Abstract
Background
Single nucleotide polymorphisms (SNPs) influence a patient's response to inhaled corticosteroids and β2-agonists, and the effect of treatment with inhaled corticosteroids is synergistic with the effect of β2-agonists. We hypothesized that use of inhaled corticosteroids could influence the effect of SNPs associated with bronchodilator response.
Objective
To assess whether, among asthma subjects, the association of SNPs with bronchodilator response is different between those treated with inhaled corticosteroids vs. those on placebo.
Methods
A genome-wide association analysis was conducted using 581 white subjects from the Childhood Asthma Management Program (CAMP). Using data for 449,540 SNPs, we conducted a gene by environment analysis in PLINK with inhaled corticosteroid treatment as the environmental exposure and bronchodilator response as the outcome measure. We attempted to replicate the top 12 SNPs in the Leukotriene Modifier Or Corticosteroid or Corticosteroid-Salmeterol (LOCCS) Trial.
Results
The combined P-value for the CAMP and LOCCS populations was 4.81E-08 for rs3752120, which is located in the zinc finger protein gene ZNF432, and has unknown function.
Conclusions
Inhaled corticosteroids appear to modulate the association of bronchodilator response with variant(s) in the ZNF432 gene among adults and children with asthma.
Clinical Implications
Clinicians who treat asthma patients with inhaled corticosteroids should be aware that the patient's genetic makeup likely influences response as measured in lung function.
Capsule Summary
Our study suggests that inhaled corticosteroids could influence the effect of multiple SNPs associated with bronchodilator response across the genome.
Keywords: asthma, bronchodilator response, lung function, inhaled corticosteroids, single nucleotide polymorphisms, zinc finger proteins, ZNF432
INTRODUCTION
Inhaled corticosteroids and β2-agonists are the two most commonly used medications for asthma. Recent evidence suggest that inhaled corticosteroid treatment can restore β2-adrenergic responsiveness of airway smooth muscle cells.1 Additional studies have shown that corticosteroids increase β2- adrenergic receptor (ADRB2) gene transcription in the lung2 and that inhaled corticosteroids potentiate β2-agonist-induced airway smooth muscle relaxation.3
Single nucleotide polymorphisms (SNPs) appear to influence a patient's response to inhaled corticosteroids and β2-agonists. For example, a significant association between 8-week response to inhaled steroids and SNPs from the corticotrophin-releasing hormone receptor 1 (CRHR1) gene, which encodes the primary receptor mediating the release of adrenocorticotropic hormone, have been demonstrated.4 One SNP, rs242941, in CRHR1 (minor allele frequency ~30%) is associated with positive treatment response to corticosteroids in adult and pediatric clinical trials (p=0.025 and 0.006, respectively).4,5 Furthermore, a CRHR1 haplotype was associated with a 2-3 times greater short-term response to inhaled steroids.4 Several ADRB2 polymorphisms, including non-synonymous changes from arginine to glycine at position 16 (A16→G) and from glutamic acid to glutamine at position 27 (E27→Q) have been associated with asthma.6 Other polymorphisms of this gene have been associated with response to inhaled [.beta]2-agonist treatment 7 and bronchodilator response (BDR).8,9 Additional genes that have been reported to be associated with BDR include corticotrophin-releasing hormone receptor 2 (CRHR2) 10, arginase 1 (ARG1)11,12.
In addition to the genetic variants found to be associated with responses to either inhaled corticosteroid alone or to β2-agonists alone, we have previously demonstrated that a non-synonymous SNP in adenylate cyclase 9 (AC9)13 predicts degree of both cellular and clinical response to β2-agonist medications and that this response was significantly enhanced in the presence of glucocorticoid medications.13 Given this preliminary data and knowledge that the effect of treatment with inhaled corticosteroids is synergistic with the effect of β2-agonists and a patient's response to both medications is genetically influenced, we hypothesized that inhaled corticosteroids could influence the effect of multiple SNPs associated with bronchodilator response across the genome.
