Abstract
Optogenetic tools enable the causal examination of how specific cell types contribute to brain circuit functions. A long-standing question is whether it is possible to independently activate two distinct neural populations in mammalian brain tissue. Such a capability would enable the examination of how different synapses or pathways interact to support computation. Here we report two new channelrhodopsins, Chronos and Chrimson, obtained through the de novo sequencing and physiological characterization of opsins from over 100 species of algae. Chrimson is 45 nm red-shifted relative to any previous channelrhodopsin, important for scenarios where red light would be preferred; we show minimal visual system mediated behavioral artifact in optogenetically stimulated Drosophila. Chronos has faster kinetics than any previous channelrhodopsin, yet is effectively more light-sensitive. Together, these two reagents enable crosstalk-free two-color activation of neural spiking and downstream synaptic transmission in independent neural populations in mouse brain slice.
INTRODUCTION
Microbial opsins, light-driven ion pumps and light-gated ion channels that can be genetically expressed in excitable cells, enable the optical activation or inhibition of the electrical activity of defined neuron types, pathways, or regions with millisecond resolution1–6. Currently, there exists an unmet need to independently drive pairs of light-driven ion channels with different colors, which would enable the independent activation of different cell populations. Many groups have used green-peaked channelrhodopsins in conjunction with blue-peaked channelrhodopsins to achieve differential spiking with yellow light7–10, but residual neural spiking crosstalk in the blue remains due to the intrinsic blue absorption by the retinal chromophore11,12. This fundamental limitation means it is currently not possible to achieve robust, temporally precise independent two-color spiking using a red-shifted channelrhodopsin alongside ChR2 in mammalian brain tissue. Previous efforts to further red-shift opsins and reduce blue light sensitivity through mutagenesis has empirically proven difficult, with little red-shifting after the first reported green-peaked channelrhodopsin, VChR19,13,14. Similarly, efforts to further increase the blue light sensitivity of ChR2 have resulted in significantly slower channelrhodopsins15–17, which elicit low temporal precision spiking in response to light pulses of up to a second or more.
One potential strategy for achieving independent two-color excitation is engineering differences in blue light sensitivity between blue- and red-light drivable channelrhodopsins, such that a low blue irradiance can drive precisely timed spikes with the blue channelrhodopsin, while only eliciting subthreshold depolarizations (not causing synaptic release) in neurons expressing the red-light drivable channelrhodopsin. To achieve these goals, we turned to the natural world, performing de novo transcriptome sequencing of 127 species of algae.
Chronos is a new blue- and green-light drivable channelrhodopsin with kinetics faster than any previous channelrhodopsin. Chrimson is a new red-light drivable channelrhodopsin with spectra 45 nm more red-shifted than any previous channelrhodopsin. Together, Chronos and Chrimson robustly mediate the independent two-color spiking of, and synaptic release from distinct neural populations in mouse brain slice. Chronos represents an excellent general-use channelrhodopsin, while Chrimson enables temporally precise experiments requiring red light, such as deep tissue targeting, or scenarios where blue light is visually distracting. To the latter end, we demonstrate through-cuticle brain stimulation and dramatic reduction in visual-system-triggered responses of Chrimson-expressing Drosophila during optogenetic control. Our paper reveals tools of fundamental importance for many new neuroscientific experimental realms, and also provides new channelrhodopsins that may serve as protein backbones for future tools.
RESULTS
Discovering novel channelrhodopsins via de novo sequencing
In recent years, a number of channelrhodopsins have been engineered for neuroscientific applications18, derived from four channelrhodopsin genes from Chlamydomonas reinhardtii or Volvox carteri. However, all currently-known natural channelrhodopsins have blue-green (430–550 nm) spectral peaks13,18–20, and engineered red-shifted channelrhodopsins such as C1V19 and ReaChR19 have peak wavelength sensitivity in the green (~545 nm) and the same spectrum as VChR17. Furthermore, existing channelrhodopsins exhibit an inverse relationship between two desired properties - high light sensitivity and fast kinetics18. We therefore sought to overcome these limitations through exploring natural channelrhodopsin genetic diversity, aiming to discover new opsins possessing unique features not found in previous channelrhodopsins.
