Abstract
The key molecular events required for the formation of Ductal Carcinoma in Situ (DCIS) and its progression to invasive breast carcinoma have not been defined. Here we show that the nuclear receptor coactivator Amplified In Breast cancer 1 (AIB1) is expressed at low levels in normal breast but is highly expressed in DCIS lesions. This is of significance since reduction of AIB1 in human MCFDCIS cells restored a more normal 3D mammary acinar structure. Reduction of AIB1 in MCFDCIS cells, both prior to DCIS development or in existing MCFDCIS lesions in vivo, inhibited tumor growth and led to smaller, necrotic lesions. AIB1 reduction in MCFDCIS cells was correlated with significant reduction in the CD24−/CD44+ Breast Cancer Initiating Cells (BCIC) population, and a decrease in myoepithelial progenitor cells in the DCIS lesions in vitro and in vivo. Loss of AIB1 in MCFDCIS cells was also accompanied by a loss of expression of NOTCH 2, 3 and 4, JAG2, HES1, GATA3, HER2 and HER3 in vivo. These signaling molecules have been associated with differentiation of breast epithelial progenitor cells. These data indicate that AIB1 plays a central role in the initiation and maintenance of DCIS and that reduction of AIB1 causes loss of BCIC, loss of components of the NOTCH, HER2 and HER3 signaling pathways and fewer DCIS myoepithelial progenitor cells in vivo. We propose that increased expression of AIB1, through maintenance of BCIC, facilitates formation of DCIS, a necessary step prior to development of invasive disease.
Introduction
There are >50,000 new cases of Ductal Carcinoma In Situ (DCIS) diagnosed every year in the USA. In DCIS lesions the lumen of the mammary ducts are filled with proliferative malignant cells that have not invaded beyond the basement membrane of the duct into the adjacent stroma. Based on epidemiology, pathology and assessment of shared genetic changes between DCIS and adjacent invasive lesions, DCIS has been identified as the precursor lesion for invasive breast cancer1-3. Extensive comparative gene expression analyses to date have not defined the critical driver pathways that are needed for the development and maintenance of DCIS and its further progression to invasive carcinoma4,5.However, DCIS and invasive disease share a similar gene expression profile, whereas the expression profile of normal breast and DCIS are significantly different6. This suggests that many of the important signaling changes that are required for the development of invasive and even metastatic disease occurred during the formation of DCIS.
AIB1 is the third member of the Nuclear Coactivator (NCOA-3) and p160 Steroid Receptor Co-activator-3 (SRC-3) family. The AIB1 oncogene is located on chromosome 20q, a region frequently amplified in breast cancer7. AIB1 is a transcriptional co-activator that promotes the transcriptional activity of multiple nuclear receptors such as the estrogen and progesterone receptors7 and a number of other transcription factors, including E2F-1, AP-1, NFκB, and STAT68. Multiple studies have shown that the AIB1 gene is amplified and overexpressed in human breast cancer9. High levels of AIB1 mRNA or protein predict significantly worse prognosis and overall survival in breast cancer patients9. Transgenic mice expressing high levels of the human AIB1 transgene developed mammary hyperplasia and tumors of the mammary gland10. Conversely, loss of AIB1 expression in the mammary gland prevents RAS- or HER2-induced mammary tumor development11,12. Transgenic mice expressing high level of an AIB1 isoform in the mammary gland resulted in the development of DCIS and fibrosis13. In the current study we determined that AIB1 is highly expressed in human DCIS samples. Based on these observations, we hypothesized that AIB1 could be important in the early stages of breast cancer notably in the development and maintenance of DCIS. We have examined the role of AIB1 in a well-described model of human DCIS, MCFDCIS.com (MCFDCIS) cells, which are a derivative of premalignant MCF-10AT cells, that when implanted in mice produce discrete DCIS lesions that rapidly progress to invasive disease14. The gross pathology of the lesions and the gene expression changes observed in the DCIS stage of MCFDCIS after implantation mirror those seen in human DCIS samples15. Using this model, we demonstrate that AIB1 is critical for the formation and maintenance of DCIS. AIB1 is required to maintain the BCIC population and the myoepithelial progenitor cell population in the MCFDCIS cells in vitro and in vivo. In parallel, AIB1 also maintains levels of NOTCH and HER2/HER3 signaling molecules in the DCIS lesions. These pathways are known to be involved in maintaining BCIC populations. Based on these data, we propose that preventive strategies aimed at reducing AIB1 in the mammary gland could be selective and effective in reducing DCIS incidence and consequently decreasing the overall incidence of invasive breast cancer.
Results
AIB1 expression in human DCIS
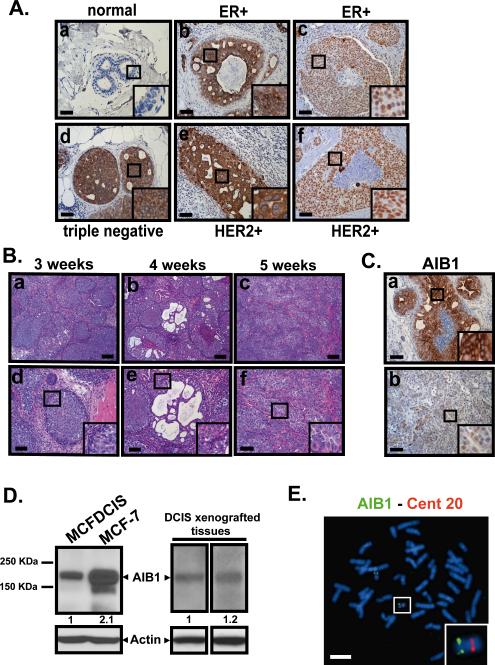
To determine the expression levels of AIB1 in human DCIS, we obtained paraffin-embedded sections of DCIS patient samples. We determined that the AIB1 levels in normal breast ducts, detected by immunohistochemistry (IHC), were very low (Fig. 1A panel a). In contrast, strong AIB1 expression was observed in the luminal cells of DCIS lesions from patients with invasive breast cancer (Fig. 1A panels b-f). Estrogen receptor positive (ER+), human epidermal growth factor receptor 2 positive (HER2+) and ER−, HER2− and progesterone receptor negative (PR−) ‘triple negative’ invasive breast cancer samples all showed strong AIB1 staining (Fig. 1A panels b-f). Strong AIB1 staining was also observed in breast tissue samples from DCIS patients that had no evidence of invasive disease (Fig. S1A panels a-d). The staining for AIB1 in the DCIS samples was high in the nuclear or cytoplasmic compartments but patterns of AIB1 subcellular distribution did not appear to be associated with a particular breast cancer subtype. Previous reports have shown that AIB1 and its isoform can traverse between different cellular compartments depending on, stage of cell cycle and cellular expression of other coregulators16-18. Overall, our IHC results indicate that AIB1 protein is highly expressed at very early stages of human breast cancer progression prior to development of Stage 1 invasive disease, and this suggests that AIB1 could function in an early stage of breast cancer formation.