METHODS
Populations and Measures
Our primary test population is composed of subjects from the Childhood Asthma Management Program (CAMP), a clinical trial that followed 1,041 asthmatic children for four to six years and randomized subjects to budesonide, nedocromil, or placebo. This study methodology and design have been described previously.13 For the current work, CAMP subjects in the budesonide arm were considered to be exposed to ICS while subjects in the nedocromil and placebo groups were not exposed to ICS. We chose to study BDR at the conclusion of the CAMP clinical trial, 48 months after randomization in order to study BDR after maximal exposure to ICS.13 Of the 581 Caucasian subjects who had available DNA for genotyping, 30% (172) were in the inhaled corticosteroid (budesonide) group, 29% (171) were in the nedocromil group, and 41% (238) were in the placebo group. For the purposes of this analysis, we combined the nedocromil and placebo groups.
Replication was conducted in subjects in the Leukotriene Modifier Or Corticosteroid or Corticosteroid-Salmeterol Trial (LOCCS).14 At the baseline visit, subjects were asked, “Over the past 6 months, on average how often did you use inhaled corticosteroids (e.g. Beclovent, Pulmicort, Flovent, etc)?” Subjects who responded “daily” or “2-6 times per week” were considered to be in the inhaled corticosteroid group, whereas subjects who responded “1-2 times per month,” “less than 1 time per month,” or “never” were placed in the non-inhaled corticosteroid group. All subjects were started on an inhaled corticosteroid for four weeks and then BDR was measured at the randomization visit. Subjects were later randomized to fluticasone, montelukast, or fluticasone/salmeterol combination therapy.
In both populations, BDR was based on prebronchodilator and postbronchodilator measurements. After two puffs (180 ug/puff) of albuterol by metered dose inhaler with spacer were administered, at least 10 minutes elapsed before the postbronchodilator spirometry was performed. BDR was calculated as the percent difference between the prebronchodilator and postbronchodilator FEV1 value [BDR=100 × (post FEV1-preFEV1/preFEV1)]. Institutional Review Board approval was obtained at each institution. Informed consent was obtained for all study participants.
Genotyping and Quality Control
Genome-wide SNP genotyping was performed on the CAMP sample using Illumina's HumanHap550 Genotyping BeadChip by Illumina, Inc (Illumina, Inc., San Diego, CA) or Illumina's Infinium HD Human610-Quad BeadChip at the Channing Laboratory. Before frequency and genotyping pruning, there were 516,512 SNPs. SNPs were excluded for missing in more than 5% of subjects (n=26,070), having MAF less than 5% (n=41,621), having Hardy-Weinberg equilibrium p-values among controls less than 0.001 (n=547). After frequency and genotyping pruning, there were 449,540 SNPs. The average genotyping completion rate for each subject was 99.8%.
Genotyping of LOCCS was conducted at the Riken Center for Genomic Medicine using the Illumina Infinium HD Human610-Quad BeadChip. We studied 12 SNPs that were identified from the screening stage in CAMP in the LOCCS population. The average genotyping completion rate for each subject was 99.7%.
Statistical Methodology
We performed a genome-wide study to examine the interaction of genetic variants and treatment with inhaled corticosteroids on bronchodilator response. In the CAMP population, we conducted a gene by environment analysis in PLINK12 with inhaled corticosteroid treatment as the environmental exposure and bronchodilator response (BDR) as the outcome measure using genome-wide data. SNPs were excluded from the analysis if they were missing in more than 5% of subjects, having minor allele frequency (MAF) less than 5%, having Hardy-Weinberg equilibrium p-values among controls less than 0.001. We then replicated our top SNPs that had P-value <1E-05 in LOCCS. We calculated combined P-values for CAMP and LOCCS using Fisher's Method.15. We adjusted for age and gender in our analyses. We conducted a stratified analysis of ICS and placebo groups using genetic linear association analysis with BDR as the outcome and adjusting for age and gender. Local association plots were created using LocusZoom (http://csg.sph.umich.edu/locuszoom).
Expression Quantitative Trait Analysis
To identify expression quantitative trait loci (eQTL) in response to ICS treatment, lymphoblastoid cell lines derived from 151 CAMP participants were treated with 10−6 M dexamethasone (a corticosteroid). After six hours, expression levels were measured using the Illumina HumanRef8 v2 BeadChip (Illumina, San Diego, CA). After arrays and probes filtering, vst transformation, and quantile normalization, 21,175 gene probes were kept. We limited eQTL analysis to 117 Caucasian individuals who were randomized to ICS treatment in the CAMP clinical trial and for whom GWAS genotyping data was available.