We de novo sequenced 127 algal transcriptomes21 and identified 61 channelrhodopsin homologs, which we subsequently synthesized and screened for photocurrents in HEK293 cells via whole cell patch-clamp (Supplementary Figs. 1–4). Of these, we selected opsins with novel characteristics for further characterization in cultured neurons (Fig. 1), focusing primarily on photocurrent, wavelength sensitivity, kinetics, and trafficking (Fig. 1 and Supplementary Figs. 5–9). To avoid selection bias, all opsins were co-transfected into neurons with a secondary tdTomato plasmid, and we selected cells based solely on the presence of cytosolic tdTomato expression (Fig. 1a and Supplementary Fig. 5a,b). This unbiased selection method was applied throughout the paper in all culture experiments, unless otherwise indicated.
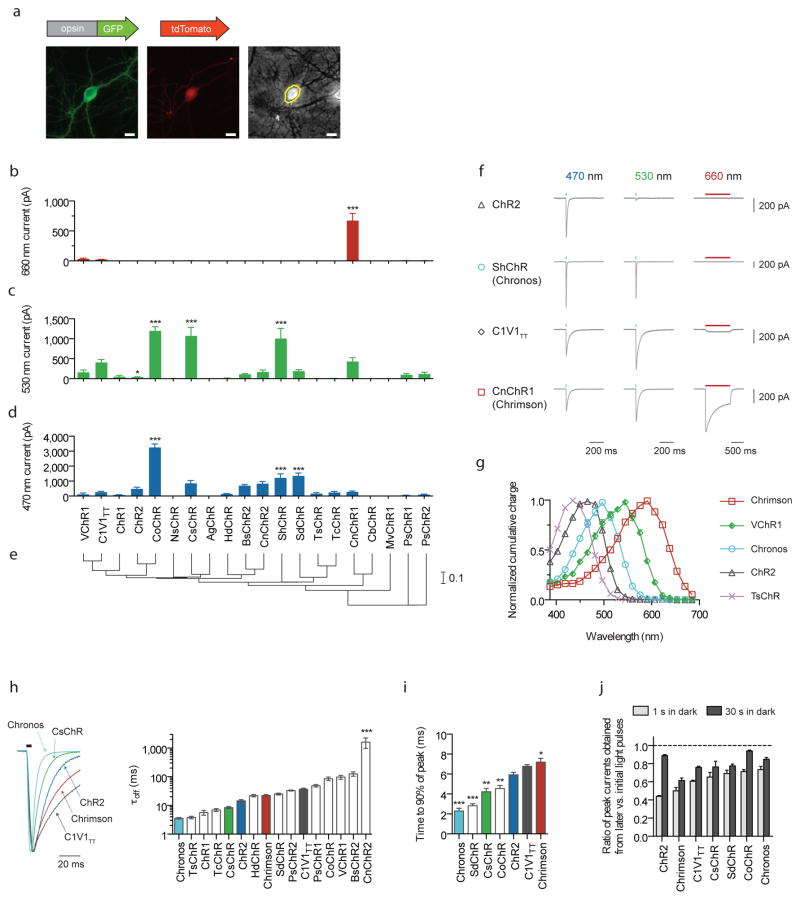
Figure 1. Novel channelrhodopsin spectral classes discovered through algal transcriptome sequencing.
