe system using the LightCycler 480 Real-Time PCR System. (B) Hierarchical clustering of methylomic profiles of CP70, CP70sps and IOSE cells (β-value 0–0.7, green and red color represents low and high methylation, respectively). (C) CONSORT diagram of the analytical strategy in all ovarian patients. (D) Selection of differential methylation probes in CP70 and CP70sps from a methylation microarray. The global differential methylation level and intensity in each probe between CP70 and CP70sps was displayed (left). After excluding probes that have a detection P > 0.01 in the assay, 973 probes show as black dots (right). Two hundred and five probes show as green dots that had at least 10% of differential methylation between CP70 and CP70sps cells. Forty-nine genes had at least 20% of differential methylation between CP70 and IOSE cells (blue dots). Sixteen genes exhibited significant differences between the two groups that were compared (red dots).
Verification of methylation status and mRNA expression of candidate genes in cell lines
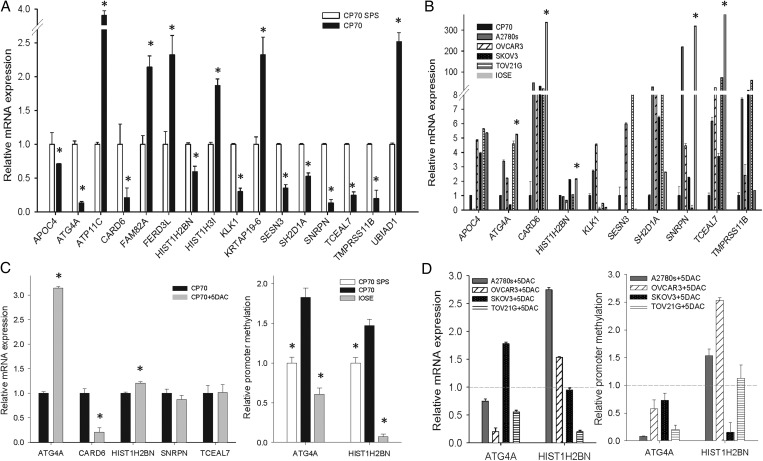
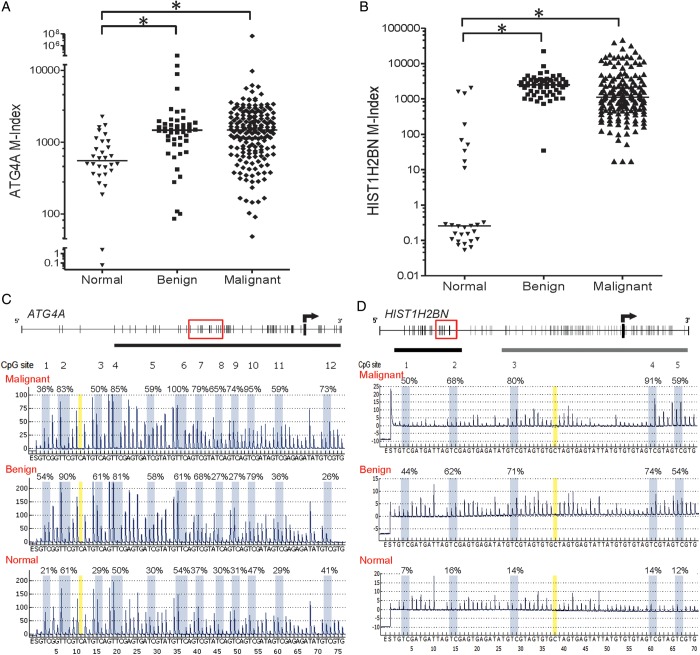
We used qRT–PCR to verify the mRNA expression of these 16 genes in CP70 and CP70sps cells (Fig. 2A). Genes with DNA hypermethylation in CP70 should have lower mRNA expression compared with CP70sps. Ten genes fulfilled this assumption. To generalize this finding, we tested the expression of these 10 genes in cancer cell lines (Fig. 2B). Genes with DNA methylation in cancer cells should have lower expression compared with the normal ovarian epithelium. Five out of the 10 genes comply with our hypothesis and were selected for further analysis. Demethylation treatment using 5-aza-2′-deoxycytidine in CP70 cells was used to verify the relevance of DNA methylation and gene expression of these five genes. ATG4A and HIST1H2BN fulfilled this criterion. Quantitative methylation analysis confirmed the higher methylation level of ATG4A and HIST1H2BN in cancer cells compared with TICs and the normal ovarian epithelium (Fig. 2C). The promoter demethylation and gene reexpression of these two genes after 5-aza-2′-deoxycytidine treatment were also verified in other ovarian cancer cell lines (Fig. 2D). The methylation status of ATG4A and HIST1H2BN was validated further in clinical samples including normal ovarian tissues and benign and malignant tumor tissues using quantitative methylation-specific PCR (qMSP) (Fig. 3A and B) and pyrosequencing (Fig. 3C and D). Both benign and malignant tumors had significantly higher methylation levels than did normal ovarian tissues.
Figure 2.
Verification of mRNA expression and DNA methylation of candidate genes in cell lines. (A) mRNA expression of candidate genes in cancer cells (CP70) relative to TICs (CP70sps). (B) mRNA expression of candidate genes in various ovarian cell lines. DNA methylation of ATG4A, CARD6, HIST1H2BN, SNRPN and TCEAL7 in cancer cells have lower expression than the normal ovarian epithelium. (C) Reexpression of candidate genes in CP70 treated with 5-aza-2′-deoxycytidine (left). Methylation level of candidate genes in CP70, CP70sps and IOSE cells (right). (D) mRNA reexpression of candidate genes in ovarian cancer cell lines treated with 5-aza-2′-deoxycytidine. The y-axis indicates the change in mRNA expression (left). DNA demethylation of candidate genes in ovarian cancer cell lines treated with 5-aza-2′-deoxycytidine. The y-axis indicates the change in promoter methylation (right). The dotted line represents the mRNA and DNA methylation status of the original cell as a reference. *P < 0.05.
Figure 3.
The promoter methylation status of ATG4A and HIST1H2BN in ovarian tissues. (A) ATG4A, (B) HIST1H2BN determined by qMSP in ovarian tissues. The M-index indicates the DNA methylation level estimated using the formula: 10 000 × 2[(Cp of COL2A) – (Cp of gene)]. (C) ATG4A, (D) HIST1H2BN representative methylation profile determined by pyrosequencing. The C base at nucleotide numbers 11 and 38 (marked in yellow) is a negative control for the assurance of complete bisulfite conversion. *P < 0.05.