To detect SNPs associated to the variations of gene expression levels, we applied a general linear model (GLM), in which the predictor is the genotype of a SNP using additive coding and the outcome variable is the pre-processed expression level of a gene probe from dexamethasone-treated cell lines. For each gene probe, we considered all SNPs within 50 KB of both end of the gene in the cis-eQTL analysis.
RESULTS
Descriptive Statistics
Table I provides baseline demographic characteristics measured in our study population. Our initial study population included a total of 808 Caucasian subjects, including 581 Caucasian subjects from CAMP. The replication population included 227 Caucasian subjects from LOCCS who had available genotype information. The mean age was 8.87 years [SD 2.14 years] in CAMP and 40.8 years [14.1 SD] in LOCCS. Sixty percent of the CAMP population was male while 37% of the LOCCS population was male. The mean bronchodilator response while on inhaled corticosteroids was 8.7% [SD 8.1%] in CAMP and 6.5% [SD 6.8%] in LOCCS.
Table I.
Baseline demographics of the CAMP and LOCCS populations.
| N=808 | CAMP (n=581) | LOCCS (n=228) |
|---|---|---|
| Mean [SD] or Percent (n) | ||
| Age in years [SD] (range) | 8.9 [2.1] (5.2−13.2) | 40.8 (14.1) (15−76) |
| Treatment group | ||
| Budesonide (ICS) | 172 (30%) | 76 (33%) Fluticasone (ICS) |
| Nedocromil | 171 (29%) | 83 (36%) Fluticasone/salmeterol |
| Placebo | 238 (41%) | 69 (30%) Montelukast |
| Gender, | ||
| Male | 347 (60%) | 85 (37%) |
| Female | 243 (40%) | 143 (63%) |
| PreFEV1 at follow up, | 1.91 [0.53] | 2.89 [0.78] |
| Bronchodilator Response at follow up | 0.097 [0.081] | 0.065 [ 0.068] |
| FEV1 percent predicted at follow up | 95.68% [13.56%] | 88% [16.7%] |
Gene by Environment Analysis
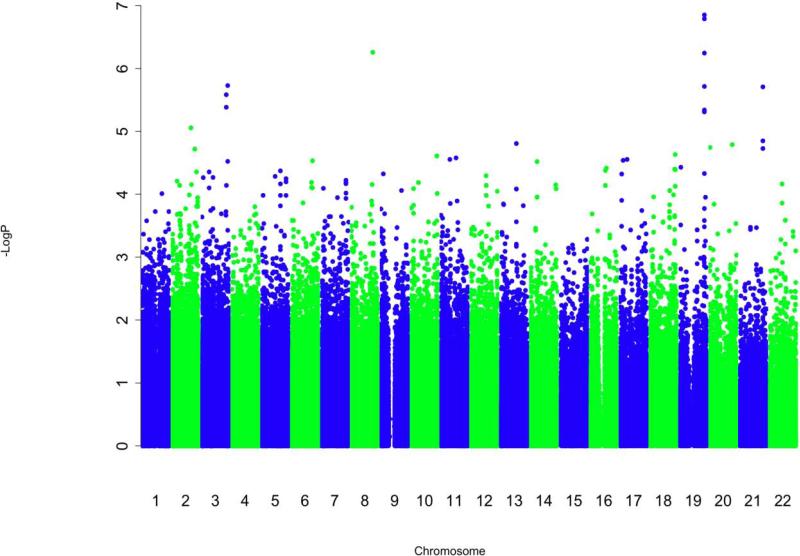
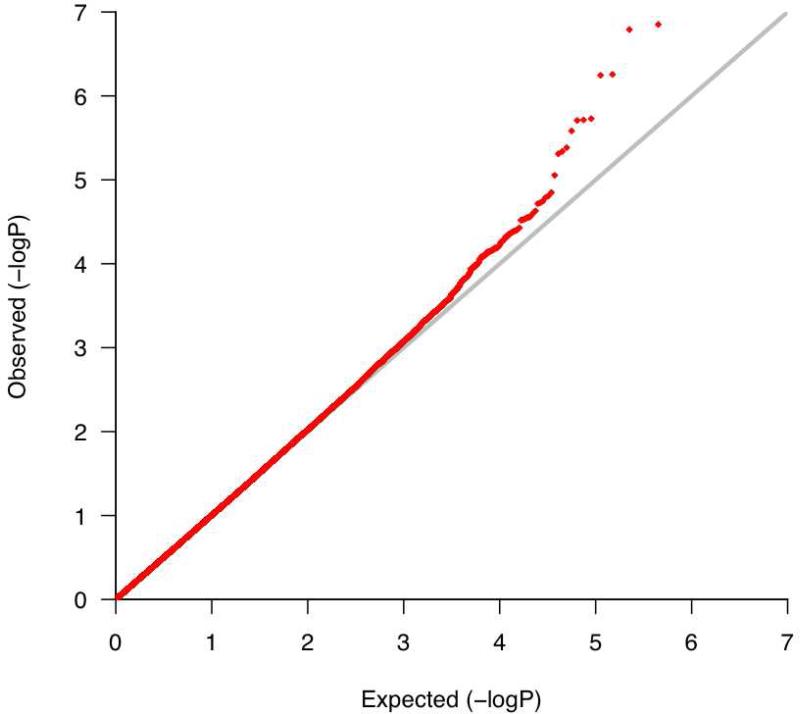
The corresponding quantile-quantile (Q-Q) plot (Figure 1 in Online Repository) demonstrates that the SNPs with the lowest P-values deviate from what is expected for a null distribution, suggesting that some may reflect true associations with bronchodilator response that are modulated by exposure to inhaled corticosteroids. The genomic inflation factor was 1.03, demonstrating minimal population stratification. The Manhattan plot (Figure 1) for this analysis shows that the regions of SNPs that are most significantly associated with bronchodilator response while accounting for inhaled corticosteroid treatment are in chromosomes 19 and 8.
Figure 1.