Fig. 1. AIB1 expression in DCIS.
(A) Representative images of IHC staining using AIB1 antibody (5E11, Cell Signaling Technology, 1:75) in normal breast (a) and human comedo DCIS associated with invasive lesions obtained from ER+ (b,c), triple negative (d) and HER2+ specimen (e,f). (B) H&E stained images of tissue sections from MCFDCIS xenograft tumors at 3, 4 and 5 weeks. (a-c) scale bar = 0.2 mm. (d-f) scale bar = 0.1 mm. (C) Representative images of IHC staining for AIB1 in MCFDCIS xenograft tumor tissues. Scale bar= 0.1 mm. (D) Western blot analysis using AIB1 (5E11, Cell Signaling Technology, 1:1000) and β-actin (C4 Millipore, 1:3000) antibodies in protein lysates from MCFDCIS and MCF-7 cells in vitro (left) and from MCFDCIS tumor tissues (right). (E) Representative metaphase spread of MCFDCIS cells hybridized with bacterial artificial chromosome clones (RP11-1151C1 and RP11-976F15) containing sequences of the AIB1 gene located at 20q12. Note two copies of the AIB1 gene (green signals) and centromeric control probe for chromosome 20 (RP11-90H19)(red signals). Scale bar= 20 μm. Insets outline a zoom-in of the framed areas.
To determine the role of AIB1 in early stage breast cancer we utilized the MCFDCIS cell model of breast cancer14. When implanted subcutaneously MCFDCIS cells give well defined DCIS lesions that progress to invasive disease within 3-5 weeks19(Fig. 1B). The ductal lesions that develop after 3 weeks implantation of MCFDCIS cells resemble high grade comedo DCIS (Fig. 1B panels a, b, d, e) and both luminal and myoepithelial cell layers of DCIS lesions are generated by the progenitor MCFDCIS cells14(Fig. 1B). Invasive carcinoma develops from the DCIS lesions between weeks 3-5 after implantation of cells (Fig. 1B panel c and f). Similar to the human DCIS samples strong AIB1 staining was found in the MCFDCIS lesions 3-weeks after implantation (Fig. 1C upper panel and Fig. S1B). The presence of AIB1 protein was confirmed by Western blot analysis of MCFDCIS cells in vitro (Fig. 1D left) and in MCFDCIS tumor extracts (Fig. 1D right). AIB1 was also detected in the invasive lesions that developed from the MCFDCIS cells in vivo although the staining was less intense than in the DCIS lesions (Fig. 1C lower panel). FISH analysis revealed that the MCFDCIS cells do not have amplified copies of either AIB1 (Fig. 1E) or HER2 (Fig. S1C). The MCFDCIS lesions are PR-negative (Fig. S1D) and have low levels of ERα (Fig. S2A and B). MCFDCIS tumor growth is estrogen-independent since it is unaffected by ovariectomy (Fig. S2C and D) or by treatment of mice with the estrogen antagonist fulvestrant (Fig. S2E-G). These observations are consistent with a previous classification of the MCFDCIS lesions as a basal form of human DCIS15.
AIB1 has a role in the maintenance of DCIS multi-acinar structures in a 3D matrix
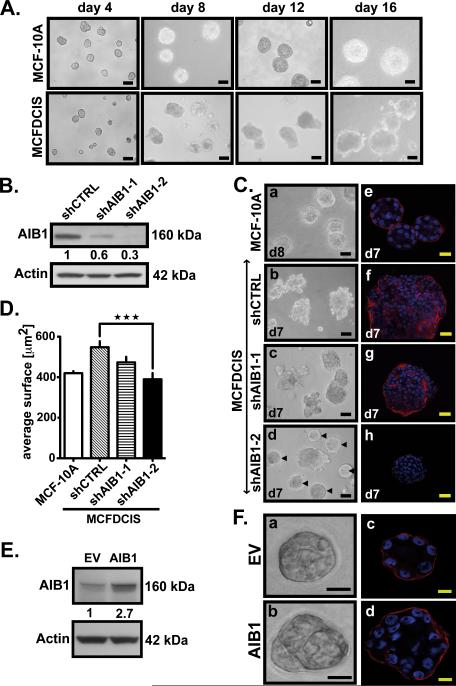
We first examined the impact of changing AIB1 levels in mammary acinar 3D structures. Normal mammary epithelial cells (MEC) cells, such as MCF-10A, grown in Matrigel™, recapitulate features of normal breast, including polarized acini, and deposition of basement membrane characterized by laminin V. After a number of days of growth, the acini develop a hollow lumen characteristic of the lumen seen in mammary acini in vivo20(Fig. 2A upper panel and Fig. 2C panels a and e). The overexpression of oncogenes, such as HER2, disrupts normal mammary acinar structure and instead larger, multi-acinar structures are observed, with filled lumen and irregular deposition of basement membrane proteins21. The MCFDCIS cells grown in Matrigel™ showed multi-acinar structures, with no lumen development and with deposition of laminin throughout the multi–acinar structure (Fig. 2A lower panel, Fig. 2C panels b and f). To determine the impact of AIB1 on maintenance of the DCIS phenotype in Matrigel™, we infected MCFDCIS cells with a lentiviral vector expressing two distinct AIB1 shRNAs (shAIB1-1 and shAIB1-2) that lowered the endogenous AIB1 levels by 40% and 70% respectively (Fig. 2B). Reduction in AIB1 levels after shAIB1-1 infection showed some changes in laminin V distribution to the periphery of the acini (Fig. 2C panel g), but with the more effective shRNA AIB1-2 a more normal spheroid acinar structure was observed (Fig. 2C panels d and h). The average spheroid size of MCFDCIS shAIB1-2 spheroids was also significantly smaller than MCFDCIS shCTRL and similar to the average size seen in MCF-10A (Fig. 2D). Consistent with this, overexpression of AIB1 in MCF-10A (Fig. 2E) caused loss of polarization and an increase in highly disorganized acini in 3D culture (Fig. 2F panel b and d). Interestingly, in the MCFDCIS shAIB1 spheroids we observed Caspase 3-positive apoptotic cells especially in the center of the sphere (Fig. S3A) and an increased expression of the apoptotic marker BIM EL and the cell cycle inhibitor p21 (Fig. S3B). Changes in expression of these molecules have been associated with resumption of a more normal mammary acinar structure with the hollow lumen being created by increased apoptosis20. Overall, the data indicate that high AIB1 levels in the MCFDCIS cells are required to maintain a disorganized multi-acinar structure in Matrigel™ and reduction of AIB1 levels can partially restore normal mammary acinar structure in MCFDCIS cells.