(a) Representative GFP (left), tdTomato (middle) and phase contrast (right) images of a tdTomato and opsin-GFP fusion transfected neuron. Yellow line indicates the mask boundary used to quantify soma fluorescence. Scale bar is 10 μm. (b–d) Maximum photocurrents in cultured neurons in response to far-red (660 nm), green (530 nm) and blue (470 nm) light; blue and green photon fluxes were matched, with illumination conditions defined as follows: 1 s pulse at 10 mW/mm2 for red, 5 ms pulse at 3.66 mW/mm2 for green, and 5 ms pulse at 4.23 mW/mm2 for blue. n = 5 – 12 cells for channelrhodopsins with nonzero photocurrent for at least one color; n = 2 – 6 cells for opsins that did not exhibit any photocurrent. See Supplementary Fig. 6 for individual cell data. Plotted is mean ± standard error of the mean (s.e.m) throughout. (e) Phylogeny tree based on transmembrane helix alignments. Scale is # of substitutions per site. (f) Representative voltage-clamp traces in cultured neurons as measured under the screening conditions in b–d (the longer red light pulse was used to ensure we did not miss any red-sensitive channelrhodopsins in our screen). (g) Channelrhodopsin action spectra (HEK293 cells; n = 6 – 8 cells; measured using equal photon fluxes, ~2.5 × 1021 photons/s/m2). (h–j) Channelrhodopsin kinetic properties as measured in cultured neurons. Off-kinetics (h) were measured under the same conditions as b–d; on- (i) and recovery-kinetics (j) were measured with 1 s pulse at 5 mW/mm2. All opsins were illuminated near their respective peak wavelength, which was either blue or green for all opsins except Chrimson, which was characterized at 625 nm (n = 5 – 12 cells for all kinetic comparisons). (h) τoff is the monoexponential fit of photocurrent decay. (i–j) Raw traces are shown in Supplementary Fig. 7, and further details in Supplementary Fig. 8. (j) Peak current recovery ratios determined from three 1 s light pulses, with the first pulse response used as the baseline for peak current recovery ratio calculations for both second (1 s in dark after first pulse) and third pulse response (30 s in dark after second pulse). Opsin/genus/species names are: VChR1 (Volvox carteri), C1V1TT (VChR1/ChR1 chimaera), ChR1 (Chlamydomonas reinhardtii), CsChR (Chloromonas subdivisa), AgChR (Asteromonas gracillis-B), ChR2 (Chlamydomonas reinhardtii), CoChR (Chloromonas oogama), NsChR (Neochlorosarcina sp.), ShChR (Stigeoclonium helveticum; also called Chronos), MvChR1 (Mesostigma viride), SdChR (Scherffelia dubia), TsChR (Tetraselmis striata), TcChR (Tetraselmis cordiformis), BsChR (Brachiomonas submarina), CnChR (Chlamydomonas noctigama; also called Chrimson), HdChR (Haematococcus droebakensis), CbChR (Chlamydomonas bilatus-A), PsChR (Proteomonas sulcata). Statistics for panels b, c, d, h, and i: *P < 0.05, **P < 0.01 and ***P < 0.001; ANOVA with Dunnett’s post hoc test, with ChR2 as the reference in d, h, i, and C1V1TT as the reference in b and c. Plotted is mean ± standard error of the mean (s.e.m.) throughout.
We assessed wavelength sensitivity and photocurrent amplitude, using ChR2 as a blue (470 nm) reference, and C1V1TT9 as a green (530 nm) and far-red (660 nm) reference (Fig. 1b–f). Of the 20 opsins screened in neurons, we found four previously unknown channelrhodopsins from the species Chloromonas oogama (CoChR), Chloromonas subdivisa (CsChR), Stigeoclonium helveticum (ShChR) and Scherffelia dubia (SdChR) which bore either significantly higher blue photocurrents vs. ChR2 (P < 0.001; ANOVA with Dunnett’s post hoc test used for all multi-way comparisons; Fig. 1d) or significantly higher green photocurrents vs. C1V1TT (P < 0.001; Fig. 1c). Additionally, we discovered the first reported yellow-peaked channelrhodopsin CnChR1, from the species Chlamydomonas noctigama, with 660 nm far-red-light photocurrents of 674 ± 120 pA (values throughout are mean ± s.e.m., n = 11 cells), significantly higher (~30x, P < 0.0001, Fig. 1b,f) than C1V1TT. Based on its spectral sensitivity, we nicknamed this molecule “Chrimson”. With a spectral peak at 590 nm, Chrimson is 45 nm more red-shifted than any other previously known channelrhodopsin (Fig. 1g, Supplementary Figs. 5c,d and 9).