Manhattan Plot of –log10 (P-value) for the analysis in CAMP. This figure demonstrates that multiple SNPs in chromosome 19 may be associated with bronchodilator response and modulated by inhaled corticosteroid treatment.
Table II shows the top SNPs (gene by environment interaction P-value < 1E-05) in CAMP and LOCCS. We attempted to replicate these 12 SNPs in the LOCCS population. The strongest associations are on chromosome 19. One combined P-value for the CAMP and LOCCS populations reached genome-wide significance: 4.81 × 10−8 for rs3752120, which was in linkage disequilibrium with rs3450 (R2=0.82 in CAMP and 0.75 in LOCCS, combined P=7.56 × 10−9) and rs12460587 (R2=0.84 in CAMP and 0.84 in LOCCS, combined P=2.43 × 10−9). These three SNPs are in or near ZNF432 gene.
Table II.
Results of Gene by Environment Analysis.
| CHR | SNP | Beta Estimate for SNP | SE | Beta Estimate for ICS Exposure | SE2 | MAF | HWE p value | A1 | A2 | CAMP P_GXE | LOCCS P_GXE | Combined P_GXE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 19 | rs10411428 | 0.042 | 0.0099 | −0.027 | 0.0087 | 0.41 | 0.17 | T | C | 1.41x10−7 | 0.00036 | 1.24x10−9 |
| 19 | rs11666341 | 0.042 | 0.0098 | −0.027 | 0.0087 | 0.45 | 0.17 | A | G | 1.62x10−7 | 0.00036 | 1.42x10−9 |
| 19 | rs12460587* | 0.035 | 0.0097 | −0.029 | 0.0085 | 0.33 | 0.22 | G | T | 5.69x10−7 | 0.00018 | 2.43x10−9 |
| 19 | rs3450 | 0.031 | 0.0099 | −0.030 | 0.0082 | 0.18 | 0.72 | C | T | 1.93x10−6 | 0.00017 | 7.56x10−9 |
| 19 | rs3752120** | 0.032 | 0.0107 | −0.031 | 0.0090 | 0.15 | 0.21 | T | C | 4.58x10−6 | 0.00050 | 4.81x10−8 |
| 19 | rs2288884 | 0.033 | 0.0107 | −0.031 | 0.0090 | 0.34 | 0.40 | T | C | 4.91x10−6 | 0.00050 | 5.14x10−8 |
| 8 | rs6469488 | 0.072 | 0.0170 | −0.032 | 0.0127 | 0.07 | 0.79 | G | A | 5.54x10−7 | 0.77 | 6.70x10−6 |
| 21 | rs4919929 | 0.062 | 0.0153 | −0.031 | 0.0122 | 0.07 | 0.42 | T | C | 1.96x10−6 | 0.79 | 2.22x10−5 |
| 3 | rs9868563 | 0.099 | 0.0190 | −0.012 | 0.0138 | 0.06 | 0.10 | T | C | 2.62x10−6 | 0.60 | 2.25x10−5 |
| 3 | rs4686399 | 0.041 | 0.0094 | −0.015 | 0.0068 | 0.29 | 0.41 | G | A | 1.87x10−6 | 0.87 | 2.33x10−5 |
| 3 | rs1889261 | 0.084 | 0.0180 | −0.018 | 0.0134 | 0.06 | 0.76 | A | G | 4.14x10−6 | 0.45 | 2.66x10−5 |
| 2 | rs4233808 | 0.041 | 0.0100 | −0.019 | 0.0089 | 0.16 | 0.69 | G | A | 8.84x10−6 | 0.72 | 8.20x10−5 |
This table summarizes the results of testing and replication in CAMP and LOCCS using the additive model sorted by combined population-based P values. The beta estimate for each SNP and for ICS exposure are provided. CHR=chromosome. SNP=single nucleotide polymorphism. SE= standard error. ICS=inhaled corticosteroid.