Fig. 2. Decrease in AIB1 levels results in a reversion of MCFDCIS acini to more normal acinar structures in 3D culture.
(A) Representative phase-contrast images of MCF-10A cells (upper) and MCFDCIS cells (lower) cultivated on Matrigel™ at day 4, 8, 12 and 16. Acini were analyzed and photographed using an Olympus IX-71 Inverted Epifluorescence Scope and DP controller Software version 3.1.1.267. Scale bar= 0.1 mm. (B) Western blot analysis of AIB1 and β-actin expressions in MCFDCIS cells infected with control or AIB1 shRNAs. (C) Representative phase-contrast images of MCF-10A acini at day 8 (a) and MCFDCIS acini infected with lentiviral shRNA control or AIB1 shRNA at day 7 (b) to (d). Normal-appearing acinar structures are indicated by arrows in (d). Scale bar= 0.1 mm. Representative immunofluorescence pictures of MCF-10A acini at day 8 (e) and MCFDCIS acini expressing control (f) or AIB1 shRNAs at day 7 (g, h) stained with laminin V antibody (Millipore, 1:100)(red signals) and with Dapi (blue signals), and analyzed using an Olympus FV300 Confocal microscope equipped with Fluoview 300 Software. Scale bar= 20 μm. (D) Average size of MCF-10A and MCFDCIS acini determined by microscopic area measurements (Nuance software ; n=100 ; ***, p<0,0001 vs. control). (E) Western blot analysis using AIB1 and β-actin antibodies in protein lysates from MCF-10A cells infected with a lentivirus containing AIB1 or control vector. AIB1 protein levels indicated under the western blot were quantified using ImageJ 1.33u software and normalized to β-actin. (F) Representative phase-contrast (a,b) and immunofluorescence images (c,d) of MCF-10A acini overexpressing AIB1 (b,d) or not (a,c) at day 10. Acini were stained for Laminin V (red signals) and with Dapi (blue signals). Scale bar= 10 μm.
AIB1 is required for the formation of DCIS lesions in vivo
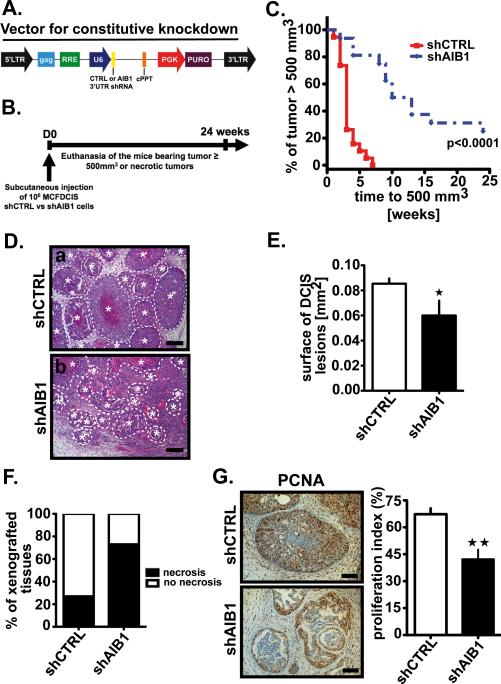
We next determined if the loss of AIB1 in MCFDCIS cells could affect MCFDCIS tumor development in vivo. To examine this, we infected the MCFDCIS cell line with a lentiviral vector (Fig. 3A). In these pooled selected cells, AIB1 mRNA and protein levels are constitutively depleted (Fig. S4A and B). Cells were injected subcutaneously into nude mice and the tumors monitored for up to 24 weeks or to the time that they exceeded the size of 500 mm3 (Fig. 3B). The control-infected cells grew rapidly and all of the tumors had reached 500 mm3 volume by 7 weeks after implantation (Fig. 3C). In contrast the AIB1 shRNA infected cells grew slowly and 20% of the tumor injection sites had not developed lesions>500 mm3 by 24 weeks (Fig. 3C). Tumors from the control animals had the characteristic comedo DCIS lesions (Fig. 3D upper panel). DCIS lesions were also observed in the shAIB1 cells, but they were of a significantly smaller area (Fig. 3D lower panel and Fig. 3E), with less overall prevalence (Fig. S4C), and more necrosis than the control (Fig. 3F). We also observed a decrease in the number of proliferative cells in the MCFDCIS AIB1 shRNA lesions as determined by the quantitation of overall PCNA staining (Fig. 3G) and the percentage of p21-positive cells (Fig. S4D). These data indicate that MCFDCIS cells with low AIB1 levels will not progress to invasive tumors as rapidly as the control cells. Furthermore, the DCIS lesions that develop in the presence of lower levels of AIB1 will be smaller and more necrotic.
Fig. 3. AIB1-depleted MCFDCIS cell xenografts show increased necrosis, decreased proliferation and delayed tumor formation.
(A) Lentiviral vector scheme for constitutive control and AIB1 shRNAs directed at human 3’ UTR sequences. CAG, CMV early enhancer/chicken β actin promoter; RRE: Rev responsive element; U6, PolIII promoter; cPPT: Central polypurine tract helps; PGK: phosphoglycerate kinase ; Puro: puromycin resistant gene. (B) Schematic representation of the constitutive AIB1 knockdown MCFDCIS xenograft mouse model. Tumor volume was measured twice weekly with a caliper. Mice were euthanized by CO2 inhalation when the tumor size reached 500 mm3, were necrotic or 24 weeks after cell implantation. (C) Kaplan-Meier curves showing the percentage of tumors with size ≥ 500 mm3. Control tumors (shCTRL, n=19), AIB1-deficient tumors (shAIB1, n=16). Logrank test, p<0.0001 vs. control. (D) Representative H&E stained images of tissue section of shCTRL (a) and shAIB1 (b) xenograft tumors. Lesions containing proliferating MCFDCIS cells and surrounded by a thick basement membrane and a layer of myoepithelial cells were defined as DCIS lesions (highlighted by the dotted line ✶). Scale bar=0.2 mm. (E) Measurement of the DCIS lesion surface area in representative shCTRL or shAIB1 xenografted tissue sections using Image J software. Mean * SEM (shCTRL, n=15; shAIB1, n=11; *, p<0.01 vs. control). (F) Percent of tumors exhibiting macroscopic necrosis in AIB1 deficient (shAIB1, n= 11) or control (shCTRL, n=15) tumors. (p<0.01 vs. control by chi-square test). (G) Representative IHC pictures of control or AIB1-deficient tumor sections stained with PCNA antibody (Dako, 1:13,000)(left) and proliferation index determined by measuring the percentage of positive cells per field in at least five fields non-overlapping using Photoshop CS3 software (right). Scale bar= 0.1 mm. Mean ± SEM (shCTRL, n=15; shAIB1, n=11; **, P<0.001 vs. control).