Kinetic parameters and spiking performance
The ability to optically evoke spikes necessitates that channelrhodopsins possess not only photocurrents sufficient to depolarize the neuron cell membrane above its spike threshold, but also on-, off-, and recovery kinetics fast enough to precisely control spike timing and fidelity18,22. Previously published green- and red-light drivable channelrhodopsins have relatively slow off-kinetics, which limits their utility for high frequency neural activation18,19. We characterized the kinetic properties of opsins with comparable or higher green photocurrents than C1V1TT and found only CsChR and ShChR had faster turn-on, turn-off, and recovery kinetics (Fig. 1h–j). With a turn-on of 2.3 ± 0.3 ms (n = 8 cells) and a turn-off of 3.6 ± 0.2 ms (n = 7 cells), the S. helveticum channelrhodopsin ShChR possesses the fastest reported kinetics to date (Supplementary Fig. 3): we therefore nicknamed this molecule “Chronos”.
We assessed Chronos’ green light (530 nm) spiking fidelities at various irradiances and frequencies in cultured neurons (Fig. 2a–c, Supplementary Figs. 10 and 11). As expected from its fast kinetic properties, Chronos-mediated optical spiking perfectly replicated electrically driven spiking between 5 to 60 Hz (Supplementary Fig. 11). In contrast, CsChR only reliably drove spikes up to 20 Hz, and C1V1TT could not reliably drive spikes above 10 Hz even at the highest expression level (Supplementary Figs. 10 and 11). It has previously been noted that slow off-kinetics or poor recovery kinetics of channelrhodopsins can cause depolarization block or reduce photocurrents over sustained pulse trains18. Consistent with this observation, the spike failures we observed at high frequencies for CsChR and C1V1TT primarily occurred later in the pulse train (Supplementary Fig. 10e).
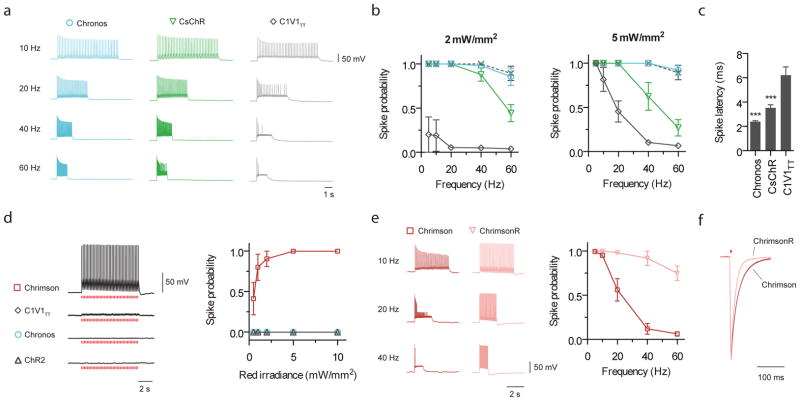
Figure 2. Comparison of optical spiking in cultured neurons.
(a–c) Side-by-side comparison of green light-driven spiking fidelity. All green light spiking protocols used a train of 40 pulses, 2 ms pulse width, at 530 nm, and at the indicated powers (n = 5 – 8 cells for each opsin). (a) Representative green light driven spiking traces at the indicated frequencies at 5 mW/mm2. (b) Green light driven spike probability over a range of frequencies. Dashed line in b is the electrical spiking control from Chronos-expressing neurons (electrical control consisted of a train of 40 pulses at the indicated frequencies; each current injection pulse was 5 ms long and was varied from 200 – 800 pA depending on each neuron’s spike threshold). (c) Spike latencies calculated for 5 Hz trains at 5 mW/mm2 (latency is defined as the time between light pulse onset to the spike peak). (d–f) Comparison of red light (625 nm) spiking. (d) Representative current-clamp traces of red light response and spike fidelity (n = 5 – 8 cells for each opsin; 5 ms pulses, 5 Hz, 5 mW/mm2). (e) Comparison of wildtype Chrimson and Chrimson K176R mutant (a.k.a. ChrimsonR) high frequency red light spiking (n = 10 cells for Chrimson, n = 4 cells for ChrimsonR; 40 pulse train, 2 ms pulse width, 5 mW/mm2). (f) Representative off-kinetics traces for Chrimson vs. ChrimsonR. Statistics for panel c: *P < 0.05, **P < 0.01 and ***P < 0.001; ANOVA with Dunnett’s post hoc test with C1V1TT.
