Abstract
Muscle contraction is regulated by tropomyosin movement across the thin filament surface, which exposes or blocks myosin-binding sites on actin. Recent atomic structures of F-actin-tropomyosin have yielded the positions of tropomyosin on myosin-free and myosin-decorated actin. Here, the repositioning of α-tropomyosin between these locations on F-actin was systematically examined by optimizing the energy of the complex for a wide range of tropomyosin positions on F-actin. The resulting energy landscape provides a full-map of the F-actin surface preferred by tropomyosin, revealing a broad energy basin associated with the tropomyosin position that blocks myosin-binding. This is consistent with previously proposed low-energy oscillations of semi-rigid tropomyosin, necessary for shifting of tropomyosin following troponin-binding. In contrast, the landscape shows much less favorable energies when tropomyosin locates near its myosin-induced “open-state” position. This indicates that spontaneous movement of tropomyosin away from its energetic “ground-state” to the open-state is unlikely in absence of myosin. Instead, myosin-binding must drive tropomyosin toward the open-state to activate the thin filament. Additional energy landscapes were computed for disease-causing actin mutants that distort the topology of the actin-tropomyosin energy landscape, explaining their phenotypes. Thus, the computation of such energy landscapes offers a sensitive way to estimate the impact of mutations.
Keywords: actin, coiled-coil, energy landscape, myosin, muscle regulation, thin filaments
Introduction
Tropomyosin together with the troponin complex is responsible for Ca2+-dependent regulation of muscle contraction [1–3]. Tropomyosin, a 42 nm long coiled-coil, forms head-to-tail connections producing a continuous superhelical strand that wraps around the actin helix of the thin filament (Fig. 1a). Its path is defined by periodic electrostatic interactions between tropomyosin pseudo-repeat domains and each of seven successive actin subunits along the thin filament (Fig. 1b). In turn, these interactions then specify the binding of the troponin complex to the thin filament [1–7].
Figure 1.
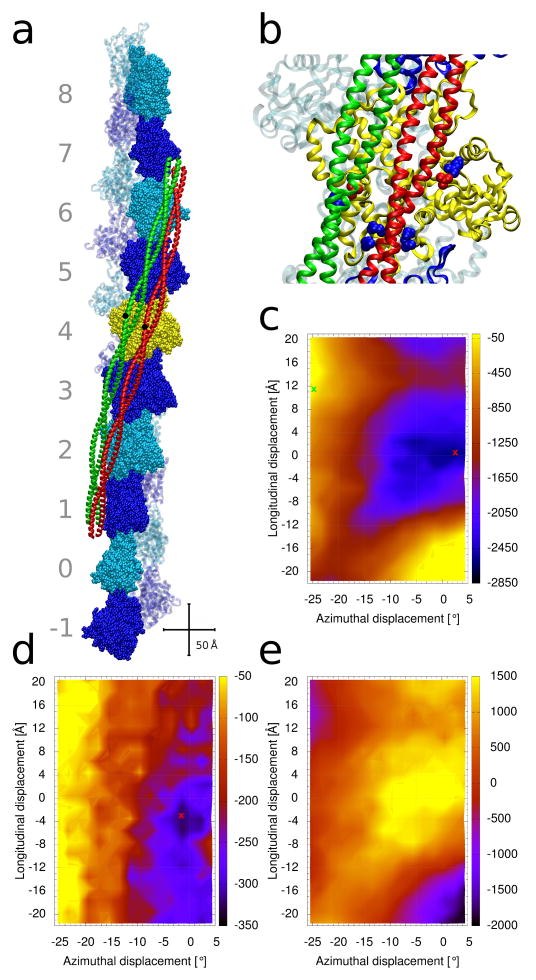
Tropomyosin positions on the surface of F-actin. (a) Structure of F-actin complexed with tropomyosin in either the troponin and myosin-free A-position (colored red) [6], or in the S1-decorated M-position (green) [12]. Ten actin-pairs (each pair shown in space-filling and ribbon representations) are colored alternatively in dark-blue and cyan. The pairs are numbered from −1 to 8. The actin filament is oriented with its pointed end facing up; hence, the N-terminus of tropomyosin also faces up. Centrally located tropomyosin residue 125 is highlighted in black as a reference point to indicate the relative sliding of tropomyosin between positions. Crisscrossing scale bars – 50 Å. (b) Enlarged view of the central yellow actin subunit in (a), showing some of the acidic and basic actin residues of actin which form attractive interactions with tropomyosin when it is in the myosin-free A-position (red and blue spheres, respectively, which highlight residues D25 and R28 (top right pair), K147, K326, K328 (bottom cluster)). D311 and K315 (left) interact with tropomyosin in the M-position (green) [7,12]. (c–e) Energy landscapes, with the energy levels (in kcal/mol) shown by the color scales on their right. At each point of the landscape, tropomyosin was repositioned longitudinally and azimuthally (see Methods) and the energy of the complex was optimized. The [0, 0] position corresponds to the tropomyosin position described by the Li et al. [6] (red ribbon location in panel (a)). (c) Contributions from the Coulombic interaction energy. The energy minimum is indicated by a red cross. The open M-state position of tropomyosin [12] is indicated by a green cross. (d) Corresponding total potential energy and (e) the contribution from the solvation energy (see Methods).