AIB1 is required for the maintenance of DCIS lesions in vivo
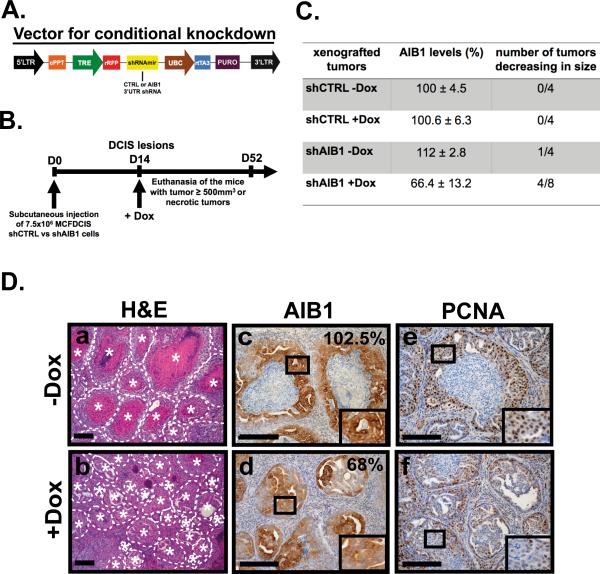
We next asked whether reduction of AIB1 in MCFDCIS cells would impact on the maintenance of DCIS lesions once they have formed. We developed a MCFDCIS derivative line infected with lentivirus that harbored the control scrambled shRNA or shRNA AIB1-2 as well as the turbo red fluorescent protein (tRFP) under the control of a tetracycline inducible promoter (Fig. 4A). Thus, doxycycline (Dox) induction of tRFP and shRNA expression can be detected in MCFDCIS cell culture (Fig. S5A) and by in vivo imaging (Fig. S5B). The administration of Dox causes reduction of AIB1 mRNA and protein in vitro (Fig. S5C and D). After implantation of these cells subcutaneously into nude mice, DCIS lesions were allowed to develop for 2 weeks and then doxycycline was included in the diet (Fig. 4B). The level of AIB1 expressed in the tumors was determined by IHC. The AIB1 levels in the shCTRL tumors was unaffected by doxycycline treatment (Fig. 4C). All the MCFDCIS control cells grew rapidly irrespective of doxycycline treatment (Fig. 4C). In contrast induction of AIB1 shRNA in MCFDCIS by doxycycline resulted in reduced AIB1 protein expression in MCFDCIS shAIB1 tumors (Fig. 4C and D panel d) and a decrease in size in 50% of the tumors (Fig. 4C). The tumors that developed from the AIB1-depleted MCFDCIS cells had smaller and more necrotic lesions (Fig. 4D panel b) as well as an overall decrease in number of PCNA-positive cells compare to the controls (Fig. 4D panel f). Overall the data in both the constitutive and conditional AIB1 shRNA expressing systems suggest that AIB1 is required for the establishment and maintenance of DCIS.
Fig. 4. Conditional depletion of AIB1 alters DCIS lesions and causes a reduction or disappearance of DCIS xenograft tumors.
(A) Lentiviral vector scheme. cPPT: central polypurine tract ; TRE: tet-inducible promoter ; rRFP: turbo RFP gene; UBC: human ubiquitin C promoter; rtTA3: reverse tet-transactivator; Puro: puromycin resistant gene. (B) Experimental scheme of using a conditional AIB1 knockdown MCFDCIS xenograft mouse model. Mice were fed with a normal or doxycycline-containing diet (200 mg/kg) after the establishment of DCIS lesions (2 weeks after injection). Fourteen days after doxycycline treatment, the mice were imaged using a CRi Maestro Imaging System. Mice were euthanized when the tumor size reached greater than 500 mm3, were necrotic or at 52 days after cell implantation. (C) Summary table of AIB1 levels shown as percentage of the shCTRL group (mean value set at 100%) and number of tumors decreasing in size after doxycycline treatment. Mean ± SEM (shCTRL –Dox, n=4; shCTRL +Dox, n=4; shAIB1 –Dox, n=4 ; shAIB1 +Dox, n=8). (D) Representative cross-sections of H&E staining (a, b), AIB1 staining (c, d) and PCNA staining (e, f) in xenografted tumor tissues from MCFDCIS shAIB1 cells treated with doxycycline (b, d, f) or not (a, c, e). DCIS lesions are highlighted by the dotted line and |. Percentage of AIB1 positive-cells is indicated for each AIB1-stained tissue section (upper right corner). Insets outline a zoom-in of the framed areas. Scale bar= 0.2 mm.
Loss of AIB1 causes reduction in the BCIC population of MCFDCIS and affects MCFDCIS cell differentiation
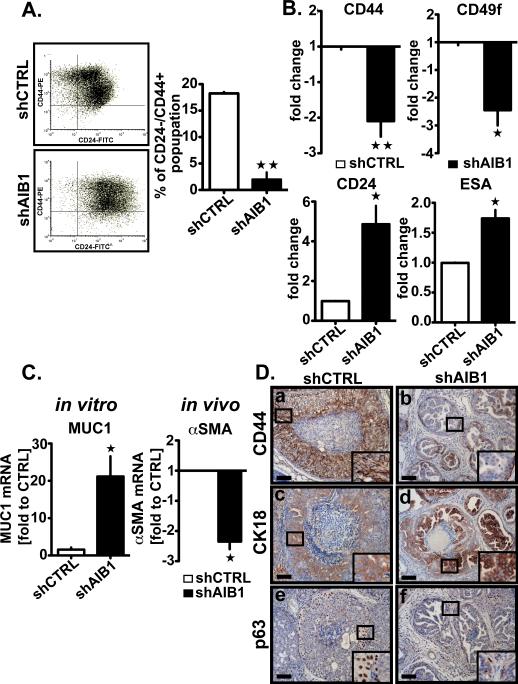
To investigate how AIB1 affects initiation and progression of DCIS, we compared global gene expression changes in MCFDCIS cells +/−AIB1 shRNA in vitro using cDNA array analysis. Notable among the most significant expression changes between the two groups were genes that are differentially expressed in BCIC such as CD2422 and members of the NOTCH signaling pathway (DLL1 and DLL3)23(Table S1). Luminal progenitor marker expression, such as Mucin 1 (MUC 1)24, was also increased (Table S1). To determine if these expression changes impacted the population of BCIC, we FACS sorted MCFDCIS +/− AIB1 shRNA cells for CD24−/CD44+ (Fig. 5A). Reduction of AIB1 levels caused the percentage of CD24−/CD44+ cells in the MCFDCIS population to drop substantially from 18% to <3 % (Fig. 5A). Consistent with a loss of BCIC, the CD44 and CD49f levels were also significantly reduced in MCFDCIS AIB1shRNA cells (Fig. 5B upper panel). In contrast, the level of expression of CD24 and ESA on their cell surface was increased (Fig. 5B lower panel). There was also an increase in overall expression of MUC1 mRNA in the AIB1shRNA MCFDCIS cells (Fig. 5C left panel). CD24 and ESA, like MUC1 are considered luminal cell markers and the gain in their expression after AIB1 reduction suggested that there were changes in relative levels of progenitor luminal vs. progenitor myoepithelial cells in MCFDCIS population. Consistent with a role for AIB1 in BCIC maintenance, overexpression of AIB1 in MCF-10A cells led to increased sphere formation and increased expression of CD44 and CD49f (Fig. S6 A and B).