located in ZNF841.
located in ZNF432.
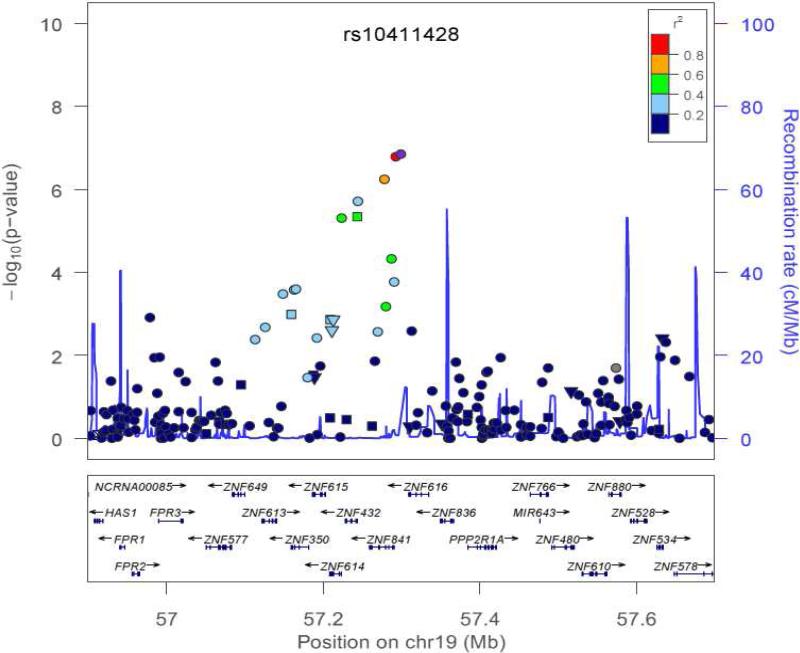
Figure 2 depicts the effect of treatment with ICS versus no ICS and genotype on the outcome of BDR for rs3752120. The x-axis shows the number of genotypes. There are 3 possibilities for the number of copies of rs3752120: 0 copy, 1 copy, or 2 copies. The Y-axis shows the BDR. This figure demonstrates that having two copies of the mutant allele and not being treated with ICS produces a higher BDR than having two mutant alleles and being treated with ICS. In addition, rs2288884 is located near a second gene on chromosome 19, ZNF614 (combined P=5.14 ×10−8) and rs11666341 is located near ZNF841 (combined P=1.42 × 10−9). Figure 3 depicts a plot of the regional association results from our genome-wide association study on chromosome 19, in the region of rs10411428. The plot demonstrates the magnitude of association of SNPs in this region in addition to the pairwise linkage disequilibrium patterns associated with rs10411428. Multiple SNPs in this region are in linkage disequilibrium and are associated with bronchodilator response while modulated by ICS.
Figure 2.
Depiction of the effect of treatment with inhaled corticosteroids (ICS) versus no ICS and genotype on the outcome of bronchodilator response (BDR) for rs3752120. The x-axis shows the number of genotypes. There are 3 possibilities for the number of copies of rs3752120: 0 copy, 1 copy, or 2 copies. Y axis shows the BDR. This figure demonstrates that having two copies of the mutant allele and not being treated with ICS produces a higher BDR than having two mutant alleles and being treated with ICS.