The azimuthal positions assumed by tropomyosin on the surface of the thin filaments are modulated by Ca2+, troponin and myosin-binding [4–7]. At low Ca2+ concentration, troponin constrains tropomyosin in a “blocking-position” over the myosin-binding sites on actin, thereby inhibiting the acto/myosin-crossbridge cycle and consequently relaxing muscles. Rising Ca2+ concentration and Ca2+-binding to troponin releases the constraint, and tropomyosin moves off the myosin-binding site to initiate crossbridge - thin filament interaction. However, tropomyosin is only likely to move completely away from the myosin-binding site to a wholly “open-position” on actin following myosin activation [8]. During this process, the movement of semi-rigid tropomyosin propagates to neighboring actin monomers along thin filaments and leads to a cooperative full-activation of the thin filament and contraction [1–8]. Taken together with kinetic characterization of the system, these structural insights led to formulation of a “three-state model” for thin filament regulation and the naming of the so-called low-Ca2+ “blocked”, high-Ca2+ “closed”, and myosin-induced “open” states, also known as the B-, C- and M-states [8–12].
Electron microscopy and 3D reconstruction provide the average azimuthal position of tropomyosin near to the myosin-blocking site on troponin- and myosin-free F-actin [6,10,13]. This “A-state” position [7] of tropomyosin on actin-tropomyosin filaments is virtually the same as that of B-state tropomyosin in troponin-regulated filaments except that tropomyosin’s positional variance on actin is greater when troponin and myosin are absent [7,13]. Nevertheless, the average A-state tropomyosin position near to the outer edge of actin could be determined without confounding contributions from TnT running alongside tropomyosin. However, the low resolution of the reconstructions precluded determination of the longitudinal position of tropomyosin along F-actin and hence a complete atomic model needed for landscape determinations. Li et al. [6] overcame this obstacle by developing an atomic model of the A-state by using a local computational search that optimizes the electrostatic interaction energy between actin and tropomyosin, and then showed that this structural model was consistent with the density profile of EM reconstructions. The high-resolution cryo-EM reconstructions by Behrmann et al. [12] later gave an all-atom structure of the M-state position of tropomyosin [7,8], which described “rigor-bonded” myosin at the end of the cross-bridge cycle inducing tropomyosin movement to the extreme inner edge of the actin surface. Hence, high resolution structural models are available that place tropomyosin on actin at or near to its end-states during regulatory switching.
Low-energy barriers are thought to separate the different structural states of tropomyosin on F-actin, with the underlying energy landscape possibly partitioned so that distinct energy minima would correspond to each regulatory-state position [10,11]. The absence of steric obstructions on actin limiting tropomyosin movement would be consistent with such low-energy transitions [4,7]. However, despite the recent advances in developing atomic models characterizing the actin-tropomyosin complex [6,12], the pathway(s) taken by tropomyosin during the regulatory transition remain unknown, leaving uncertainty about the switching mechanisms needed to operate the three-state model. To address this issue, we have now extended and refined the previous computational search over all regions of actin likely to be explored by tropomyosin between the A- and the M-positions. An “all-atom” model has been used to compute the energy landscape of tropomyosin on the F-actin surface and to determine the positions of energy minima in this landscape. Given that the two atomic structures available locate tropomyosin at or near to what likely are its end point positions on the F-actin surface (Fig. 1a,b) [7], low energy pathways between these minima should then specify the motions of tropomyosin between the different regulatory-states.
The energy landscape was obtained by first generating all combinations of longitudinal displacements (i.e. parallel to the central axis of F-actin) and azimuthal rotations (around the central axis of F-actin) of tropomyosin, and then optimizing the energy of each of these configurations of the tropomyosin/F-actin complex. Contrary to expectation, only a single significant energy minimum is found on the energy landscape. It is located within a broad but shallow energy basin centered on the A-position. Thus, unconstrained by troponin or myosin, tropomyosin presumably can explore the surrounds of the energy minima at relatively low-energy cost. The breadth and flatness of this basin explains how troponin can shift tropomyosin from the low-Ca2+ B-state at one edge of the basin towards the other edge of the basin to yield the C-state at high Ca2+. In contrast, the myosin-induced M-state position for tropomyosin is well outside of the energy basin in an unfavorable high-energy region of the landscape. Thus, tropomyosin is only likely to move to the M-position following strong myosin-binding to F-actin. The energy landscape has no deep valleys, which means that distinct minima and connecting pathways for the tropomyosin transitions between A-, B-, C-, and M-positions are not well defined in absence of troponin and myosin. Therefore, both troponin and myosin must change the contours of the actin - tropomyosin energy landscape for muscle regulation to operate properly. Troponin is likely to reshape the landscape in relaxed muscle to trap tropomyosin in the B-state position and bias tropomyosin to the C-state position in Ca2+-activated filaments. In addition, the strong binding of myosin to actin must completely reconfigure the actin-tropomyosin energy landscape to fully activate the thin filament.