DNA methylation of ATG4A and HIST1H2BN as a prognostic factor
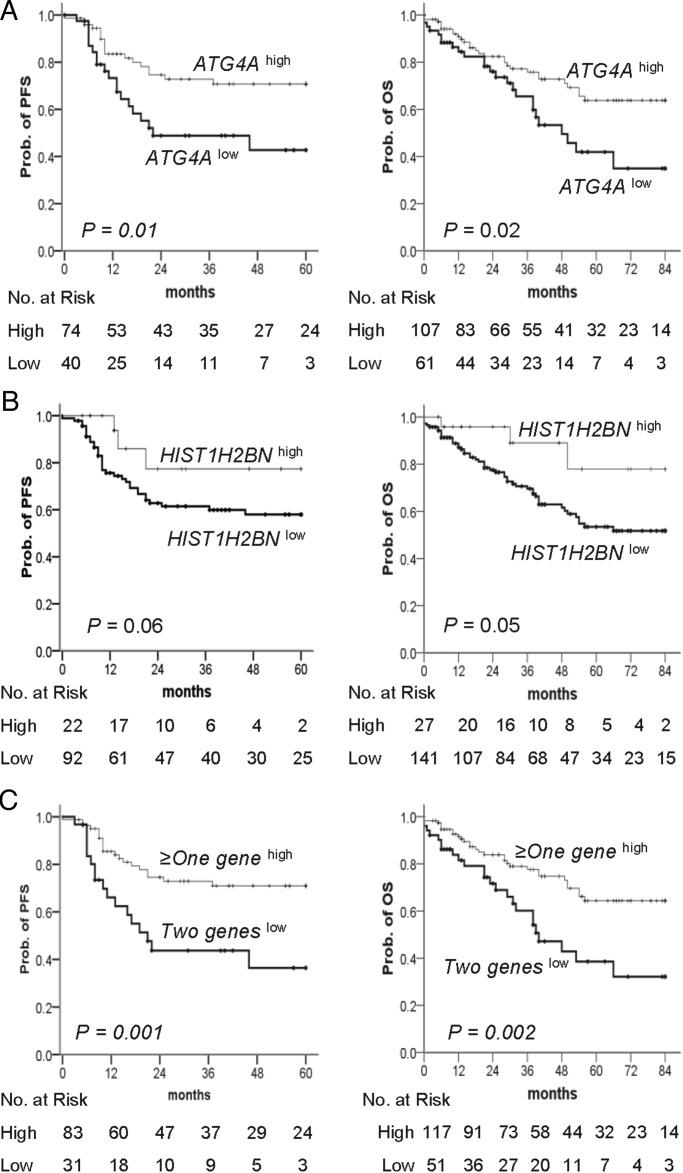
To evaluate the levels of DNA methylation as a prognostic biomarker, we prepared a CONSORT diagram including all EOC patients in the quantitative DNA methylation analysis (Fig. 1C). The methylation status of ATG4A correlated significantly with the clinical stage (P = 0.002). The methylation status of HIST1H2BN did not correlate with age, clinical stage, histological grade or subtype; however, the cisplatin resistance was significantly associated with low methylation of HIST1H2BN (P = 0.01). Forty-nine of the 61 patients (80.3%) with low methylation of ATG4A had advanced-stage disease; this percentage was significantly >57.0% (61/107) in patients with high methylation levels (Table 1).
Table 1.
Patient characteristics and clinicopathological features by ATG4A and HIST1H2BN methylation status
| ATG4A |
HIST1H2BN |
ATG4A/HIST1H2BN | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Methylation status | High (N = 107; 63.7%) | Low (N = 61; 36.3%) | P-value | High (N = 27; 16.1%) | Low (N = 141; 83.9%) | P-value | High (N = 117; 69.6%) | Low (N = 51; 30.4%) | P-value |
| Characteristics | |||||||||
| Age (years) | |||||||||
| Mean, range | 54.0 (19–90) | 54.3 (18–85) | 0.90 | 57.2 (39–79) | 53.5 (18–90) | 0.20 | 54.9 (19–90) | 52.2 (18–85) | 0.23 |
| FIGO stage | |||||||||
| Early (I, II) | 46 (43.0) | 12 (19.7) | 0.002* | 11 (40.7) | 47 (33.3) | 0.46 | 49 (41.9) | 9 (17.6) | 0.002* |
| Late (III, IV) | 61 (57.0) | 49 (80.3) | 16 (59.3) | 94 (66.7) | 68 (58.1) | 42 (82.4) | |||
| Grade | |||||||||
| G1/G2 | 50 (46.7) | 21 (34.4) | 0.12 | 13 (48.1) | 58 (41.1) | 0.50 | 54 (46.2) | 17 (33.3) | 0.12 |
| G3 | 57 (53.3) | 40 (65.6) | 14 (51.9) | 83 (58.9) | 63 (53.8) | 34 (66.7) | |||
| Histology | |||||||||
| Serous type | 71 (66.4) | 44 (72.1) | 0.44 | 18 (66.7) | 97 (68.8) | 0.83 | 79 (67.5) | 36 (70.6) | 0.69 |
| Other types | 36 (33.6) | 17 (27.9) | 9 (33.3) | 44 (31.2) | 38 (32.5) | 15 (29.4) | |||
| Platinum response | |||||||||
| Sensitive | 75 (70.1) | 40 (65.6) | 0.54 | 24 (88.9) | 91 (64.5) | 0.01* | 85 (72.6) | 30 (58.8) | 0.08 |
| Resistant | 32 (29.9) | 21 (34.4) | 3 (11.1) | 50 (35.5) | 32 (27.4) | 21 (41.2) | |||
*Significantly correlated with outcome, P < 0.05.
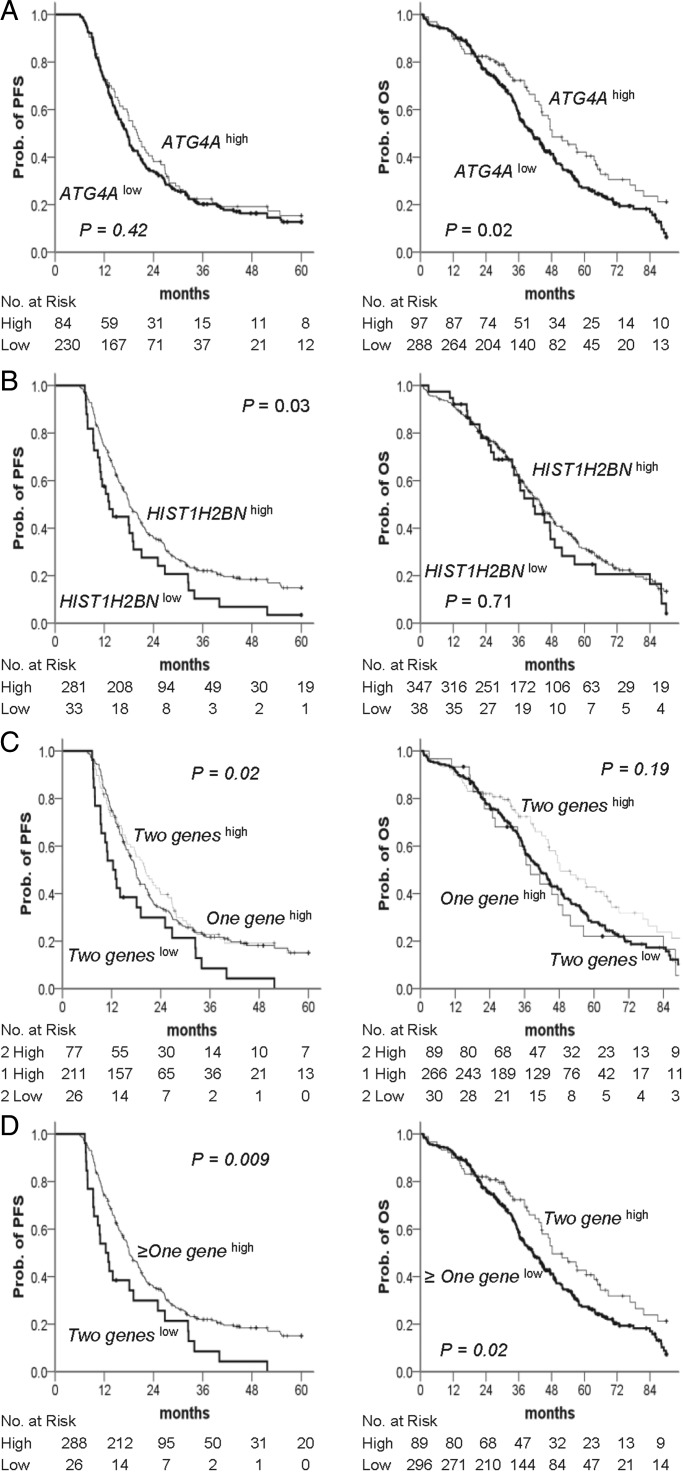
The prognostic significance of the DNA methylation identified was tested. The results of the univariate Cox regression analysis for progression-free survival (PFS) and OS are presented in the Supplementary Material, Table S2. As expected, FIGO stage and grade were associated with PFS and OS. Low methylation of ATG4A was significantly associated with poor PFS (hazard ratio (HR), 2.3; 95% confidence interval (CI), 1.2–4.4) and OS (HR, 1.9; 95% CI, 1.1–3.3). The prognosis of patients with low methylation of HIST1H2BN was associated with worse OS (HR, 3.1; 95% CI, 1.0–9.7). The Kaplan–Meier analysis for the PFS and OS of cancer patients revealed that, compared with patients with high methylation of ATG4A or HIST1H2BN, patients with low methylation had a significantly shorter PFS (Fig. 4A left and B left; P = 0.01 and 0.06, respectively) and were more likely to die (Fig. 4A right and B right; P = 0.02 and 0.05, respectively) within the follow-up period. There was a good discrimination of PFS and OS between low and high methylation status of combined ATG4A and HIST1H2BN in cancer patients (Fig. 4C; P < 0.01). In the multivariate Cox proportional-hazards regression analysis, after adjusting for the related factors, methylation of HIST1H2BN was an independent predictor of PFS and OS. Patients with low methylation of HIST1H2BN had an HR of 4.5 (95% CI, 1.4–14.8) for PFS and 4.3 (95% CI, 1.3–14.0) for OS (Table 2). Although the low methylation of ATG4A was a significant predictor of recurrence and death in the univariate analysis, this effect was no longer evident in the multivariate analysis. However, the adjusted HRs for PFS and OS in the low methylation status of both ATG4A and HIST1H2BN versus the more than one genes high methylation patients were 1.8 (95% CI, 1.0–3.6) and 1.7 (95% CI, 1.0–3.0), respectively (Table 2).