Fig. 5. Depletion of AIB1 reduces the tumor initiating cell subpopulation and affects the differentiation of MCFDCIS cells in vitro and in vivo.
(A) Representative fluorescence-activated cell sorting scatter plots of CD44+/CD24− cells (left), and the average percentage of the CD44+/CD24-subpopulation in shCTRL and shAIB1 MCFDCIS cells (right). Data were analyzed using a Facstar-Plus Dual Laser flow cytometer (Becton Dickinson) equipped using FlowJo (version 7.6.1). Mean ± SEM (n=3; **, p<0.001 vs. control). (B) Expression of tumor initiating cells markers CD44 (upper left) and CD49f (upper right), and luminal epithelial cells markers, CD24 (lower left) and ESA (lower right) analyzed by flow cytometry in MCFDCIS cells infected with lentiviral shCTRL or shAIB1. Mean ± SEM (n=3; *, p<0.01 vs. Control ; **, p<0.001 vs. Control). (C) qRT-PCR analysis of MUC1 mRNA levels in MCFDCIS cells depleted or not of AIB1 (left) and αSMA mRNA levels in the constitutive AIB1 knockdown MCFDCIS xenograft mouse model (right). Mean ± SEM (n=3; *, p<0.01 vs. control). (D) Representative IHC images of tissue section of xenografted tumors constitutively depleted or not of AIB1 stained with CD44 (SPM521, Thermo Scientific, 1:250)(a,b), CK18 (Clone E431-1, Thermo Scientific Pierce, 1:200)(c,d) and p63 (4A4, Santa Cruz Biotechnology, 1:200)(e,f) antibodies. Insets outline a zoom-in of the framed areas. Scale bar= 0.1 mm.
To determine if the changes in BCIC cell populations were observed after AIB1 reduction in vivo we stained MCFDCIS tumors obtained from the constitutive and conditional mouse models for CD44, CK18 and p63 expression (Fig. 5D and Fig. S6C). A significant reduction in overall CD44 staining in the smaller lesions that developed in MCFDCIS AIB1 shRNA cells implanted in vivo is also consistent with BCIC loss (Fig. 5D panel b and Fig. S6C panel b). Interestingly, in the MCFDCIS AIB1 shRNA lesions, there was a gain in vivo in the CK18 staining, a marker of differentiated luminal epithelium (Fig. 5D panel d, Fig. S6C panel d, and Fig. S6D). In contrast, myoepithelial progenitor cells were significantly reduced in AIB1 shRNA, cells, indicated by loss of overall expression of αSMA mRNA in vitro (Fig. 5C right panel) and loss of p63-positive cells in vivo (Fig. 5D panel f and Fig. S6E and F).
In summary, the loss of AIB1 in MCFDCIS cells in vitro and in vivo reduces the BCIC and myoepithelial progenitor populations and also prevents the progression of the remaining luminal cells to invasive cancer.
Disruptions in NOTCH and HER signalings are observed in MCFDCIS AIB1 shRNA cells and tumors
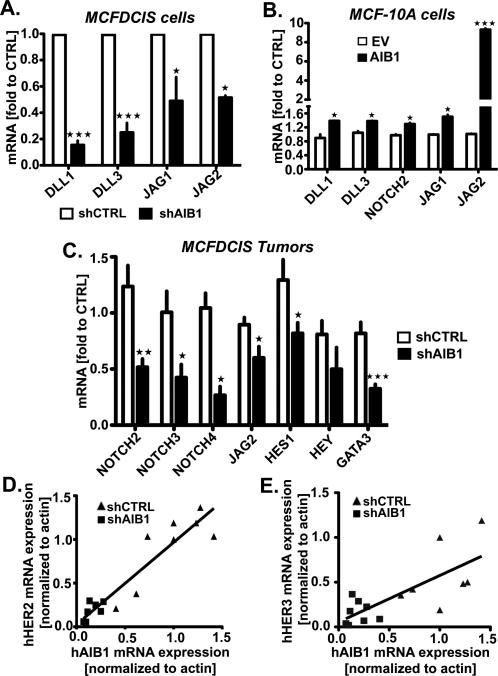
cDNA array analysis of gene expression changes induced by loss of AIB1 in MCFDCIS cells indicated that NOTCH ligands (DLL1 and DLL3) were significantly downregulated (Table S1). This was of interest since the NOTCH signaling pathways has been shown to be involved in BCIC cell maintenance and differentiation23. We therefore examined the impact of reduction in AIB1 expression in MCFDCIS cells in vitro and in vivo on the gene expression pattern of a panel of NOTCH signaling molecules. Downregulation of DLL1, DLL3, JAG 1 and JAG2 mRNA levels was confirmed by real-time PCR in MCFDCIS cells upon shRNA AIB1 expression (Fig. 6A). Consistent with this, overexpression of AIB1 in MCF-10A results in an increase in DLL1, DLL3, NOTCH 2,JAG1 and JAG 2 mRNA levels (Fig. 6B). In the MCFDCIS tumors we also observed JAG2, NOTCH 2, 3 and 4 mRNA levels downregulated (Fig. 6C) as well as the NOTCH target genes HES1 and GATA3 (Fig. 6C). Previous reports have indicated that HER2 and HER3 control the level of NOTCH signaling in BCIC23. Thus it was possible that changes in AIB1 levels were affecting HER2 and HER3 signaling and this in turn was altering expression of members of the NOTCH pathway. We determined that HER2 and HER3 mRNA and protein were detectable by PCR and Western blot analysis in MCFDCIS cells and tumors (Fig. S7A and Fig. 6D and E). A significant reduction in HER2 and HER3 mRNA and protein levels are observed in the MCFDCIS tumors after reduction in AIB1 levels (Fig. 6D and E, and Fig.S7 C-F) while EGFR protein levels are unchanged (Fig. S7G). Furthermore, a clear positive correlation between AIB1 mRNA and HER2 and HER3 mRNA levels was found in the tumors (Fig. 6D and E). Consistent with this, we demonstrated that HER2 mRNA and protein are upregulated in MCF-10A cells infected with lentivirus expressing AIB1 (Fig. S7H). In summary, reduction in AIB1 levels in MCFDCIS tumors in vivo leads to loss of HER2, HER3 as well as loss of NOTCH signaling pathway members. These changes in gene expression are paralleled by a significant loss in BCIC population and myoepithelial progenitor cells.