Materials and methods
All-atom model
The energy landscapes were computed for a model system consisting of a single tropomyosin molecule bound to one of the helical strands of a 20 subunit-long model of F-actin (Fig. 1a). The Oda et al. [14] structure of F-actin was used. A canonical model was used for tropomyosin [6,15], whose superhelical shape matches the helical twist of F-actin perfectly [6,7,15,16], and whose side chains are oriented as in high-resolution crystal structures (previously described in Li et al. [17]). The tropomyosin pseudorotation angle (pseudorotation of the coiled-coil determines which face of tropomyosin interacts with the F-actin) was taken from Li et al. [6] and Behrmann et al. [12]. When tropomyosin polymerizes into a cable on F-actin, the N- and C-terminal ends of tropomyosin form an overlap complex of about 10 residues [3,15,16]. Since a single tropomyosin molecule was used here, these end-end overlaps cannot form. Previous MD simulations have shown that the “free ends” of single tropomyosin molecules are not stably linked to F-actin [16]. To avoid artifactual contributions from the coiled-coil ends, the tropomyosin was truncated here at both the N- and C-termini by 20 residues. This was necessary for two reasons: 1. As mentioned, the terminal residues of single tropomyosin molecules are not bound to F-actin as canonical coiled-coils and display large positional variance [16]; hence, these residues could not be realistically modeled and assessed; 2. Calculation of solvation energy at the free ends is not meaningful, and would confound energy determination. No attempts were made to model end-to-end bonded tropomyosin, because of uncertainties about the structure of the junctional domains of full-length tropomyosin and their overlap geometry on actin filaments. Tropomyosin was positioned at a constant radius of 39 Å from the central F-actin axis, which is the radius derived from thin filament fiber diffraction studies [15]. Changing the radial position of tropomyosin had little qualitative effect on the energy gradients measured, except of course that the Coulombic terms diminished at higher radii.
Grid underlying the landscape
The landscape was computed over a combination of longitudinal and azimuthal positions of tropomyosin on F-actin, covering a range of 42 Å in the longitudinal direction and a 30° azimuthal rotation around the filament, i.e. over the flat interfaces of actin subunits over which tropomyosin traverses [7] (Fig. 1c). The entire tropomyosin molecule was rigidly moved longitudinally in 2 Å increments and rotated azimuthally in 2° steps, thus producing 352 different positions on F-actin. The position chosen here as [azimuth=0, longitud=0] corresponds to the Li et al. model of F-actin-tropomyosin [6], which served as reference structure [6]. The structure of the M-position [12] is then located at coordinates [−24.5°, +11.5 Å] (see Fig. 1c). The axis of tropomyosin’s superhelical shape was always placed parallel to the central axis of the F-actin. An important factor observed is that the internal conformation of tropomyosin is essentially the same in the A- [6] and M-structures [12], which means that there is no measurable pseudorotation of the coiled-coil between these two extreme states, suggesting that tropomyosin slides over the actin surface between the A- and M- end-states [12]. Given the semi-rigid nature of tropomyosin, this is not surprising [7]. Therefore, it was assumed here that tropomyosin does not undergo significant pseudo-rotation in the transitions examined, so that the same pseudorotation angle was used in all positions of the “grid”. (In contrast, if helically curved tropomyosin rolled between different azimuthal positions to a significant degree, it would need to pseudorotate to maintain continuous contact with F-actin; i.e., it would need to change conformation in concert with such a rolling rotation in order to keep its initial superhelical shape and still stay in continuous contact with the F-actin helix.)
Minimization of structures on the grid
Each individual F-actin-tropomyosin structure for each grid position in the search was subjected to energy minimization using Steepest Decent (SD) followed by Conjugate Gradient (CG) methods until the potential energy gradient did not exceed 0.05 kcal/mol. This differs from the protocol used in Li et al. [6], where the grid search was performed only locally and without minimizing the energy at each grid-point.