Figure 3.
Region of association near rs10411428 to bronchodilator response while modulated by treatment with inhaled corticosteroids. The x-axis denotes position along chromosome 19. The y-axis denotes –Log10 (P) corresponding to 1000GP imputed data P-values between the SNP, rs10411428, to each SNP in the plot is denoted in colors and was computed according to 1000GP June 2010 CEU data. Plot was created using LocusZoom.22
We also conducted individual regression models for BDR as an outcome for the ICS group alone and for the placebo group alone, while adjusting for age and gender. Our results are depicted in Table III and show that the beta estimates are in opposite directions for the ICS and placebo groups, suggesting that ICS modulates the effect of SNPs on bronchodilator response in a direction distinct from placebo. Analysis of microarray data from lymphoblastoid cell lines from a subset of CAMP subjects determined that the variant rs11666341 is associated with variable gene expression of ZNF432 (p=0.046). Results are presented in Table 1 of the Online Repository. Cells from subjects who were homozygous for the major allele, rs11666341, had lower expression levels under dexamethasone-treated conditions (Figure 2 in Online Repository).
Table III.
Individual regression models for bronchodilator as an outcome stratified by ICS or placebo groups in CAMP.
| ICS group (n=172) | Placebo group (n=409) | |||
|---|---|---|---|---|
| SNP | Beta-estimate | P | Beta-estimate | P |
| rs4233808 | 0.041 | 8.25E−05 | −0.020 | 0.026 |
| rs9868563 | 0.099 | 7.13E−07 | −0.012 | 0.37 |
| rs1889261 | 0.084 | 5.98E−06 | −0.018 | 0.19 |
| rs4686399 | 0.04 | 3.28E−05 | −0.014 | 0.040 |
| rs6469488 | 0.072 | 2.71E−05 | −0.032 | 0.012 |
| rs2288884 | 0.032 | 0.0032 | −0.030 | 0.00096 |
| rs3752120 | 0.033 | 0.0030 | −0.030 | 0.00096 |
| rs3450 | 0.031 | 0.0019 | −0.029 | 0.00047 |
| rs12460587 | 0.035 | 0.00041 | −0.028 | 0.0012 |
| rs11666341 | 0.042 | 3.86E−05 | −0.025 | 0.0042 |
| rs10411428 | 0.042 | 4.12E−05 | −0.026 | 0.0034 |
| rs4919929 | 0.061 | 0.00011 | −0.032 | 0.0094 |
DISCUSSION
Our study has several key findings. Treatment with inhaled corticosteroids appears to modify the effect of SNPs on bronchodilator response. Our analysis, was conducted in a pediatric population, and replicated in an adult population, suggesting that our results are generalizable across age groups. Secondly, we have identified a region of association on chromosome 19 that contains multiple zinc finger protein genes. Variation in this region could influence the effect of inhaled steroids on bronchodilator response. Finally, synergistic effects observed between inhaled corticosteroids and β2 agonists caused by specific genes.
Previous studies have suggested that the anti-inflammatory effects of corticosteroids increase airway response to β2 agonists by up-regulating β2 adrenergic receptor expression and increasing cyclic AMP production by airway epithelial cells. 2,16,17 This leads to the synergistic effect that steroids have on bronchodilator response to β2 agonists.2,16,17 A study by Jin et al assessed for effect modification by use of inhaled corticosteroids and examined whether SNPs in the dual-specificity phosphatase 1, DUSP1 gene, which are associated with BDR, are modified by concurrent use of ICS medication. The authors found that the DUSP1 polymorphisms did modulate the effect between ICS use and BDR.18 Thus, our study is consistent with previous studies in suggesting that genetic factors could mediate the relationship between inhaled corticosteroid use and BDR. The clinical implications of a variant that predicts BDR while on ICS are unclear. Patients with a variant that leads to higher BDR while on ICS may benefit by preserving or restoring smooth muscle function; on the other hand, patients with the variant may need to avoid ICS because a higher BDR while on ICS may signify the ICS is not working. Further in vitro and in vivo studies are necessary to elucidate the clinical implications of our results.