Figure 4.
Kaplan–Meier plots of the probability of PFS and OS in ovarian cancer patients. (A–C) PFS (left) and OS (right) stratified by the methylation status of (A) ATG4A and (B) HIST1H2BN are shown for ovarian cancer patients as estimated by Kaplan–Meier curves and the log-rank test. Straight line: high methylation; bold line: low methylation. (C) Two geneslow is defined as low methylation of both genes and ≥one geneshigh as high methylation of more than one gene. The clinical endpoint is indicated on the y-axis. PFS, progression-free survival; OS, overall survival; Prob., probability.
Table 2.
Multivariate Cox regression analysis for PFS and OS of ovarian cancer patients
| Event | PFS |
OS |
||||
|---|---|---|---|---|---|---|
| Variable | Adjusted HRa (95% CI) | Adjusted HRb (95% CI) | ||||
| Age (years) | 1.0 (0.9, 1.0) | 1.0 (0.9, 1.0) | 1.0 (0.9, 1.0) | 1.0 (0.9, 1.0) | 1.0 (0.9, 1.0) | 1.0 (0.9, 1.0) |
| Methylation status | ||||||
| ATG4A (low versus high) | 1.4 (0.7, 2.6) | 1.4 (0.8, 2.4) | ||||
| HIST1H2BN (low versus high) | 4.5 (1.4, 14.8)* | 4.3 (1.3, 14.0)* | ||||
| ATG4A/HIST1H2BN (low versus high) | 1.8 (1.0, 3.6)* | 1.7 (1.0, 3.0) | ||||
| FIGO stage (late versus early) | 10.7 (3.0, 38.3)* | 14.5 (4.1, 51.4)* | 10.6 (3.0, 37.8)* | 17.6 (4.0, 77.3)* | 23.2 (5.3, 102.1)* | 17.3 (3.9, 76.0)* |
| Grade (G3 versus G1/G2) | 1.8 (0.8, 4.0) | 1.7 (0.8, 3.9) | 1.8 (0.8, 3.9) | 1.3 (0.6, 2.6) | 0.9 (0.3, 1.8) | 1.2 (0.6, 2.5) |
HR, hazard ratio; CI, confidence interval.
aThe HR adjusted by gene methylation status, stage and grade.
bThe HR adjusted by age, gene methylation status, stage and grade.
*Significantly correlated with outcome, P < 0.05.
Using similar methods, we based our analysis on ovarian cancer samples and performed a survival analysis of the methylation status of the two genes in high-grade serous ovarian cancer samples using The Cancer Genome Atlas (TCGA) data portal. This dataset consisted of 385 cases and was used as an independent validation set in our analysis. The results of the Cox proportional hazard model for recurrence are presented in Supplementary Material, Table S3. FIGO stage, HIST1H2BN methylation status and ATG4A/HIST1H2BN methylation status were independently correlated with recurrence (all P < 0.05). Specifically, patients with a low HIST1H2BN methylation level had a significantly higher risk of recurrence than did patients with a high methylation level (HR, 1.5; 95% CI, 1.0–2.2; P = 0.04). The risk of disease recurrence in patients with low ATG4A/HIST1H2BN methylation level was 1.7 times higher than that observed in patients with high methylation levels (95% CI, 1.1–2.5; P = 0.02). The results of the Cox proportional hazard model for variables that influenced the risk of death are presented in Supplementary Material, Table S4. Age at diagnosis, methylation status and stage were the main predictors of death (all P < 0.05). Compared with patients with a low ATG4A methylation level, the HR for death in patients with a high methylation level was 1.4 (95% CI, 1.0–1.9; P = 0.03). Moreover, patients with low methylation of ATG4A and HIST1H2BN were more likely to die within the follow-up period (HR, 1.4; 95% CI, 1.0–1.9; P = 0.03). Patients with Stage IV disease were two to three times as likely as patients with Stage II to have disease recurrence or death (HR, 2.4; 95% CI, 1.2–4.6; P = 0.009 and HR, 3.0; 95% CI, 1.4–6.7, respectively). The PFS status for the 314 patients included in these analyses is presented in Figure 5C (left). Among the patients in the group with low methylation of both genes, half had recurrence at 12 months of follow-up, whereas the patients in the group with high methylation of both genes exhibited recurrence at 20 months. The difference in PFS status at the end of follow-up between groups with low methylation of both genes and high methylation in more than one gene was statistically significant (Fig. 5D, left; P = 0.009). Figure 5C (right) shows the OS status according to gene methylation. The median OS was 37 months for the group with low methylation of both genes and 48 months for the group with high methylation of both genes. This difference in OS between groups with high methylation of both genes and low methylation of more than one gene was statistically significant (Fig. 5D right; P = 0.02).
Figure 5.
Kaplan–Meier plots of the probability of PFS and OS in TCGA ovarian cancer patients. (A–D) PFS (left) and OS (right) stratified by the methylation status of (A) ATG4A and (B) HIST1H2BN are shown for ovarian cancer patients as estimated by Kaplan–Meier curves and the log-rank test. Straight line: high methylation; bold line: low methylation. (C) Two geneslow is defined as low methylation of both genes, one geneshigh as high methylation of any one gene, and two geneshigh as high methylation of both genes. (D) Two geneslow are defined as low methylation of both genes, ≥ one geneshigh as high methylation of more than one gene and ≥ one geneslow as low methylation of more than one gene. The clinical endpoint is indicated on the y-axis. PFS, progression-free survival; OS, overall survival; Prob., probability.