Energy and solvation
All calculations were performed with the CHARMM program version c35b2 [18] using the CHARMM27 force-field [19,20], as described previously [16]. The energy of non-bonded interactions was evaluated using atom based cutoffs with a switching function for inter-atomic distances from 16 Å to 20 Å. All atoms in actin and tropomyosin were allowed to move during the minimization. Implicit water solvation was implemented according to the Generalized Born model [21] using the GBSW protocol [22], with a 150 mM salt concentration, 50 points for angular integration, 0.005 kcal/(mol·Å2) for the non-polar surface tension coefficient and default values for all other GBSW parameters. The potential energy resulting from the minimizations served to build the energy landscapes over the grid. For some landscapes, the total energy was decomposed into the GBSW solvation term, or into the Coulombic electrostatic term, whose changes are mostly due to the interaction between the tropomyosin and the F-acin (the contribution from the van der Waals interactions were negligible).
Results and Discussion
The energetics of tropomyosin movement over the surface of F-actin was explored by determining the interaction energy terms for tropomyosin located at different positions on actin. To accomplish this, a matrix of uniformly distributed tropomyosin positions spanning the blocked to the open thin filament states was generated with tropomyosin placed at a 39 Å radius from the central axis of F-actin as found in situ [15]. The F-actin—tropomyosin structures were then energy minimized and respective energy terms extracted for each new position of tropomyosin on F-actin (Fig. 1c–e). The effect of troponin on the energy landscape was not evaluated in this work, as requisite structures are not available.
Coulombic energy landscape
At a super-helical radius of 39 Å, tropomyosin hovers over the surface of F-actin at a distance of 8 to 10 Å. Therefore, the interactions between actin and tropomyosin are primarily electrostatic in nature [3,4,6,15]. The energy landscape of the Coulombic interactions is shown in Fig. 1c. The position of lowest energy (red crosses in Fig. 1c and Supplementary Item Fig. S1) is found at [2°, 0 Å]. This is very near to the Li et al. [6] position ([0, 0]) determined previously, despite the use here of a solvation potential during the minimizations. This position is characterized by a close pairing of ~26 oppositely charged residues between tropomyosin and actin subunits and is consistent with previous suggestions that electrostatic interactions form between actin and tropomyosin [6]. Indeed, we and others [3,6,7,23,24] have proposed that multiple acidic residues on tropomyosin interact with Arg147, Lys326, Lys328 and Arg28 and basic ones with Asp25 on successive actin monomers along F-actin (Fig. 1b). It is not surprising that electrostatic interactions dominate tropomyosin binding to F-actin, since tropomyosin has a high net negative charge of −54e at neutral pH.
The Coulombic landscape surrounding position [0, 0] displays a broad (15° wide, 15 Å high) energy basin surrounding the energy minimum. In marked contrast, the position at the M-state configuration (green cross in Fig. 1c) has a much less favorable electrostatic energy. Nor is there a local energy minimum within the elevated energy plateau surrounding the M-position. Thus, the M-state is clearly unfavorable electrostatically in absence of myosin and troponin. This reflects the fact that, in the M-position, F-actin presents a low number of positively charged residues (~11) to tropomyosin, compared to that in the basin surrounding the A-position [7,12].
Energy landscapes of individual actin subunits
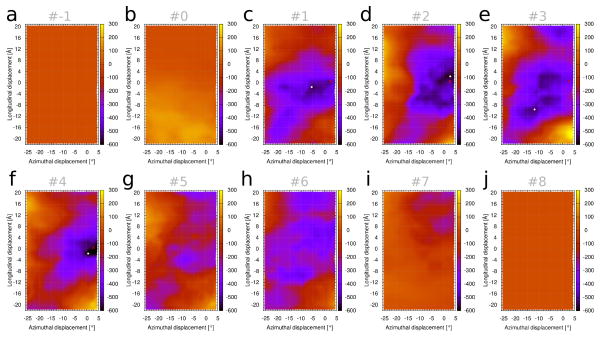
Figure 1c represents the Coulombic energy landscape of tropomyosin interacting with all 10 actin-pairs in our model structure. It results from the composite electrostatic contributions from all seven tropomyosin pseudo-repeats, each one interacting mainly with one of the successive actin subunits along F-actin. However, the amino acid sequences of the seven pseudo-repeats are not identical. Thus, their binding to actin subunits, and hence their individual contributions to the landscape, may vary. To test this, we dissected the Coulombic landscape of Figure 1c, focusing on the local contributions made by single actin subunits (Fig. 2). It shows that the landscapes measured separately over individual actin subunits 1, 2, 3 and 4 (which respectively interact with tropomyosin pseudo-repeats 7, 6, 5 and 4, see Fig. 1a), have deeper and more localized energy minima than those of actin units 5 and 6 (interacting with tropomyosin pseudo-repeats 2 and 3). Hence, the former pseudo-repeats contribute more to the overall preference of the [0, 0] region than the later. Note that much of the so-called α-zone [3] in the N-terminus of tropomyosin pseudo-repeat 1 (containing charged residues that may interact with actin) was truncated from the present tropomyosin structure (as described in Methods), and thus this domain was excluded from the analysis. While energy minima associated with tropomyosin repeats 7, 6, 5 and 4 were discretely localized, the exact position of the respective minima (white dots in Fig. 2) varied in each of the individual landscapes. Moreover, the boundaries of these localized minima were more circumscribed than the energy basin in the composite landscape (Fig. 1c). When summed, the individual basins seen in Figure 2 yield the outlines of the composite map (see Supplementary Item Fig. S2). Hence, the shape of the overall energy basin for full-length tropomyosin on F-actin is determined by a collection of electrostatic contacts that show regional differences. The fact that the individual minima are spread over a wide region may be a design feature of tropomyosin, in order to yield a wider composite basin, which as mentioned below is essential to allow the displacement of tropomyosin from the B-state to the C-state upon Ca2+ binding of troponin. Moreover, regional differences in pseudo-repeat – actin binding strength may facilitate development of preferred target regions for initial myosin crossbridge – actin interaction (discussed below).