We have identified a region on chromosome 19 that appears to influence the effect of inhaled steroids on asthma. We focused on the gene that coincides with the peak, the zinc finger protein gene, ZNF432, which is located on chromosome 19. The results from the eQTL analysis suggested that the variant rs11666341 is associated with variable gene expression of ZNF432, further supporting the finding that ZNF432 modulates the effect of inhaled corticosteroids in adults and children with asthma. Cells from subjects who were homozygous for the major allele had lower expression levels under dexamethasone-treated conditions, supporting our hypothesis that this variant modulates bronchodilator response. While the function of ZNF432 is unknown, other zinc fingers have been found to play a role in asthma. For example, Zfp35 (ZNF271) appears to influence the pathogenesis of airway inflammation and hyperresponsiveness in asthma by controlling Th2 cell generation and Th2cytokine expression.19 Many zinc finger proteins have been demonstrated to be involved in transcription.20ZNF432 has been found to be associated with inflammatory bowel disease, at P= 8.3 × 10−5.21 Thus, further study of these zinc finger proteins may provide insight on the mechanisms by which inhaled corticosteroids influence bronchodilator response.
Despite the strengths of our study, a few limitations deserve mention. First, our study is relatively small, with only 808 subjects in total. However, we did not have access to data for larger populations because few pharmacogenetic trials have genetic data on subjects who are taking both inhaled corticosteroids and have short acting bronchodilator response measurements. We relied on patient report of inhaled corticosteroid use in our replication population. It is possible that subjects who are randomized to an inhaled corticosteroid group in a clinical trial such as CAMP may be more likely to be taking inhaled corticosteroids than subjects who report taking inhaled corticosteroids that are prescribed by their physicians during the run-in period of a trial such as LOCCS. In neither trial is ICS use directly monitored. Furthermore, we were unable to conduct functional studies to study whether corticosteroids modulate the zinc finger proteins we identified; nevertheless the eQTL analyses do support our findings.
In conclusion, inhaled corticosteroids appear to influence the effect of genetic information on bronchodilator response. A zinc finger protein, ZNF432, appears to modulate the effect on inhaled corticosteroids on bronchodilator response in adults and children with asthma.
Supplementary Material
ACKNOWLEDGMENTS
We thank all subjects for their ongoing participation in this study. We acknowledge the CAMP investigators and research team, supported by NHLBI, for collection of CAMP Genetic Ancillary Study data, and the American Lung Association Asthma Clinical Research Centers (ALA-ACRC) investigators and research team for data from the LOCCS trial. The LOCCS trial was supported by GlaxoSmithKline (a monetary gift to the ALA-ACRC Data Coordination Center (DCC) by providing drugs and appropriate placebos) whose employees played no role in the conduct of the trial or its data analysis or interpretation. All work on data collected from the CAMP Genetic Ancillary Study was conducted at Channing Laboratory of the Brigham and Women's Hospital under appropriate CAMP policies and human subject's protections. CAMP is supported by U01 HL076419, U01 HL65899, P01 HL083069, and T32 HL07427 from the National Heart, Lung, and Blood Institute, National Institutes of Health.
Funding: This work is supported by NHLBI via a K08 HL088046 (PI: Wu), U01 HL065899 (PI: Weiss and Tantisira).