Fig. 6. AIB1 regulates NOTCH and HER signaling pathways in MCFDCIS xenografts.
(A) qRT-PCR analysis of DLL1, DLL3, JAG1, and JAG2 in MCFDCIS cells deficient (n=3) or not (n=3) in AIB1, (B) DLL1, DLL3, NOTCH2, JAG1 and JAG2 in MCF-10A overexpressing AIB1 (n=3) or not (n=3), and (C) NOTCH2, NOTCH3, NOTCH4, JAG2, HES1, HEY, and GATA3 in shCTRL (n=15) or shAIB1 (n=11) MCFDCIS xenografted tissues. Mean ± SEM (n≥3; *, p<0.01 vs. control; **, p<0.001 vs. control; ***, p<0.0001 vs. control). (D) Scatter plot representation of the correlation between human AIB1 (hAIB1) and human HER2 (hHER2) mRNA expression or (E) human HER3 (hHER3) mRNA expressions analyzed by qRT-PCR in xenografted tumors from MCFDCIS shCTRL (n=15) or shAIB1 (n=11). The solid lines show the positive linear regression fit. Goodness of fit (A, r2=0,8771; B, r2 0,5519).
Discussion
This is the first report that defines a role for AIB1 in the development and maintenance of human DCIS. Although AIB1 is known to be involved in the proliferation of a number of different epithelial tumors25, the role of AIB1 in the maintenance of BCIC has not been reported previously. The loss of the BCIC population in MCFDCIS tumors with reduced levels of AIB1, most likely leads to the reduced size of DCIS lesions and the loss of overall MCFDCIS tumor burden due to increased necrosis. While increased necrosis is often seen in the lumen of DCIS lesions in vivo, the necrosis we observed was more associated with the inability to sustain the DCIS cells thus inhibiting their progression to invasive lesions. Interestingly reduction of AIB1 after formation of DCIS lesions also causes formation of smaller lesions, reduced tumor burden and increased necrosis. This suggests that sustaining the BCIC population is required throughout DCIS development, not just after initial implantation of the cells in vivo. This has important therapeutic implications since it suggests that a reduction of AIB1 or inhibition of AIB1 signaling pathways involved in the maintenance of BCIC would hamper development of DCIS. It should be noted that only 20-30% of human DCIS is thought to progress to invasive disease26 and this raised the question of the role of AIB1 in progression of DCIS to invasive disease. Although MCFDCIS cells can give rise to invasive lesions in vivo, they have low motility in a Boyden chamber invasion assay (Fig. S8A) and low invasion potential in an endothelial layer in an ECIS assay compared to invasive MDA-MB-231 cells (Fig. S8B). It is also known that MCFDCIS cells have low cell velocity in the wound healing assay27. Consistent with this low motile potential of MCFDCIS, no micrometastases were observed after tumor formation in vivo (Fig. S8C). We found that reduction in AIB1 did not affect these minimal phenotypes of MCFDCIS cells in either the in vitro invasion or motility assays (Fig. S8A and B). However, AIB1 may have an additional role in epithelial cell invasion in the context of stroma that is not apparent in these assays. Since AIB1 is highly expressed in the patient DCIS samples that we have examined to date, and its loss prevents MCFDCIS invasive tumor development, we can conclude that AIB1 overexpression is necessary primarily for development and maintenance of DCIS through preserving the BCIC population, but further genetic changes are most likely necessary for the development of the invasive phenotype in a subset of DCIS.
Our analysis of gene expression controlled by changes in AIB1 levels in MCFDCIS cells revealed that members of the NOTCH pathways are regulated by AIB1. This was confirmed by analysis of the in vivo samples although interestingly the patterns of changes in NOTCH signaling molecules in vivo did not parallel the changes seen in vitro. Although NOTCH1 was not regulated by AIB1 in vitro or in vivo, notably NOTCH 2, 3 and 4, and JAG 2 were significantly downregulated in vivo. We conjecture that the regulation of NOTCH by AIB1 in vivo requires crosstalk with stromal components that are vital to the maintenance of DCIS in vivo. NOTCH signaling has been implicated in both restricting mammary stem cell expansion and has been associated with commitment of mammary stem cells to the luminal lineage, leading to hyperplasia and tumor formation28. In implanted MCFDCIS in which AIB1 was lowered we did observe a decrease in hyperplasia and a decrease in the number of tumors although we did not see eradication of luminal cells which might be expected with a significant reduction in NOTCH signaling. Interestingly, recent reports have demonstrated that the NOTCH pathway can be controlled by HER family signaling in breast cancer stem cells29 and in DCIS30. AIB1 is required for HER2 mediated mammary tumor formation in vivo11,12 and high levels of AIB1 and HER2 have been associated with worse prognosis and tamoxifen resistance in human breast cancer9. Overexpression of HER2 is seen in significant portion of DCIS cases estimated at 50-80%31. However, an association between AIB1 and HER2 in the development of DCIS and its progression to invasion has not been established. In our gene expression analysis of +/−AIB1 in MCFDCIS cells in vitro, major changes in expression of HER family members HER1, 2 and HER3 were not observed. However, in vivo a reduction in AIB1 led to loss in HER2 and HER3 expression, again suggesting the importance of stromal crosstalk in determining levels of gene expression in DCIS. HER2 signaling can regulate mammary stem cell renewal32 and loss of HER2 signaling controlled by loss of AIB1 could explain the significant loss of BCIC in vivo. Interestingly, HER3 has recently been shown to play a role in maintaining luminal epithelium cells and loss of HER3 signaling causes a switch to mammary basal epithelium gene expression patterns33. Thus, the loss of HER3 signaling in vivo may also play a role in overall loss of luminal cells, although it would not explain the decrease in the myoepithelial progenitor layers in the AIB1 depleted lesions. We tried to rescue BCIC loss caused by reduced AIB1 with overexpressed HER2 but were unable to increase HER2 expression in the shAIB1 cells (Fig. S7I), presumably because the CMV promoter regulating HER2 expression in the lentiviral vector is also dependent on AIB1. Nevertheless, the data in Fig. 6 and Fig. S7 strongly implicate a connection between AIB1, HER2/3 and NOTCH in development and maintenance of DCIS.