Figure 2.
Coulombic interaction (in kcal/mol) between single actin subunits (shown Fig. 1a) and tropomyosin. The energy landscape of Figure 1c was decomposed so that each panel (a–j) shows the contribution from one actin subunit (numbered as in Fig. 1a), interacting with the partner pseudo-repeat of tropomyosin which is closest to that actin subunit. By convention, actin subunit and tropomyosin pseudo-repeat numbering are opposite, hence actins 1 to 7 are represented by (c) pseudo-repeat 7 (C-terminal end), (d) pseudo-repeat 6, (e) pseudo-repeat 5, (f) pseudo-repeat 4, (g) pseudo-repeat 3, (h) pseudo-repeat 2, (i) pseudo-repeat 1 (N-terminal repeat). White dots indicate the location of the lowest minima for the decomposed landscapes, and red crosses the position of the “composite” minimum for the whole system, as in Figure 1c. In panels (a), (b) and (j), the tropomyosin is not in close contact with the respective actin subunit, and hence there is very little energy variation in those landscapes.
Effect of solvation
Using an implicit solvation method [21] allows an accounting of the screening of the electrostatic interactions by water. The resulting total potential energy mapping produced the landscape shown in Figure 1d. Its low energy minimum is centered on [0.5°, −3.6 Å], located close to the Coulombic landscape minima [2°, 0 Å], near to the A-state location. Once again, the myosin-induced M-position (at [−24.5°, +11.5 Å]) is in a region of much higher energy than the A-state. Thus the landscape accounting for solvation confirms the view that the M-state is rarely populated in absence of myosin. The total energy landscape shows a pattern of obliquely oriented stripes (Fig. 1d) representing increasingly unfavorable energies as tropomyosin moves from the A- to the M-state. Interestingly, the pathway orthogonal to these stripes equates to the course taken by tropomyosin when it moves orthogonally across actin.
Note that the solvation energy term by itself yields a landscape (Fig. 1e) that is the negative of the Coulombic landscape (compare Figs. 1c and 1e). This is expected, since the screening of a Coulombic interaction always has a sign opposite to that of the interaction itself. Consequently, the effect of the solvent is to partially disfavor the A-position and to somewhat facilitate the movement towards the M-position. A practical consequence is that increasing the ionic strength of the solvent (which increases the shielding effect) will shift the equilibrium constant between states, where a higher ionic strength makes the M-state less unfavorable.
Tropomyosin movement on thin filaments
The energy landscapes (both for the total energy and the Coulombic term) display a single energy basin, whose minimum lies much closer to the low-Ca2+ B-position (near the [0, 0] point) than it does to the high-Ca2+ C-state identified in EM reconstructions, which is 15–20° away. The landscapes show no evidence of multiple distinct minima that might correspond to the other regulatory-state positions, nor are there energy valleys on the landscapes that would correspond to preferred connecting pathways. These observations suggest that other factors, namely the binding of myosin and troponin to actin or to tropomyosin, alter the energy landscape to trap and stabilize tropomyosin structurally in new energy minima that cannot be seen in the absence of these proteins. The amplitude of local azimuthal oscillation of thin filament tropomyosin [10,11,25,26], when undampened by troponin or myosin, depends on the depth and breadth of the corresponding basin of the energy landscape. The propensity for local chatter in the azimuthal position of tropomyosin is favored by the dual possibility of tropomyosin forming and then breaking interactions with specific actin residues as some tropomyosin pseudo-repeats make and others sever their locally preferred electrostatic contacts: e.g. with residue 326, 328 and 147 (see Fig. 1b) when tropomyosin is positioned in the middle of the basin, or with residues 25 and 28 when at the outer edge of the basin. The degree of such tropomyosin “vibration” [25], in any one regulatory state, may differ among different tropomyosin isoforms and thus influence cooperative switching of thin filament activity [10].