Abbreviations
- AC9
adenylate cyclase 9
- ADRB2
β2-adrenergic receptor
- BDR
bronchodilator response
- CAMP
Childhood Asthma Management Program
- CRHR1
corticotrophin-releasing hormone receptor 1
- ICS
inhaled corticosteroid
- eQTL
expression quantitative trait loci
- Q-Q
quantile-quantile
- LOCCS
Leukotriene Modifier Or Corticosteroid or Corticosteroid-Salmeterol Trial
- SNP
single nucleotide polymorphism
- SPATS2L
spermatogenesis associated serine-rich 2-like gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brieva JL, Danta I, Wanner A. Effect of an inhaled glucocorticosteroid on airway mucosal blood flow in mild asthma. Am J Respir Crit Care Med. 2000;161:293–6. doi: 10.1164/ajrccm.161.1.9905068. [DOI] [PubMed] [Google Scholar]
- 2.Mak JC, Nishikawa M, Barnes PJ. Glucocorticosteroids increase beta 2-adrenergic receptor transcription in human lung. Am J Physiol. 1995;268:L41–6. doi: 10.1152/ajplung.1995.268.1.L41. [DOI] [PubMed] [Google Scholar]
- 3.Mendes ES, Horvath G, Campos M, Wanner A. Rapid corticosteroid effect on beta(2)-adrenergic airway and airway vascular reactivity in patients with mild asthma. J Allergy Clin Immunol. 2008;121:700–4. doi: 10.1016/j.jaci.2007.10.038. [DOI] [PubMed] [Google Scholar]
- 4.Tantisira KG, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13:1353–9. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 5.Mougey EB, et al. Pharmacogenetics of asthma controller treatment. Pharmacogenomics J. doi: 10.1038/tpj.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reihsaus E, Innis M, MacIntyre N, Liggett SB. Mutations in the gene encoding for the beta 2-adrenergic receptor in normal and asthmatic subjects. Am J Respir Cell Mol Biol. 1993;8:334–9. doi: 10.1165/ajrcmb/8.3.334. [DOI] [PubMed] [Google Scholar]
- 7.Israel E, et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004;364:1505–12. doi: 10.1016/S0140-6736(04)17273-5. [DOI] [PubMed] [Google Scholar]
- 8.Drysdale CM, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci U S A. 2000;97:10483–8. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman EK, et al. Family-based association analysis of beta2-adrenergic receptor polymorphisms in the childhood asthma management program. J Allergy Clin Immunol. 2003;112:870–6. doi: 10.1016/s0091-6749(03)02023-2. [DOI] [PubMed] [Google Scholar]
- 10.Poon AH, et al. Association of corticotropin-releasing hormone receptor-2 genetic variants with acute bronchodilator response in asthma. Pharmacogenet Genomics. 2008;18:373–82. doi: 10.1097/FPC.0b013e3282fa760a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himes BE, et al. Genome-wide association analysis in asthma subjects identifies SPATS2L as a novel bronchodilator response gene. PLoS Genet. 8:e1002824. doi: 10.1371/journal.pgen.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Litonjua AA, et al. ARG1 is a novel bronchodilator response gene: screening and replication in four asthma cohorts. Am J Respir Crit Care Med. 2008;178:688–94. doi: 10.1164/rccm.200709-1363OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tantisira KG, Small KM, Litonjua AA, Weiss ST, Liggett SB. Molecular properties and pharmacogenetics of a polymorphism of adenylyl cyclase type 9 in asthma: interaction between beta-agonist and corticosteroid pathways. Hum Mol Genet. 2005;14:1671–7. doi: 10.1093/hmg/ddi175. [DOI] [PubMed] [Google Scholar]
- 14.Peters SP, et al. Randomized comparison of strategies for reducing treatment in mild persistent asthma. N Engl J Med. 2007;356:2027–39. doi: 10.1056/NEJMoa070013. [DOI] [PubMed] [Google Scholar]
- 15.Fisher RA. Statistical methods for research workers. Oliver and Boyd; Edinburgh: 1950. p. xv.p. 354. [Google Scholar]
- 16.Adcock IM, Stevens DA, Barnes PJ. Interactions of glucocorticoids and beta 2-agonists. Eur Respir J. 1996;9:160–8. doi: 10.1183/09031936.96.09010160. [DOI] [PubMed] [Google Scholar]
- 17.Aksoy MO, et al. Glucocorticoid effects on the beta-adrenergic receptor-adenylyl cyclase system of human airway epithelium. J Allergy Clin Immunol. 2002;109:491–7. doi: 10.1067/mai.2002.122154. [DOI] [PubMed] [Google Scholar]
- 18.Jin Y, et al. Dual-specificity phosphatase 1 as a pharmacogenetic modifier of inhaled steroid response among asthmatic patients. J Allergy Clin Immunol. 126:618–25. e1–2. doi: 10.1016/j.jaci.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitajima M, et al. Enhanced Th2 cell differentiation and allergen-induced airway inflammation in Zfp35-deficient mice. J Immunol. 2009;183:5388–96. doi: 10.4049/jimmunol.0804155. [DOI] [PubMed] [Google Scholar]
- 20.Deng Y, et al. ZNF552, a novel human KRAB/C2H2 zinc finger protein, inhibits AP-1- and SRE-mediated transcriptional activity. BMB Rep. 43:193–8. doi: 10.5483/bmbrep.2010.43.3.193. [DOI] [PubMed] [Google Scholar]
- 21.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.