We next examined red light (625 nm) evoked spiking fidelities. Consistent with the earlier photocurrent screening, Chrimson was the only opsin capable of red light-driven spiking with 5 ms pulses (Fig. 2d): these irradiances and pulse widths resulted in Chronos depolarizations of less than 1.5 mV (Supplementary Fig. 12g). However, Chrimson’s slow tau-off of 21.4 ± 1.1 ms (n = 11 cells) in conjunction with its poor recovery kinetics caused depolarization block and channelrhodopsin inactivation at frequencies exceeding 10 Hz (Fig. 2e, Supplementary Fig. 12a–d). We therefore optimized these parameters via mutagenesis, and identified the K176R mutant, denoted as ChrimsonR, which sped up the off-kinetics to 15.8 ± 0.4 ms (n = 5 cells) without altering the red-shifted action spectrum (Fig. 2f, Supplementary Fig. 12e,f). This kinetic improvement enables fast, reliable red-light driven spiking at frequencies of at least 20 Hz in both cultured neurons and acute cortical slice (Fig. 2e and Supplementary Fig. 13), comparable to the blue-light spiking performance of the commonly used ChR2 (H134R)18,23. We additionally found ChrimsonR to be capable of reliably eliciting spikes in cortical slice using >20 millisecond pulses of far-red light (735 nm) (Supplementary Fig. 13), which may be useful for in vivo scenarios desiring light penetration without visual drive.
Chrimson in Drosophila
While optogenetic tools have become widely used in mammalian behavioral experiments, such tools have found more limited use in Drosophila experiments24–28, due to strong innate behavioral artifacts induced by stimulation light reaching photosensitive regions29. Optogenetics has therefore typically been used in constrained circumstances in which light is delivered to peripheral organs while being blocked from reaching the eyes26, in blind flies24, or under circumstances in which undesired side effects of visible-light stimulation do not significantly impact experimental interpretation30. Other technologies such as thermogenetics are often used despite their significantly slower timecourse31,32. We conjectured that red-light-activation with Chrimson would extend the reach of optogenetic tools to Drosophila behavioral experiments.
We first used the Drosophila larval neuromuscular junction (NMJ) to examine the reliability of light triggered action potentials in fly axons expressing Chrimson. Larval muscles have passive membrane properties, so excitatory junction potentials (EJPs) at the larval NMJ accurately reflect spiking in motor axons32. In Chrimson-expressing larvae, both 470 nm and 617 nm triggered EJPs, even at short (1–2 ms) light pulse durations and low intensities (0.06–0.14 mW/mm2) (Fig. 3a,b). In response to light pulse durations over 2 ms, Chrimson activation triggered long lasting barrages of EJPs (Fig. 3a,b). Long wavelength red light (720 nm) also triggered EJPs (Fig. 3c); however, high intensity light (1 mW/mm2) and long light pulse durations (40–160 ms) were required for robust activation of the NMJ. As a control, we tested a commonly used ChR2 fly33 and examined responses to 470 nm and 617 nm light pulses. As in previous work, 470 nm light triggered EJPs in ChR2-expressing animals, but only after relatively long light pulses (16 ms); 617 nm light pulses did not trigger EJPs in ChR2 animals (Supplementary Fig. 14).
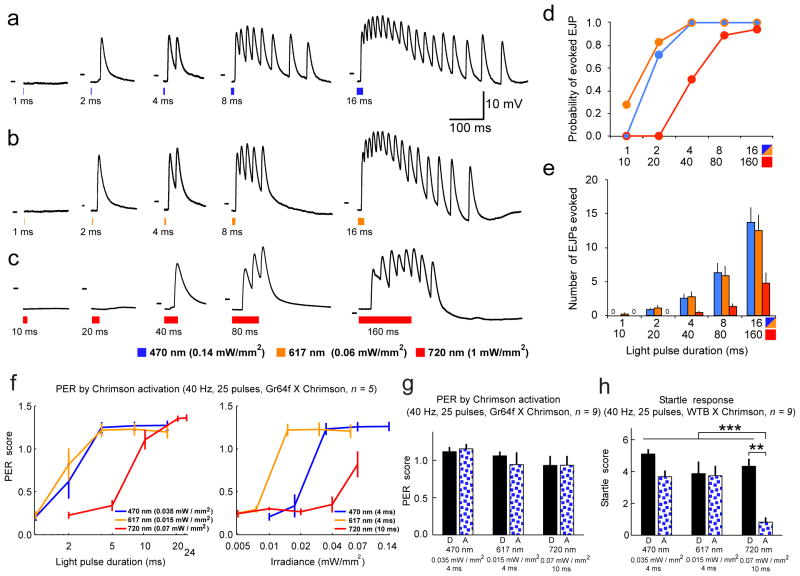
Figure 3. Chrimson evokes action potentials in larval Drosophila motor neurons, and triggers stereotyped behavior in adult Drosophila.