We expect that Ca2+ binding to cardiac and skeletal muscle troponin will promote a ~15 to 20° tropomyosin movement towards the “left” edge of the energy basin, away from its global minimum near to the myosin-blocking B-position [7]. Hence, Ca2+-saturated troponin needs both to release its constraining influence on B-state tropomyosin and also promote tropomyosin movement to the closed C-state position, as suggested by previous structural studies [13]. Thus, it appears that Ca2+-saturated troponin changes the actin-tropomyosin energy landscape such that the minimum is now closer to the C-state. While the azimuthal positions of tropomyosin on F-actin in A-, B-, C- and M-states are known [7], the axial alignment of C-state tropomyosin on F-actin is not. Hence, the C-state coordinates in our energy landscapes cannot be assigned. Moreover, there is no secondary energy minimum in our landscape maps that might serve as a guidepost to locate a potentially discrete C-state position.
Mutations distort the actin-tropomyosin energy landscape
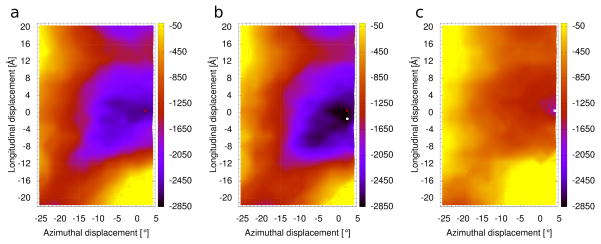
Disease-causing point mutations of thin filament-linked proteins frequently occur at residues on the interface between F-actin and tropomyosin. We anticipate that in many of these cases the mutations will distort the normal thin filament energy landscape described above. For example, the actin mutation, D292V, associated with congenital fiber type disproportion (CFTD) [32], removes a negative charge on actin that is adjacent to basic residues K326, K328 and R147, all thought to be needed to bind acidic residues on tropomyosin [3,6,23,24,32]. This would tend to strengthen corresponding intermolecular electrostatic interactions further and thus stabilize tropomyosin in the blocking or closed state to strongly inhibit actin-myosin interaction, as is observed experimentally [32]. As proof-of-concept, we tested this possibility by calculating the energy landscape for wild-type tropomyosin on the mutant F-actin. Indeed, the electrostatic energy landscape shows a considerably deeper energy basin (Fig. 3b) than exhibited by the wild-type filament, consistent with tropomyosin being strongly trapped in a blocking state. The total energy landscape (see Supplementary Item Fig. S3) also has a narrow but deep basin with a discrete minimum centered on the blocking state position, and thus tropomyosin may be less prone to oscillate away from the blocking position. An example where converse of such energetics occurs is seen with the K326N actin mutation, linked to the development of stiff, hypercontractile muscles [32]. Computation of the energy landscape for this mutant (Fig. 3c) shows that the Coulombic interaction energy is reduced over the whole landscape. In particular, the energy difference between the A- and M-states is much smaller than in the wild-type. This would facilitate a shift of tropomyosin towards the open-state of the filament, and can explain the increased Ca2+-sensitivity and hyper-contractility of affected muscles. These two examples show how energy landscape determination offers a sensitive means of predicting imbalances in thin filament regulation. We plan to apply this approach as part of a continuing comprehensive study of known actin and tropomyosin mutations affecting muscle regulation. The method is particularly useful to study mutations that are not easily assessed experimentally.
Figure 3.
Coulombic interactions (kcal/mol) landscape for actin mutants. (a) Wild-type actin (same as Fig. 1c). (b) D292V actin. (c) K326N actin. White dots indicate the location of the minima for the mutant filament landscapes and red crosses the position of the minimum for wild-type, as in Figure 1c.
Muscle activation
The present study indicates that actin subunits 6 and 5 (Fig. 1a) interact with tropomyosin pseudo-repeats 2 and 3 more weakly than subunits that bind to repeats 4, 5, 6 or 7. Consequently, the initial weak binding of myosin heads to thin filaments during the actin-myosin cross-bridge cycle may first favor interactions with actins 5 and 6. Indeed, tropomyosin segments 2 and 3 would offer less resistance to myosin-induced displacement, and thus likely be favored target zones for initial myosin head association during muscle activation. This would be consistent with observations that such target zones localize between periodically distributed troponin core domains associated with tropomyosin pseudo-repeat 4 and 5 [27,28]. Once strongly bound to F-actin, myosin interaction dramatically changes the energy landscape seen by tropomyosin and confines tropomyosin in the M-state position [12]. In fact, myosin associates with actin-tropomyosin with a Ka of ~107–108 M−1 [29,30] and will compete favorably for sites on actin that otherwise support weaker tropomyosin-actin interactions in the B- and C-states (Ka of ~3 × 103 M−1) [31]. The resulting displacement of tropomyosin from the blocked and closed position by such strong myosin-binding to actin (“rigor binding”) and away from the energy basin determined here is observed experimentally as mentioned above [8,12]. After relocating to the open-state site, the tropomyosin M-position is stabilized because favorable local tropomyosin-myosin interactions form [12] and because tropomyosin is trapped in a structural well, bounded on one side by myosin and on the other by the protruding inner edge of actin subdomains 3 and 4 [7,12].