The expression of ATG4A increased stem properties
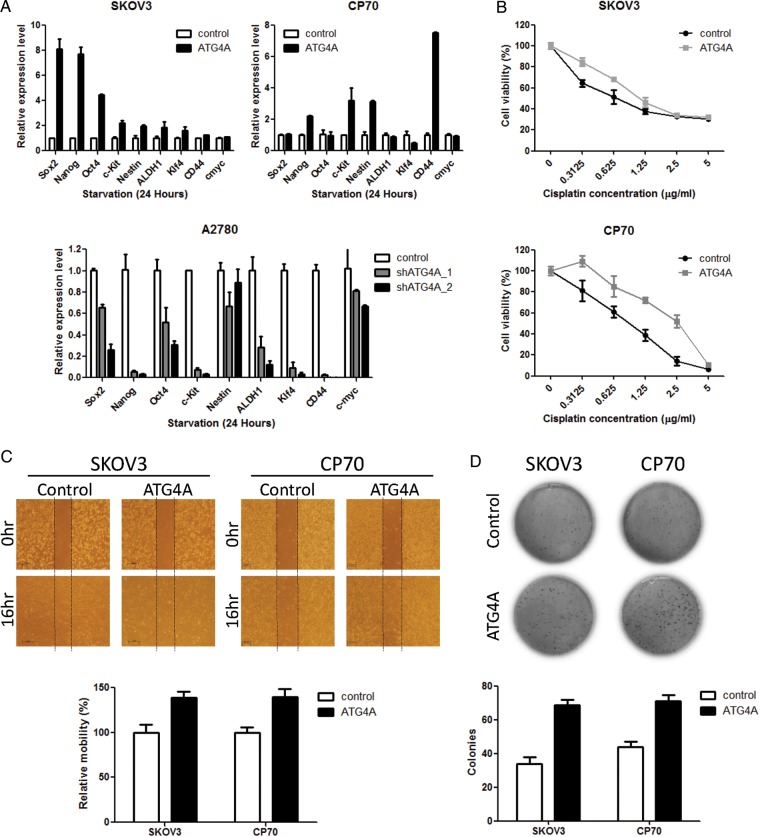
As shown in Figure 2A, high expression of ATG4A was observed in OTICs (CP70sps), whereas it was epigenetically silenced in ovarian cancer cells (CP70). The low methylation of ATG4A in CP70sps and high methylation of ATG4A in CP70 cells are shown in Figure 2C. In addition, the high methylation of ATG4A was related to poor prognosis in ovarian cancer patients (Figs 4 and 5). Thus, the relevance of ATG4A/HIST1H2BN in TICs is that these two genes are differentially methylated and expressed in TICs compared with differentiated cancer cells. These differences may represent the enrichment of TICs and confer different malignant behaviors on ovarian cancer cells in patients, leading to different outcomes. To address the biological function of ATG4A in ovarian cancer, we expressed ATG4A ectopically in the ovarian cancer cell lines SKOV3 and CP70, and knocked it down in A2780 cells (Supplementary Material, Fig. S1A). We observed that ATG4A increased the stem properties and malignant phenotype of cells, including increasing the expression of stem markers (Fig. 6A), drug resistance (Fig. 6B, Supplementary Material, Fig. S1C), cell migration (Fig. 6C) and anchorage-independent growth (Fig. 6D).
Figure 6.
Effects of ATG4A on stem markers, drug resistance, cell migration and anchorage-independent growth in ovarian cancer cell lines. (A) The mRNA expression of the target genes was detected after 24 h of serum starvation. The expression of genes was measured by qRT-PCR and normalized to GAPDH levels. The relative levels of expression after qRT-PCR were compared with the vector control. Each bar represents the average of triplicate reactions ± SD. (B) The effect of ATG4A on the chemosensitivity of SKOV3 and CP70 cells to cisplatin was determined by MTS cytotoxicity assay. Cells were seeded on 96-well plates and treated with various concentrations of cisplatin for 3 days after 24 h of serum starvation. Cell numbers were determined by MTS assay and compared with untreated cells. Each bar represents the average of triplicate reactions ±SD. (C) The migration abilities of SKOV3 and CP70 cells were assayed after 24 h of serum starvation by wound-healing assay. Images were taken immediately after scratching the cultures (0 h) and 16 h later. The quantitation of wound healing was measured by ImageJ. The increased area of control was set as 100%. Data are presented as means ± SD. Three independent experiments were performed in triplicate. (D) The anchorage-independent growth of SKOV3 and CP70 cells was determined by soft agar assay after serum starvation for 24 h. The expression of ATG4A increased colony formation significantly in SKOV3 and CP70 cells. Each bar represents the average of triplicate reactions ±SD.
DISCUSSION
Predictive and prognostic biomarkers offer a potential means of personalizing cancer medicine. Translation from molecular insights to clinical stratification of patients may help to identify subgroups of patients responsive to current treatments and to diminish the unnecessary side effects in patients with predictive poor responses (22) EGFR mutations for lung cancer (23) and HER2-directed therapy for breast cancer patients (24) are two examples of current personalized therapy. However, investigations of biomarkers including ‘BRCAness’ and ‘PI3Kness’ for personalizing therapy in EOC have not been successful so far (25).
Earlier reports using CpG island microarrays to interrogate thousands of CpG loci revealed a methylation signature comprising 112 loci that predicted PFS in advanced ovarian cancer patients (9,26). Epigenetic modifications at specific CpG sites correlate with PFS and OS in ovarian cancer patients treated with conventional chemotherapeutics (8,16,27–29). Experiments using demethylation demonstrated resensitization of platinum-resistant ovarian cancer cells to commonly used drugs (30,31). Recent trials of the demethylation agents, azacitidine and decitabine in ovarian cancer patients demonstrated reversal of platinum resistance (16,32). The number of demethylated genes in tumor tissues after treatment with decitabine and carboplatin was greater in patients with PFS >6 months (16), indicating the demethylation effects of decitabine in vivo. Here, we show that our systemic methylation profiling identified and verified candidate gene DNA methylation during repopulation by OTICs. We found hypomethylation of ATG4A and HIST1H2BN in OTICs; in addition, methylation distinguished the prognosis of patients treated with conventional therapies.
HIST1H2BN, located on human chromosome 6 (6p21–p22), encodes a member of the histone H2B family known as the Histone H2B-type 1-N protein in humans (33). This is linked to histone H1 that interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. The promoter of HIST1H2BN has putative p53-binding sites, and p53 has been speculated to repress HIST1H2BN gene expression in head and neck squamous cell carcinomas (34). The etiology of ovarian cancer is not known. However, dysfunction of p53 is detrimental, and the latest TCGA report indicated that it is mutated in up to 96% of ovarian cancers (35). The mutation of cells at the stem cell niche in the tubo-ovarian junction promotes the development of high-grade serous cancer (36). As there are no functional studies of HIST1H2BN in cancer biology, we speculate that the mutation or deletion of p53 (35) may result in HIST1H2BN gene dysregulation and contribute to cancer progression. ATG4A encodes a cysteine protease and is involved in autophagy. Decreased levels of autophagy have been described in some malignant tumors (37). Women carrying variant alleles of ATG4A have a reduced risk of ovarian cancer (38). This study is the first report of HIST1H2BN and ATG4A methylation in cancers.
The present study compared differential methylation between TICs and cancer cells, rather than between cancer cells and noncancerous cells. Genes previously shown to display aberrant methylation in ovarian cancer cells may not be involved in the differentiation of TICs, and so may not be detected in the present study. In addition, the present study used the 27K Illumina bead array, which contains only 14 475 genes and targets promoter regions. Genes not included in the array cannot be analyzed, and sequences outside promoter regions are also missed. New versions of the array such as the 450K, which contains a genome-wide distribution of promoters, may provide more information. Non-targeted methods such as methylation DNA captured deep sequencing technology may reveal more comprehensive methylation profiles of intergene regions or gene bodies.
An increasingly accepted hypothesis for treatment failure is the failure to eradicate TICs. Several OTICs have been isolated based on side population (SP) analysis (39), spheroid culturing (40), ALDH1 activity (41) or cell-surface protein markers, such as CD117, CD133, CD24, CD44 and MyD88 (40,42). Although investigated extensively in recent years, the clinical relevance of this stem-like cancer cell theory remains elusive. However, enrichment of ovarian cancer stem cell markers in tissues correlates with poor prognosis (41,43). An embryonic stem cell gene expression signature is associated with poorly differentiated human solid tumors and with poor clinical outcome (44). The expression profile of 11 gene signatures related to a putative stem cell renewal protein, Bmi-1, in primary tumors could be a consistent predictor of a short PFS, distant metastasis and death after therapy in patients diagnosed with various cancer types (45). These findings suggest that a stem-enriched phenotype is a prognostic indicator of the poor outcome in ovarian cancer patients.