Previous studies in transgenic mice indicated that overexpression of AIB1 can lead to DCIS13 and we now show that human DCIS has high expression of AIB1 at the protein level. 5-10% of breast cancers harbor an AIB1 amplicon7 although high AIB1 levels can be observed in the absence of gene amplification in up to 60% of breast cancer. AIB1 protein levels are regulated by multiple post-transcriptional mechanisms that control steady state levels of AIB1 during the transformation process8. AIB1 levels in cells can be downregulated by estrogen34. However, loss of estrogen regulation does not seem to be the explanation of high AIB1 in DCIS since we observed high AIB1 levels in ER+ and ER− DCIS as well as HER2+ and HER2-DCIS. A previous study examining ERα knock out mice demonstrated that ER signaling was necessary for the formation of DCIS35. Our group also published a study in which development of DCIS and invasive mammary cancer was correlated with over expression of AIB1 with ERα in transgenic mice13. The present study extends these observations demonstrating that AIB1 over-expression can also contribute to the development of the basal form of DCIS. Thus the contribution of AIB1 over-expression to DCIS development and maintenance is not breast cancer sub-type specific. In conclusion, regardless of the mechanism of upregulation of AIB1 in human breast epithelium, our data indicate a critical role for this event in the development and maintenance of DCIS lesions in vivo through preservation of BCIC in part through NOTCH, HER2 and HER3 signaling. Our data suggest that selective degradation of AIB1 in early breast cancer lesions could be a useful therapeutic approach to prevent the development and maintenance of DCIS, thus reducing the overall incidence of invasive breast cancer.
Materials and Methods
Human Tissue Samples and Mice
De-identified formalin-fixed, paraffin-embedded tissues from normal breast reduction mammoplasty and DCIS were obtained from Georgetown University tissue bank shared resource. Athymic nude mice were obtained from Harlan Laboratories, were maintained in the Georgetown University's animal facility and animal experiments were conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee.
Cell lines
We thank Dr Fred R. Miller of the Karmanos Cancer Institute for the gift of the MCFDCIS and MCF-10A cell lines. These cells were maintained as described previously36,37.
Histology analysis
Hematoxylin and eosin, immunohistochemistry and immunofluorescence analyses were performed on paraffin-embedded 5 μm sections using standard protocols described elsewhere12.
Western Blot
Whole-cell extracts were isolated from cultured MCFDCIS, MCF-7 and MCF-10A cells or DCIS tumor samples, and immunoblot assays were performed as described previously38.
Fluorescence In Situ Hybridization (FISH) analysis
FISH hybridization and analysis were performed in the MCFDCIS cells using a standard protocol previously described39,40.
Short Hairpin RNA Constructs and lentivirus Infection
For gene down-regulation studies, short hairpin RNAs (shRNAs) against AIB1 gene (shRNA#1 5’GCAGTCTATTCGTCCTCCATA3’, shRNA#2: 5’TGGTGAATCGAGACGGAAACA 3’,) were cloned into PLKO.1 lentiviral vector (Addgene) as described in41 or into XhoI and EcoRI restriction sites in Doxycycline-inducible lentiviral expression vector pTRIPZ (Open Biosystems). A scrambled shRNA was used as a control (Addgene). For gene overexpression, the coding sequence of human AIB1 was cloned into AscI and PacI restriction sites in the lentiviral expression vector pCDF1-MSC2-EF1-Puro (System Biosciences). Lentivirus production and infection were performed as described elsewhere36,42.
Xenografts
MCFDCIS xenograft models were generated as detailed in Fig. 3B and 4B following the protocol described presviously15.
Analysis of three-dimensional Matrigel™ cultures by Bright-field and Confocal Microscopy
3D culture assays were performed as previously described36,37. For confocal microscopy, immunostaining of the acinar structures was carried out as described elsewhere43.
Quantitative RT-PCR
Real-time quantitative RT-PCR analyses of total cellular RNA from MCFDCIS cells, MCF-10A cells and DCIS xenografted tissues was carried out as previously described41. All of the primers used for quantitative RT-PCR are given in Table S2. The results were calculated by the comparative CT method, with relative transcript levels determined as 2-ΔΔCT and were normalized using actin gene44.
Flow Cytometry Analysis
MCFDCIS cells were stained with the following antibodies: anti-CD24-488, anti-CD44-PE, anti-CD49f-APC, anti-ESA-488, and anti-CD10-PE (BioLegend). FACS analysis was performed in the Lombardi Comprehensive Cancer Center Flow Cytometry Shared Resource.
Statistics
All experiments have been carried out at least three times. Statistical differences and linear regression analysis were performed with GraphPad Prism software (Graph-Pad Software, Inc., San Diego, CA, USA). The significance of changes was assessed by the application of an unpaired Student's t-test, with p<0.05 considered statistically significant unless stated otherwise.