Larval motor axons expressing Chrimson fires in response to blue, orange, and red light pulses. (a–c) Intracellular recordings from m6 muscles in 3rd instar larvae expressing Chrimson in motor neurons. Responses to 470 nm, 617 nm, and 720 nm light pulses of indicated power and increasing duration are shown. Short (1–2 ms pulses) of either 470 nm or 617 nm light trigger single excitatory junction potentials (EJPs), longer pulses (4–16 ms) evoke barrages of EJPs. Long duration, high intensity 720nm light pulses also trigger EJPs. Dashes in each subpanel indicate −50 mV. (d) Probability of light-evoked EJPs after 1, 2, 4, 8, 16 ms pulses in response to 470 nm and 617 nm light and after 10, 20, 40, 80, 160 ms pulses in response to 720 nm light. Sample size: n = 6 muscles in 3 animals, for all larvae experiments. (e) Mean ± s.e.m number of EJPs evoked in response to light pulses. (f–h) Behavioral response of flies to light (n = 5 flies in each case for the adult fly experiments). (f) Proboscis extension reflex (PER) of flies (pUAS-Chrimson-mVenus in attP18/w-;Gr64f-Gal4/+;Gr64f-Gal4/+, shown as Gr64f x Chrimson) to 25 pulses of lights at 470 nm, 617 nm, 720 nm (see Methods for PER scoring). (g) Left: PER of Gr64f x Chrimson flies to pulsed lights in darkness (D) or in a visual arena with flowing blue random dots (A). Right: Startle response of control flies (pUAS-Chrimson-mVenus in attP18/+;+/+;+/+, shown as WTB x Chrimson) to the same visual stimuli as left. Startle score is defined as the number of moving legs after stimulation. ***P < 0.001, **P < 0.01.
In adult flies, we expressed Chrimson in sweet taste receptors (Gr64f-Gal4, expressed in the proboscis and legs34) and measured the proboscis extension reflex (PER) elicited by light stimulation at different wavelengths and intensities (Supplementary Video 1 and Supplementary Fig. 15). At 470 nm and 617 nm, PER were robust at very low light intensities (0.02 mW/mm2 and 0.015 mW/mm2 respectively, Fig. 3f). Flies also responded to an intensity of 720 nm light substantially lower than that required in the larvae (0.07 mW/mm2, 10 ms, Fig. 3f, Supplementary Video 2). 720 nm is believed to be outside of the fly photoreceptor light absorption spectra35,36, suggesting that Chrimson could be used without inducing visually driven behavioral artifacts. However, control flies showed a clear startle response to 720 nm stimuli in darkness (Fig. 3h and Supplementary Video 3). Nevertheless, we reasoned that the saliency of 720 nm light would drop if it were presented along with other visual stimuli at a wavelength well within the sensitivity of photoreceptors, as might be expected during visual behavior experiments in adult flies. As expected, the startle response was efficiently inhibited when we introduced flowing blue random dots during 720 nm stimulation (reduction from 93.2% nonzero startle responses out of 44 valid trials in darkness, to 22.2% out of 45 in arena, see Methods for statistics), while PER of Gr64f x Chrimson flies was preserved (Fig. 3g, Supplementary Videos 4 and 5). With a far-red-shifted activation spectrum, Chrimson also allows direct brain stimulation with intact cuticle, while animals freely behave. We expressed Chrimson in a set of antennal lobe projection neurons (PNv-1; VT03194-Gal4) innervating the V-glomerulus, which is known to respond to CO2 and induces an avoidance response when activated37. Indeed, in this non-visual paradigm, flies in a circular arena reliably avoided quadrants lit by weak red light (617nm, 0.015mW/mm2; Supplementary Fig. 16 and Supplementary Video 6; paired t-test: P = 0.007, see Methods for details), whereas wild-type flies did not show a response (P = 0.502).