The role of DNA methylation in cancer stem cells and its relevance to clinical outcome have not been investigated. A recent bioinformatic report showed that hypermethylation of a subset of stem cell polycomb group genes and hypomethylation of a subset of genes originally heavily methylated in embryonic stem cells define a poor prognostic signature of ovarian cancers (21). The present study demonstrates that a hypomethylated signature of OTICs in tumor tissues indicates a poor prognostic outcome in ovarian cancer patients. This is the first evidence of the potential clinical prognostic significance of DNA methylation changes during OTIC repopulation. We used both a methylomic approach and bioinformatics, and validated the results using different methods in cell lines and in an independent set of tissues from patients. We found that ATG4A and HIST1H2BN, which were not previously known to have stem cell functions, were hypomethylated in OTICs compared with cells from bulky tumors. Hence, we speculate that the hypomethylated signature in OTICs may represent an enriched stem-like phenotype that confers a poor clinical outcome. To test this hypothesis, we analyzed some functions of ATG4A in ovarian cancer cell lines. We found that ATG4A increased stem properties, including the expression of stem markers, and suspended the growth of ovarian cancer cells. The expression of ATG4A also promoted migration and chemoresistance in ovarian cancer cells. These additional experiments support the important roles of ATG4A in ovarian cancer biology and may have clinical significance. In addition, the stringent method combining SP analysis and spheroid formation to enrich OTICs has led to the discovery of novel compounds targeting OTICs. Further elucidation of the functions of ATG4A and HIST1H2BN in OTICs may lead to the development of new therapeutics. We also analyzed the two genes in the TCGA Data Portal to validate the ability of DNA hypomethylation to predict poor prognosis. The TCGA portal is a large database accumulated from a large number of research studies. Some researchers have used this database to analyze the molecular basis of cancer. The Cancer Genome Atlas Research Network published their results regarding ovarian cancer in 2011 (35), and some reports have provided TCGA methylomics data (28,46) that indicate the presence of gene methylation in patients with a poor prognosis. However, these reports lack quantitative verification of methylation; thus, a more comprehensive assessment of methylation in ovarian cancer is needed.
In this study, we took the advantage of TIC concept using SP spheres of CP70, which is a subclone of A2780. A2780 was thought as an adenocarcinoma cell line. CP70 is a selected chemoresistant subclones. Our previous xenograft study confirmed the growth of a poorly differentiated adenocarcinoma after injection of CP70 and CP70 SP spheres. Two recent reports using genomic profiles revealed that A2780 is more likely endometrioid type (47,48). Although A2780 is not exactly the same genomic profile of high-grade serous type, this nature does not compromise our findings since we discovered these two genes from the concept of OTICs. The clinical significance of this hypomethylation signature of OTICs derived from this concept was validated in our dataset containing different histotypes and in TCGA dataset comprising high-grade serous-type cancers. The functions relevant to stem properties were also verified in vitro. We believe that histotype is important in many aspects, but, does not compromise our findings in the present study.
In conclusion, ovarian cancer patients with low methylation of ATG4A and HIST1H2BN genes have poor prognosis. This poor prognosis is related to the population of TIC phenotypes. Further prospective clinical trials are required to prove that the methylation of these genes is a prognostic biomarker using current therapeutic modalities. The application of this DNA methylation as a predictive biomarker for epigenetic therapy may help in the delivery of personalized medicine for ovarian cancer patients.
MATERIALS AND METHODS
Cell lines and OTIC enrichment
We cultured A2780, CP70, OVCAR3, SKOV3, TOV21G and IOSE cells with and without a demethylation agent and isolated the DNA and RNA from these cells. The method used for the enrichment and characterization of OTICs using SP spheroids (sps) has been reported previously (40). In brief, for SP analysis, 108 cells were harvested and stained with 50 g/ml Hoechst 33342 (Sigma, St. Louis, MO, USA). Hoechst-stained cells were subjected to fluorescence-activated cell sorting using a FACSAria (BD Biosciences, Franklin Lakes, NJ, USA) for collection of dye-excluding SP cells. The dye-exclusion phenotype, identified by membrane efflux, was confirmed with an ABCG2-specific inhibitor, GF120918. The cells were cultured immediately in Ultra-Low attachment plates (Corning, Corning, NY, USA) with serum-free DMEM-F12 medium containing 0.4% bovine serum albumin (Sigma), 5 μg/ml insulin (Sigma), 20 ng/ml human recombinant epidermal growth factor (Invitrogen, Carlsbad, CA, USA) and 10 ng/ml basic fibroblast growth factor (Invitrogen) until spheres were grown. SP cells were then observed for the stem cell phenotype of tumor sphere formation (49,50) under suspension-cultured conditions, and these spheroid-forming cells were then referred to as SP spheroid (sps) cells. In the differentiation studies, CP70sps cells were cultured in standard coated dishes (Corning) with normal media and 10% fetal bovine serum.
To generate cells that express ATG4A stably, 3 × 105 SKOV3 and CP70 cells were plated into each 10 cm culture dish and starved the day before transfection. Immediately before transfection, cells were washed gently in 1× PBS, and 4 ml of Opti-MEM I reduced-serum medium (Invitrogen) was added into each dish. Lipofectamine 2000 (7.5 μl; Invitrogen) was diluted in 500 μl of Opti-MEM I medium and incubated for 5 min at room temperature (Solution A). The pcDNA3.1/ATG4A plasmid, pLKO/shATG4A or empty plasmid (3 μg) was added to 0.5 ml of Opti-MEM I medium and incubated for 5 min (Solution B). Solutions A and B were immediately mixed and incubated for an additional 20 min at room temperature. After incubation, the mixture (total volume of 1 ml) was added directly to the cells and incubated at 37°C for 5 h. The transfection medium mixture was replaced with fresh culture medium. After 2 days, the cells were divided equally into three 10 cm culture dishes with complete culture medium. The following day, cells were cultivated with fresh culture medium containing 400 μg/ml of Geneticin (G418) (Invitrogen), which was replaced every 3 days.
The ATG4A short hairpin RNA (shRNA) was prepared and maintained according to the protocol provided by the National RNAi Core Facility, Academia Sinica. The sequences of shRNA-scramble, shRNA-ATG4A (KD1) and shRNA-ATG4A (KD2) are 5′-CCTAAGGTTAAGTCGCCCTCG-3′, 5′-CCCGGAAAGAAATAGAACAAT-3′ and 5′-CCTGGGCATAAACCAAATCAA-3′.
Quantitative RT–PCR analysis
The expression of ATG4A and other target genes in stable cell lines was examined by qRT-PCR analysis. Fifty-fold diluted cDNA (4 μl) was amplified in a total volume of 25 μl containing 2× RT² SYBR Green qPCR Master Mixes (SABiosciences, Frederick, MD, USA) and 0.2 μm of each primer at 95°C for 10 min, followed by 45 cycles of 95°C for 15 s, the annealing temperature for 30 s and extension at 72°C for 30 s; a melting curve was obtained. No RT (reverse transcription without reverse transcriptase) was used as a negative control. The relative gene expression level was determined by comparing the threshold cycle (Ct) of the test gene against the Ct value of GAPDH in a given sample.