By changing the energy landscape over the actin surface, myosin-binding drives tropomyosin to the open-state position, which is an energetically unfavorable tropomyosin location when myosin is absent. Thus, the very favorable actin-myosin interaction energetics is coupled to the less favorable tropomyosin transition over the actin surface. Conversely, the opposite will occur following detachment of myosin heads from F-actin. In this case, tropomyosin would be expected to spontaneously revert to its minimal energy position away from the open-state conformation, thus, facilitating relaxation. Hence, Ca2+ binding to troponin followed by myosin-attachment to actin primes the thin filament to become activated, while on the other hand myosin-detachment from actin in concert with Ca2+-dissociation from troponin favors filament off-switching. The possibility that tropomyosin rapidly “snaps-back” to its low energy position once myosin dissociates during muscle relaxation is consistent with results of time-resolved fiber diffraction studies on intact muscles [28].
Summary
The energy landscape of tropomyosin on F-actin shows that there is one fairly broad energy minimum that describes the energetics of the tropomyosin-F-actin system. No obvious low energy pathway connects regulatory states of the filament. Trapping tropomyosin in the blocked, closed, and open positions of the thin filament thus requires additional structural and energetic input from the binding of troponin and myosin that deforms the actin-tropomyosin energy profile. As we have indicated here, we expect that many mutations affecting actin, myosin, tropomyosin and troponin that lead to cardiac and skeletal muscle dysfunction [32,33] may, in fact, act by distorting the thin filament energy landscape, causing muscle regulatory imbalances and failure.
Supplementary Material
Enlarged view of the local region surrounding the actin-tropomyosin Coulombic energy minimum shown in Figure 1c. Note that the color scale here covers a more limited range of values than in Figure 1c.
Coulombic interactions representing the sum of the individual landscapes in Figure 2.
The total energy landscape for tropomyosin interacting with the D292V actin mutant. Note the narrow and relatively deep energy minimum, which will limit tropomyosin oscillation away from its blocking position.
Highlights.
An energy landscape was calculated for tropomyosin on the surface of F-actin.
The energy minimum locates tropomyosin near its myosin-blocking position on F-actin.
Tropomyosin’s myosin-induced open-position is near to the landscape’s energy maximum.
Myosin-binding drives tropomyosin uphill over F-actin to activate thin filaments.
Mutations distort the energy landscape in ways that explain phenotypic traits.
Acknowledgments
We thank Drs. Xiaochuan (Edward) Li, Stefan Raunser, Steven Marston and Edyta Małolepsza for insightful discussions and helpful suggestions. These studies were supported by NIH grant R37HL036153 (to W.L.). The Massachusetts Green High Performance Computing Center and the IWR (University of Heidelberg) provided computational resources.
Abreviations
- EM
electron microscopy
- GBSW
Generalized Born with a simple switching
- MD
Molecular Dynamics
- myosin S1
myosin subfragment 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 2.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 3.Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv Protein Chem. 2005;71:121–159. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]
- 4.Poole KJ, Lorenz M, Evans G, Rosenbaum G, Pirani A, Tobacman LS, Lehman W, Holmes KC. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol. 2006;155:273–284. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Lehman W, Craig R. Tropomyosin and the steric mechanism of muscle regulation. Adv Exp Med Biol. 2008;644:95–109. doi: 10.1007/978-0-387-85766-4_8. [DOI] [PubMed] [Google Scholar]
- 6.Li XE, Tobacman LS, Mun JY, Craig R, Fischer S, Lehman W. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys J. 2011;100:1005–1013. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehman W, Orzechowski M, Li XE, Fischer S, Raunser S. Gestalt-binding of tropomyosin on actin during thin filament activation. J Muscle Res Cell Motility. 2013 doi: 10.1007/s10974-013-9342-0. (in press) [DOI] [PMC free article] [PubMed]
- 8.Vibert P, Craig R, Lehman W. Steric-model for activation of muscle thin filaments. J Mol Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 9.McKillop DF, Geeves MA. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehman W, Hatch V, Korman V, Rosol M, Thomas L, Maytum R. Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J Mol Biol. 2000;302:593–606. doi: 10.1006/jmbi.2000.4080. [DOI] [PubMed] [Google Scholar]