Supplementary Material
AKNOWLEDGEMENTS
This work was funded by the Department of Defense Breast Cancer Research Program BC098103 (VO) and by NIH/NCI grants CA113477 (ATR). This project was also supported by Award Number P30CA051008 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Defense.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Radford DM, Phillips NJ, Fair KL, Ritter JH, Holt M, Donis-Keller H. Allelic loss and the progression of breast cancer. Cancer Res. 1995;55:5180–5183. [PubMed] [Google Scholar]
- 2.Stratton MR, Collins N, Lakhani SR, Sloane JP. Loss of heterozygosity in ductal carcinoma in situ of the breast. J Pathol. 1995;175:195–201. doi: 10.1002/path.1711750207. [DOI] [PubMed] [Google Scholar]
- 3.Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, et al. In situ analyses of genome instability in breast cancer. Nature Genetics. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 4.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–375. [PubMed] [Google Scholar]
- 6.Espina V, Liotta LA. What is the malignant nature of human ductal carcinoma in situ? Nature Reviews. 2011;11:68–75. doi: 10.1038/nrc2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 8.York B, O'Malley BW. Steroid receptor coactivator (SRC) family: masters of systems biology. J Biol Chem. 2010;285:38743–38750. doi: 10.1074/jbc.R110.193367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lahusen T, Henke RT, Kagan BL, Wellstein A, Riegel AT. The role and regulation of the nuclear receptor co-activator AIB1 in breast cancer. Breast Cancer Res Treat. 2009;116:225–237. doi: 10.1007/s10549-009-0405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres-Arzayus MI, Font de Mora J, Yuan J, Vazquez F, Bronson R, Rue M, et al. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Kuang SQ, Liao L, Wang S, Medina D, O'Malley BW, Xu J. Mice lacking the amplified in breast cancer 1/steroid receptor coactivator-3 are resistant to chemical carcinogen-induced mammary tumorigenesis. Cancer Res. 2005;65:7993–8002. doi: 10.1158/0008-5472.CAN-05-1179. [DOI] [PubMed] [Google Scholar]
- 12.Fereshteh MP, Tilli MT, Kim SE, Xu J, O'Malley BW, Wellstein A, et al. The nuclear receptor coactivator amplified in breast cancer-1 is required for Neu (ErbB2/HER2) activation, signaling, and mammary tumorigenesis in mice. Cancer Res. 2008;68:3697–3706. doi: 10.1158/0008-5472.CAN-07-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakles RE, Shiffert MT, Díaz-Cruz ES, Cabrera MC, Alotaiby M, Miermont AM, et al. Altered AIB1 or AIB1Δ3 expression impacts ERα effects on mammary gland stromal and epithelial content. Mol Endocrinol. 2011;25:549–563. doi: 10.1210/me.2010-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. Journal of the National Cancer Institute. 2000;92:1185–1186. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 15.Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chien CD, Kirilyuk A, Li JV, Zhang W, Lahusen T, Schmidt MO, et al. Role of the nuclear receptor coactivator AIB1-{Delta}4 in the control of gene transcription. J Biol Chem. 2011;286:26813–27. doi: 10.1074/jbc.M110.216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qutob MS, Bhattacharjee RN, Pollari E, Yee SP, Torchia J. Microtubule-dependent subcellular redistribution of the transcriptional coactivator p/CIP. Mol Cell Biol. 2002;22:6611–6626. doi: 10.1128/MCB.22.18.6611-6626.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long W, Yi P, Amazit L, LaMarca HL, Ashcroft F, Kumar R, et al. SRC-3Delta4 mediates the interaction of EGFR with FAK to promote cell migration. Mol Cell. 2010;37:321–332. doi: 10.1016/j.molcel.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller FR. Xenograft models of premalignant breast disease. Journal of Mammary Gland Biology and Neoplasia. 2000;5:379–391. doi: 10.1023/a:1009577811584. [DOI] [PubMed] [Google Scholar]
- 20.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 21.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nature Cell Biology. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korkaya H, Wicha MS. Selective targeting of cancer stem cells: a new concept in cancer therapeutics. BioDrugs. 2007;21:299–310. doi: 10.2165/00063030-200721050-00002. [DOI] [PubMed] [Google Scholar]
- 24.Stingl J, Raouf A, Emerman JT, Eaves CJ. Epithelial progenitors in the normal human mammary gland. Journal of Mammary Gland Biology and Neoplasia. 2005;10:49–59. doi: 10.1007/s10911-005-2540-7. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nature Reviews. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cody HS. Sentinel lymph node biopsy for DCIS: are we approaching consensus? Ann Surg Oncol. 2007;14:2179–2181. doi: 10.1245/s10434-006-9300-9. [DOI] [PubMed] [Google Scholar]
- 27.Sung YM, Xu X, Sun J, Mueller D, Sentissi K, Johnson P, et al. Tumor suppressor function of Syk in human MCF10A in vitro and normal mouse mammary epithelium in vivo. PLoS ONE. 2009;4:e7445. doi: 10.1371/journal.pone.0007445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Korkaya H, Wicha MS. HER-2, notch, and breast cancer stem cells: targeting an axis of evil. Clin Cancer Res. 2009;15:1845–1847. doi: 10.1158/1078-0432.CCR-08-3087. [DOI] [PubMed] [Google Scholar]
- 30.Pradeep CR, Kostler WJ, Lauriola M, Granit RZ, Zhang F, Jacob-Hirsch J, et al. Modeling ductal carcinoma in situ: a HER2-Notch3 collaboration enables luminal filling. Oncogene. 2012;31:907–917. doi: 10.1038/onc.2011.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nofech-Mozes S, Spayne J, Rakovitch E, Hanna W. Prognostic and predictive molecular markers in DCIS: a review. Adv Anat Pathol. 2005;12:256–264. doi: 10.1097/01.pap.0000184177.65919.5e. [DOI] [PubMed] [Google Scholar]
- 32.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balko JM, Miller TW, Morrison MM, Hutchinson K, Young C, Rinehart C, et al. The receptor tyrosine kinase ErbB3 maintains the balance between luminal and basal breast epithelium. Proc Natl Acad Sci USA. 2012;109:221–226. doi: 10.1073/pnas.1115802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauritsen KJ, List HJ, Reiter R, Wellstein A, Riegel AT. A role for TGF-beta in estrogen and retinoid mediated regulation of the nuclear receptor coactivator AIB1 in MCF-7 breast cancer cells. Oncogene. 2002;21:7147–7155. doi: 10.1038/sj.onc.1205943. [DOI] [PubMed] [Google Scholar]
- 35.Torres-Arzayus MI, Zhao J, Bronson R, Brown M. Estrogen-dependent and estrogen- independent mechanisms contribute to AIB1-mediated tumor formation. Cancer Res. 2010;70:4102–4111. doi: 10.1158/0008-5472.CAN-09-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 37.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nature Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh A, List H-J, Reiter R, Mani A, Zhang Y, Gehan E, et al. The nuclear receptor coactivator AIB1 mediates insulin-like growth factor I-induced phenotypic changes in human breast cancer cells. Cancer Res. 2004;64:8299–8308. doi: 10.1158/0008-5472.CAN-04-0354. [DOI] [PubMed] [Google Scholar]
- 39.Cavalli LR, Man YG, Schwartz AM, Rone JD, Zhang Y, Urban CA, et al. Amplification of the BP1 homeobox gene in breast cancer. Cancer Genet Cytogenet. 2008;187:19–24. doi: 10.1016/j.cancergencyto.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavalli LR, Santos SC, Broustas CG, Rone JD, Kasid UN, Haddad BR. Assignment of the BLID gene to 11q24.1 by fluorescence in situ hybridization. Cancer Genet Cytogenet. 2008;186:120–121. doi: 10.1016/j.cancergencyto.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Otaiby M, Tassi E, Schmidt MO, Chien CD, Baker T, Salas AG, et al. Role of the nuclear receptor coactivator AIB1/SRC-3 in angiogenesis and wound healing. The American Journal of Pathology. 2012;180:1474–1484. doi: 10.1016/j.ajpath.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 43.Rosenfield SM, Bowden ET, Cohen-Missner S, Gibby KA, Ory V, Henke RT, et al. Pleiotrophin (PTN) Expression and Function and in the Mouse Mammary Gland and Mammary Epithelial Cells. PLoS ONE. 2012;7:e47876. doi: 10.1371/journal.pone.0047876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.