Methylation profiling
Genome-wide methylation profiling of 27 578 methylation sites in 14 475 genes was conducted using an Infinium HumanMethylation27 Bead array (Illumina Inc., San Diego, CA, USA). After normalization using the lumi methodology (51,52), we analyzed differential methylation according to the manufacturer's recommendation. We selected the probes provided with effective detection P-values of <0.001 and performed hierarchical clustering analysis using MultiExperiment Viewer (MeV) 4.6.2 (53), a Java application designed to allow the analysis of microarray data to identify patterns of gene expression and differentially expressed genes. We set the critical P-value to be ≤0.01 to select probes with significantly different methylation levels. Next, we filtered differential methylation of probes that provided the differential average of β-values (AVG-β) ≥ 0.1 between CP70 and CP70sps. A β-value between 0 and 1 (ratio of methylated to the sum of methylated and unmethylated sites) and the average β-value corresponded to the methylation level for each probe at one CpG site (54). The scheme of our analytical strategy aimed at identifying and verifying novel epigenetic biomarkers of higher methylation and lower gene expression levels in CP70 compared with CP70sps, as depicted in Figure 1A.
Bisulfite modification, qMSP and pyrosequencing
Genomic DNA was extracted for methylation analysis, as previously described (55). One microgram of genomic DNA was bisulfite modified using the CpGenome Fast DNA Modification Kit (Chemicon-Millipore, Bedford, MA, USA) according to the manufacturer's recommendations and redissolved in 70 ml nuclease-free water. We used qMSP to compare the promoter methylation status in ovarian tissues obtained from patients with EOC with that of benign and normal ovarian tissues, and we used pyrosequencing analysis to validate the results. Therefore, the reproducibility of technical duplicates was assessed. qMSP was performed in a TaqMan probe system using the LightCycler 480 Real-Time PCR System (Roche, Indianapolis, IN, USA) (55). The DNA methylation level was estimated as the methylation index (M-index) using the formula: 10 000 × 2[(Cp of COL2A) – (Cp of gene)] (56). Test results with Cp values for COL2A >36 were defined as detection failure. The primers for pyrosequencing were designed by PyroMark Assay Design 2.0 software (Qiagen, Hilden, Germany) to amplify and sequence bisulfite-treated DNA. The biotinylated PCR product was bound to streptavidin-Sepharose beads, washed and denatured. The sequencing primer was added to the PCR products, and pyrosequencing was performed using the PyroMark Q24 software (Qiagen) according to the manufacturer's instructions.
Patients and clinical samples
Tissue samples were collected with the informed consent of patients at the National Defense Medical Center, Taipei and the Taipei Veterans General Hospital, Taiwan. This study was approved by the Institutional Review Boards. The patients included 168 with EOC, 60 with benign ovarian tumors and 28 with normal ovarian tissue whose diagnosis included histological subtype and grade. The benign ovarian tumors included serous adenoma, mucinous adenoma and endometrioma. These specimens were obtained during surgery and were frozen immediately in liquid nitrogen and stored at −80°C until analysis. The inclusion criteria for the recruited patients were: confirmed FIGO stage and no prior radiotherapy or chemotherapy. The presence of malignant cells was confirmed by histological examination by gynecologic pathologists, who reviewed all of the specimens. The nature of which was epithelial ovarian carcinoma or primary peritoneal carcinomatosis. The exclusion criteria included borderline tumors, mixed mesodermal tumors, concurrent malignancies and pregnancy or malignancy within the previous 5 years. All EOC patients included in the study were aimed at verifying the applicability of the DNA methylation level as a prognostic marker. PFS was calculated from the time of the first operation to progressive disease. The patients who remained alive and did not have documented local recurrence were censored at their last follow-up date. Fifty-four patients presenting with persistent disease after the first-line standard treatment were excluded from the PFS analysis. OS was calculated from the time of the first operation to death because of EOC or the last follow-up.
We also used the DNA methylation data from tissue samples of ovarian cancer patients obtained from TCGA data portal as an independent validation dataset. The Level 3 methylation dataset on the Illumina HumanMethylation27 Beadchip of high-grade serous tumors was available to perform a survival analysis based on the information obtained from the TCGA data portal. After excluding some cases with missing values for several clinicopathological variables, 385 patients were included in our analysis.
Cell proliferation and chemosensitivity assay
Cell growth and chemosensitivity were assessed by MTS assay. One thousand cells were seeded in 96-well plates for 3 days after 24 h serum starvation with or without various concentrations of cisplatin or taxol. Cell numbers (expressed as cell viability in %) were determined using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay kit (Promega, Madison, WI, USA), according to the manufacturer's protocol. Relative cell numbers were assessed using a 96-well ELISA plate reader at an absorbance of 490 nm.
Wound-healing assays
Cells (1 × 105) were seeded on 6-well dishes until confluence and were then wounded using a yellow pipette tip. Three wounds were made for each sample, and the migration area was photographed and measured at 0 and 16 h. Images of each scratch were obtained, and cell migration was quantified by measuring the migrated area using Image J (57).
Soft agar assay for colony formation
A 2.5-ml base layer of agar (0.5% agar in culture medium plus 400 μg of G418) was allowed to solidify in a 6-well flat-bottomed plate before the addition of 2 ml of cell suspensions containing 10 000 cells in 0.3% agar in culture medium plus 400 μg of G418. The cell-containing layer was then solidified at room temperature for 20 min. Colonies were allowed to grow for 14 days at 37°C with 5% CO2 before imaging. Plates were stained with 0.05% crystal violet. The number of colonies in the entire dish was counted.
Statistical analysis
An adequate sample size of 62 patients per group, which was calculated based on a 15-year survival rate in patients with an FBXO32 methylation equal to 30% and a significant difference revealed by log-rank test at the alpha level of 5% (27), was necessary to achieve the power of 80%.
We evaluated the cutoff values for the methylation status of each gene to achieve the best clinical performance to discriminate patients with recurrence from those without recurrence by calculating the area under the receiver-operating characteristic curve. The M-index of HIST1H2BN and ATG4A >618 and 1080, respectively, was defined as high methylation in our dataset. The β-value of HIST1H2BN and ATG4A >0.025 and 0.23, respectively, was defined as high methylation in TCGA dataset. We applied the same cutoff values to distinguish patients who died from those who survived. We defined all methylation values as high or low for further statistical analysis. We also combined the results for the two genes into signatures of high methylation levels for more than one gene and low methylation levels for both genes. Descriptive statistics were used to analyze the population distributions. The associations between candidate gene methylation levels and clinical characteristics and differential diagnosis were analyzed by the χ2-test or Fisher's exact test and the Mann–Whitney U-test for categorical and continuous variables, respectively. The Kaplan–Meier analysis, log-rank test and Cox proportional-hazards models were used to estimate the survival distributions and to compare differences between the curves of PFS and OS. A univariate Cox regression analysis calculated the HR and a 95% CI for the risk of clinicopathological characteristics for each candidate gene. The multivariate Cox proportional-hazards model identified the independent prognostic values of age, DNA methylation status, tumor stage, grade and/or histological subtype. All analyses were two sided, and P-values of <0.05 were regarded as significant. All statistical calculations were performed using the statistical package SPSS version 20.0 for Windows (IBM Corp., Armonk, NY, USA).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Department of Health, Executive Yuan, ROC (grant number DOH101-TD-PB-111-NSC008 and DOH102-TD-PB-111-NSC103 to H.C.L.); National Science Council (grant number NSC101-2314-B-016-021 to H.C.L., grant number NSC102-2314-B-016-043 to Yu-Chi Wang); Tri-Service General Hospital (grant number TSGH-C102-008-S01 to H.C.L., grant number TSGH-C102-008-S02 to M.S.Y., grant number TSGH-C102-008-S03 to Yu-Chi Wang); Teh-Tzer Study Group for Human Medical Research Foundation (H.C.L.); and the National Institutes of Health (CA85289 and CA113001 to K.N.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Ms Hsin-Yi Lee and Ms Hui-Chen Wang, Department of Obstetrics and Gynecology, Tri-Service General Hospital, Taiwan, ROC, for technical assistance. The authors thank the external validation of our candidate genes used methylation data generated by TCGA group.