- 11.Pirani A, Xu C, Hatch V, Craig R, Tobacman LS, Lehman W. Single particle analysis of relaxed and activated muscle thin filaments. J Mol Biol. 2005;346:761–772. doi: 10.1016/j.jmb.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Behrmann E, Müller M, Penczek PA, Mannherz HG, Manstein DJ, Raunser S. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150:327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehman W, Galińska-Rakoczy A, Hatch V, Tobacman LS, Craig R. Structural basis for the activation of muscle contraction by troponin and tropomyosin. J Mol Biol. 2009;388:673–681. doi: 10.1016/j.jmb.2009.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 15.Lorenz M, Poole KJV, Popp D, Rosenbaum G, Holmes KC. An atomic model of the unregulated thin filament obtained by x-ray fiber diffraction on oriented actin-tropomyosin gels. J Mol Biol. 1995;246:108–119. doi: 10.1006/jmbi.1994.0070. [DOI] [PubMed] [Google Scholar]
- 16.Lehman W, Li XE, Orzechowski M, Fischer S. The structural dynamics of α-tropomyosin on F-actin shape the overlap complex between adjacent tropomyosin molecules. Arch Biochem Biophys. 2013 doi: 10.1016/j.abb.2013.09.011. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li XE, Holmes KC, Lehman W, Jung H, Fischer S. The shape and flexibility of tropomyosin coiled coils: implications for actin filament assembly and regulation. J Mol Biol. 2010;395:327–339. doi: 10.1016/j.jmb.2009.10.060. [DOI] [PubMed] [Google Scholar]
- 18.Brooks BR, Brooks CL, MacKerell AD, Nilsson L, Petrella RJ, Roux B, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 20.Mackerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 21.Bashford D, Case DA. Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 22.Im W, Lee MS, Brooks CL. Generalized born model with a simple smoothing function. J Comput Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 23.Barua B, Fagnant PM, Winkelmann DA, Trybus KM, Hitchcock-Degregori SE. A periodic pattern of evolutionarily-conserved basic and acidic residues constitutes the binding interface of actin- tropomyosin. J Biol Chem. 2013;288:9602–9609. doi: 10.1074/jbc.M113.451161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barua B, Winkelmann DA, White HD, Hitchcock-DeGregori SE. Regulation of actin-myosin interaction by conserved periodic sites of tropomyosin. Proc Natl Acad Sci USA. 2012;109:18425–18430. doi: 10.1073/pnas.1212754109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maytum R, Hatch V, Konrad M, Lehman W, Geeves MA. Ultra short yeast tropomyosins show novel myosin regulation. J Biol Chem. 2008;283:1902–1910. doi: 10.1074/jbc.M708593200. [DOI] [PubMed] [Google Scholar]
- 26.Lehrer SS, Geeves MA. The muscle thin filament as a classical cooperative/allosteric regulatory system. J Mol Biol. 1998;277:1081–1089. doi: 10.1006/jmbi.1998.1654. [DOI] [PubMed] [Google Scholar]
- 27.Wu S, Liu J, Reedy MC, Tregear RT, Winkler H, Franzini-Armstrong C, et al. Electron tomography of cryofixed, isometrically contracting insect flight muscle reveals novel actin-myosin interactions. PLoS One. 2010;5:9. doi: 10.1371/journal.pone.0012643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perz-Edwards RJ, Irving TC, Baumann BAJ, Gore D, Hutchinson DC, Kržič U, et al. X-ray diffraction evidence for myosin-troponin connections and tropomyosin movement during stretch activation of insect flight muscle. Proc Nat Acad Sci. 2011;108:120–125. doi: 10.1073/pnas.1014599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurzawa SE, Geeves MA. A novel stopped-flow method for measuring the affinity of actin for myosin head fragments using microgram quantities of protein. J Mus Res Cell Motility. 1996;17:669–676. doi: 10.1007/BF00154061. [DOI] [PubMed] [Google Scholar]
- 30.Maytum R, Geeves MA, Konrad M. Actomyosin regulatory properties of yeast tropomyosin are dependent upon N-terminal modification. Biochemistry. 2000;38:1102–1110. doi: 10.1021/bi000977g. [DOI] [PubMed] [Google Scholar]
- 31.Wegner A. The interaction of α,α- and α,β-tropomyosin with actin filaments. FEBS Lettr. 1980;119:245–248. doi: 10.1016/0014-5793(80)80263-8. [DOI] [PubMed] [Google Scholar]
- 32.Marston S, Memo MM, Messer A, Papadaki M, Nowak K, McNamara E, Ong R, El-Mezgueldi M, Li X, Lehman W. Mutations in repeating structural motifs of tropomyosin cause gain of function in skeletal muscle myopathy patients. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt345. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redwood CC, Robinson P. Alpha-tropomyosin mutations in inherited cardiomyopathies. J Muscle Res Cell Motility. 2013 doi: 10.1007/s10974-013-9358-5. (in press) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Enlarged view of the local region surrounding the actin-tropomyosin Coulombic energy minimum shown in Figure 1c. Note that the color scale here covers a more limited range of values than in Figure 1c.
Coulombic interactions representing the sum of the individual landscapes in Figure 2.
The total energy landscape for tropomyosin interacting with the D292V actin mutant. Note the narrow and relatively deep energy minimum, which will limit tropomyosin oscillation away from its blocking position.