Conflict of Interest statement. The patent of the application of this signature is pending. National Defense Medical Center is the owner of intellectual property. H.C.L. is the inventor.
REFERENCES
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2012. CA Cancer J. Clin. 2012;62:10–29. doi: 10.3322/caac.20138. doi:10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. doi:10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Cannistra S.A. Cancer of the ovary. N. Engl. J. Med. 2004;351:2519–2529. doi: 10.1056/NEJMra041842. doi:10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 4.Ozols R.F. Treatment goals in ovarian cancer. Int. J. Gynecol. Cancer. 2005;15(Suppl. 1):3–11. doi: 10.1111/j.1525-1438.2005.15351.x. doi:10.1111/j.1525-1438.2005.15351.x. [DOI] [PubMed] [Google Scholar]
- 5.Bristow R.E., Tomacruz R.S., Armstrong D.K., Trimble E.L., Montz F.J. Survival effect of maximal cytoreductive surgery for advanced ovarian carcinoma during the platinum era: a meta-analysis. J. Clin. Oncol. 2002;20:1248–1259. doi: 10.1200/JCO.2002.20.5.1248. doi:10.1200/JCO.20.5.1248. [DOI] [PubMed] [Google Scholar]
- 6.Ahluwalia A., Hurteau J.A., Bigsby R.M., Nephew K.P. DNA methylation in ovarian cancer. II. Expression of DNA methyltransferases in ovarian cancer cell lines and normal ovarian epithelial cells. Gynecol. Oncol. 2001;82:299–304. doi: 10.1006/gyno.2001.6284. doi:10.1006/gyno.2001.6284. [DOI] [PubMed] [Google Scholar]
- 7.Strathdee G., Appleton K., Illand M., Millan D.W., Sargent J., Paul J., Brown R. Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am. J. Pathol. 2001;158:1121–1127. doi: 10.1016/S0002-9440(10)64059-X. doi:10.1016/S0002-9440(10)64059-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su H.Y., Lai H.C., Lin Y.W., Chou Y.C., Liu C.Y., Yu M.H. An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int. J. Cancer. 2009;124:387–393. doi: 10.1002/ijc.23957. doi:10.1002/ijc.23957. [DOI] [PubMed] [Google Scholar]
- 9.Wei S.H., Balch C., Paik H.H., Kim Y.S., Baldwin R.L., Liyanarachchi S., Li L., Wang Z., Wan J.C., Davuluri R.V., et al. Prognostic DNA methylation biomarkers in ovarian cancer. Clin. Cancer Res. 2006;12:2788–2794. doi: 10.1158/1078-0432.CCR-05-1551. doi:10.1158/1078-0432.CCR-05-1551. [DOI] [PubMed] [Google Scholar]
- 10.Petrocca F., Iliopoulos D., Qin H.R., Nicoloso M.S., Yendamuri S., Wojcik S.E., Shimizu M., Di Leva G., Vecchione A., Trapasso F., et al. Alterations of the tumor suppressor gene ARLTS1 in ovarian cancer. Cancer Res. 2006;66:10287–10291. doi: 10.1158/0008-5472.CAN-06-2289. doi:10.1158/0008-5472.CAN-06-2289. [DOI] [PubMed] [Google Scholar]
- 11.Kwong J., Lee J.Y., Wong K.K., Zhou X., Wong D.T., Lo K.W., Welch W.R., Berkowitz R.S., Mok S.C. Candidate tumor-suppressor gene DLEC1 is frequently downregulated by promoter hypermethylation and histone hypoacetylation in human epithelial ovarian cancer. Neoplasia. 2006;8:268–278. doi: 10.1593/neo.05502. doi:10.1593/neo.05502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sellar G.C., Watt K.P., Rabiasz G.J., Stronach E.A., Li L., Miller E.P., Massie C.E., Miller J., Contreras-Moreira B., Scott D., et al. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat. Genet. 2003;34:337–343. doi: 10.1038/ng1183. doi:10.1038/ng1183. [DOI] [PubMed] [Google Scholar]
- 13.Teodoridis J.M., Hall J., Marsh S., Kannall H.D., Smyth C., Curto J., Siddiqui N., Gabra H., McLeod H.L., Strathdee G., et al. CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res. 2005;65:8961–8967. doi: 10.1158/0008-5472.CAN-05-1187. doi:10.1158/0008-5472.CAN-05-1187. [DOI] [PubMed] [Google Scholar]
- 14.Ibanez de Caceres I., Battagli C., Esteller M., Herman J.G., Dulaimi E., Edelson M.I., Bergman C., Ehya H., Eisenberg B.L., Cairns P. Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 2004;64:6476–6481. doi: 10.1158/0008-5472.CAN-04-1529. doi:10.1158/0008-5472.CAN-04-1529. [DOI] [PubMed] [Google Scholar]
- 15.Chien J., Staub J., Avula R., Zhang H., Liu W., Hartmann L.C., Kaufmann S.H., Smith D.I., Shridhar V. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24:5089–5100. doi: 10.1038/sj.onc.1208700. doi:10.1038/sj.onc.1208700. [DOI] [PubMed] [Google Scholar]
- 16.Matei D., Fang F., Shen C., Schilder J., Arnold A., Zeng Y., Berry W.A., Huang T., Nephew K.P. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. doi:10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balch C., Fang F., Matei D.E., Huang T.H., Nephew K.P. Minireview: epigenetic changes in ovarian cancer. Endocrinology. 2009;150:4003–4011. doi: 10.1210/en.2009-0404. doi:10.1210/en.2009-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houshdaran S., Hawley S., Palmer C., Campan M., Olsen M.N., Ventura A.P., Knudsen B.S., Drescher C.W., Urban N.D., Brown P.O., et al. DNA methylation profiles of ovarian epithelial carcinoma tumors and cell lines. PLoS ONE. 2010;5:e9359. doi: 10.1371/journal.pone.0009359. doi:10.1371/journal.pone.0009359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan C.T., Guzman M.L., Noble M. Cancer stem cells. N. Engl. J. Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. doi:10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 20.Dalerba P., Cho R.W., Clarke M.F. Cancer stem cells: models and concepts. Annu. Rev. Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. doi:10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 21.Zhuang J., Jones A., Lee S.H., Ng E., Fiegl H., Zikan M., Cibula D., Sargent A., Salvesen H.B., Jacobs I.J., et al. The dynamics and prognostic potential of DNA methylation changes at stem cell gene loci in women's cancer. PLoS Genet. 2012;8:e1002517. doi: 10.1371/journal.pgen.1002517. doi:10.1371/journal.pgen.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diamandis M., White N.M., Yousef G.M. Personalized medicine: marking a new epoch in cancer patient management. Mol. Cancer Res. 2010;8:1175–1187. doi: 10.1158/1541-7786.MCR-10-0264. doi:10.1158/1541-7786.MCR-10-0264. [DOI] [PubMed] [Google Scholar]
- 23.Mok T.S. Personalized medicine in lung cancer: what we need to know. Nat. Rev. Clin. Oncol. 2011;8:661–668. doi: 10.1038/nrclinonc.2011.126. doi:10.1038/nrclinonc.2011.126. [DOI] [PubMed] [Google Scholar]
- 24.Ross J.S., Slodkowska E.A., Symmans W.F., Pusztai L., Ravdin P.M., Hortobagyi G.N. The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist. 2009;14:320–368. doi: 10.1634/theoncologist.2008-0230. doi:10.1634/theoncologist.2008-0230. [DOI] [PubMed] [Google Scholar]
- 25.Romero I., Bast R.C., Jr Minireview: human ovarian cancer: biology, current management, and paths to personalizing therapy. Endocrinology. 2012;153:1593–1602. doi: 10.1210/en.2011-2123. doi:10.1210/en.2011-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei S.H., Chen C.M., Strathdee G., Harnsomburana J., Shyu C.R., Rahmatpanah F., Shi H., Ng S.W., Yan P.S., Nephew K.P., et al. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin. Cancer Res. 2002;8:2246–2252. [PubMed] [Google Scholar]
- 27.Chou J.L., Su H.Y., Chen L.Y., Liao Y.P., Hartman-Frey C., Lai Y.H., Yang H.W., Deatherage D.E., Kuo C.T., Huang Y.W., et al. Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab. Invest. 2010;90:414–425. doi: 10.1038/labinvest.2009.138. doi:10.1038/labinvest.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai W., Teodoridis J.M., Zeller C., Graham J., Hersey J., Flanagan J.M., Stronach E., Millan D.W., Siddiqui N., Paul J., et al. Systematic CpG islands methylation profiling of genes in the wnt pathway in epithelial ovarian cancer identifies biomarkers of progression-free survival. Clin. Cancer Res. 2011;17:4052–4062. doi: 10.1158/1078-0432.CCR-10-3021. doi:10.1158/1078-0432.CCR-10-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montavon C., Gloss B.S., Warton K., Barton C.A., Statham A.L., Scurry J.P., Tabor B., Nguyen T.V., Qu W., Samimi G., et al. Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecol. Oncol. 2012;124:582–588. doi: 10.1016/j.ygyno.2011.11.026. doi:10.1016/j.ygyno.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 30.Balch C., Yan P., Craft T., Young S., Skalnik D.G., Huang T.H., Nephew K.P. Antimitogenic and chemosensitizing effects of the methylation inhibitor zebularine in ovarian cancer. Mol. Cancer Ther. 2005;4:1505–1514. doi: 10.1158/1535-7163.MCT-05-0216. doi:10.1158/1535-7163.MCT-05-0216. [DOI] [PubMed] [Google Scholar]
- 31.Li Y., Hu W., Shen D.Y., Kavanagh J.J., Fu S. Azacitidine enhances sensitivity of platinum-resistant ovarian cancer cells to carboplatin through induction of apoptosis. Am. J. Obstet. Gynecol. 2009;200:177.e1–177e9. doi: 10.1016/j.ajog.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 32.Fang F., Balch C., Schilder J., Breen T., Zhang S., Shen C., Li L., Kulesavage C., Snyder A.J., Nephew K.P., et al. A phase 1 and pharmacodynamic study of decitabine in combination with carboplatin in patients with recurrent, platinum-resistant, epithelial ovarian cancer. Cancer. 2010;116:4043–4053. doi: 10.1002/cncr.25204. doi:10.1002/cncr.25204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marzluff W.F., Gongidi P., Woods K.R., Jin J., Maltais L.J. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. doi:10.1006/geno.2002.6850. [PubMed] [Google Scholar]
- 34.Yan B., Yang X., Lee T.L., Friedman J., Tang J., Van Waes C., Chen Z. Genome-wide identification of novel expression signatures reveal distinct patterns and prevalence of binding motifs for p53, nuclear factor-kappaB and other signal transcription factors in head and neck squamous cell carcinoma. Genome Biology. 2007;8:R78. doi: 10.1186/gb-2007-8-5-r78. doi:10.1186/gb-2007-8-5-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Network T.C.G.A.R. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. doi:10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flesken-Nikitin A., Hwang C.I., Cheng C.Y., Michurina T.V., Enikolopov G., Nikitin A.Y. Ovarian surface epithelium at the junction area contains a cancer-prone stem cell niche. Nature. 2013;495:241–245. doi: 10.1038/nature11979. doi:10.1038/nature11979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. doi:10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 38.Liang D., Meyer L., Chang D.W., Lin J., Pu X., Ye Y., Gu J., Wu X., Lu K. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010;70:9765–9776. doi: 10.1158/0008-5472.CAN-10-0130. doi:10.1158/0008-5472.CAN-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu L., McArthur C., Jaffe R.B. Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. Br. J. Cancer. 2010;102:1276–1283. doi: 10.1038/sj.bjc.6605626. doi:10.1038/sj.bjc.6605626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S., Balch C., Chan M.W., Lai H.C., Matei D., Schilder J.M., Yan P.S., Huang T.H., Nephew K.P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. doi:10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y.C., Yo Y.T., Lee H.Y., Liao Y.P., Chao T.K., Su P.H., Lai H.C. ALDH1-bright epithelial ovarian cancer cells are associated with CD44 expression, drug resistance, and poor clinical outcome. Am. J. Pathol. 2012;180:1159–1169. doi: 10.1016/j.ajpath.2011.11.015. doi:10.1016/j.ajpath.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 42.Curley M.D., Therrien V.A., Cummings C.L., Sergent P.A., Koulouris C.R., Friel A.M., Roberts D.J., Seiden M.V., Scadden D.T., Rueda B.R., et al. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells. 2009;27:2875–2883. doi: 10.1002/stem.236. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J., Guo X., Chang D.Y., Rosen D.G., Mercado-Uribe I., Liu J. CD133 expression associated with poor prognosis in ovarian cancer. Mod. Pathol. 2011;25:456–464. doi: 10.1038/modpathol.2011.170. doi:10.1038/modpathol.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Porath I., Thomson M.W., Carey V.J., Ge R., Bell G.W., Regev A., Weinberg R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. doi:10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glinsky G.V., Berezovska O., Glinskii A.B. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. doi:10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivan C., Hu W., Bottsford-Miller J., Zand B., Dalton H.J., Liu T., Huang J., Nick A.M., Lopez-Berestein G., Coleman R.L., et al. Epigenetic analysis of the Notch superfamily in high-grade serous ovarian cancer. Gynecol. Oncol. 2012;128:506–511. doi: 10.1016/j.ygyno.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anglesio M.S., Wiegand K.C., Melnyk N., Chow C., Salamanca C., Prentice L.M., Senz J., Yang W., Spillman M.A., Cochrane D.R., et al. Type-specific cell line models for type-specific ovarian cancer research. PLoS ONE. 2013;8:e72162. doi: 10.1371/journal.pone.0072162. doi:10.1371/journal.pone.0072162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Domcke S., Sinha R., Levine D.A., Sander C., Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013;4:2126. doi: 10.1038/ncomms3126. doi:10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pastrana E., Silva-Vargas V., Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. doi:10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ponti D., Costa A., Zaffaroni N., Pratesi G., Petrangolini G., Coradini D., Pilotti S., Pierotti M.A., Daidone M.G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. doi:10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 51.Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. doi:10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 52.Du P., Zhang X., Huang C.C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. doi:10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 54.Thirlwell C., Eymard M., Feber A., Teschendorff A., Pearce K., Lechner M., Widschwendter M., Beck S. Genome-wide DNA methylation analysis of archival formalin-fixed paraffin-embedded tissue using the Illumina Infinium HumanMethylation27 BeadChip. Methods. 2010;52:248–254. doi: 10.1016/j.ymeth.2010.04.012. doi:10.1016/j.ymeth.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 55.Su P.H., Lin Y.W., Huang R.L., Liao Y.P., Lee H.Y., Wang H.C., Chao T.K., Chen C.K., Chan M.W., Chu T.Y., et al. Epigenetic silencing of PTPRR activates MAPK signaling, promotes metastasis and serves as a biomarker of invasive cervical cancer. Oncogene. 2013;32:15–26. doi: 10.1038/onc.2012.29. [DOI] [PubMed] [Google Scholar]
- 56.Eads C.A., Danenberg K.D., Kawakami K., Saltz L.B., Blake C., Shibata D., Danenberg P.V., Laird P.W. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. doi:10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. doi:10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.