Principles of two-color independent neural excitation
The fundamental limitation in creating an independent two-color channelrhodopsin pair is that all opsins can be driven to some extent by blue light. Additionally, neuron-to-neuron variation in opsin expression levels and optical scattering and absorption in tissue suggest that a large difference in effective blue light sensitivity between blue and red-shifted channelrhodopsins, in addition to a large spectral separation, are both required to guarantee robust, independent spiking in mammalian brain tissue. We here systematically explored channelrhodopsins’ blue light sensitivity in cultured neurons, where unlike intact brain tissue, it is possible to precisely control light powers.
In evaluating potential blue partners, we first examined the importance of fast channelrhodopsin kinetics. When Chrimson is exposed to blue light levels as dim as 0.1 mW/mm2 over long durations, as a slow-to-activate blue channelrhodopsin might require, charge integration can result in action potentials (Fig. 4a). However, the on-kinetics of Chronos are roughly 3 times faster than those of ChR2 and 10 times faster than those of Chrimson across all blue irradiances tested (Fig. 4d), suggesting it may be possible to activate Chronos without substantially activating Chrimson.
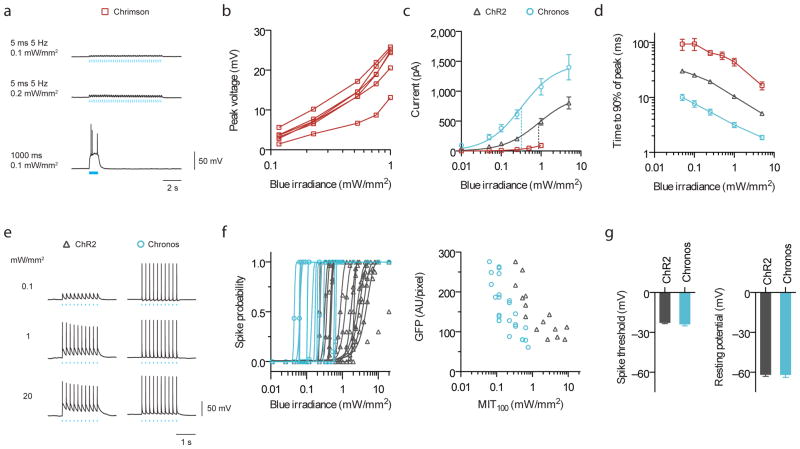
Figure 4. Characterization of channelrhodopsin blue light (470 nm) sensitivities for two-color excitation in cultured neurons.
(a) Current-clamp traces of representative Chrimson-expressing neuron under pulsed vs. continuous illumination. (b) Chrimson blue light induced crosstalk voltages vs. irradiances for individual cells under pulsed illumination (5 ms, 5 Hz, n = 5 cells). (c) Photocurrent vs. blue irradiances (5 ms pulses; n = 4 cells for Chrimson, n = 8 – 10 cells for others). Vertical dotted lines indicate halfway points up the curves for ChR2 and Chronos, as fitted, analogous to “EC50”. (d) Turn-on kinetics (1 s pulse; n = 4 – 7 cells; see Supplementary Fig. 17b,c for raw traces). (e–g) Comparison between ChR2 and Chronos spike probability over three logs of blue irradiance. All pulsed illuminations used 10 pulses, 5 Hz, 5 ms pulse width. (e) Representative spiking traces at the indicated irradiances. (f) Spike probability vs. blue light irradiance, plotted for individual Chronos- or ChR2-expressing neurons and minimum irradiance threshold for 100% spiking (MIT100) as a function of GFP fluorescence. (g) Neuron spike threshold and resting potentials (n = 16 – 23 cells).
Thus, we examined expression variation in Chrimson cells (Fig. 4b), as this variance will translate into some cells exhibiting larger blue light depolarizations than others. We found crosstalk of up to 25 mV at typical blue light powers used for ChR2 excitation (1 mW/mm2, 470 nm, 5 ms pulse). Given Chronos’ high photocurrent (Fig. 1d), high light sensitivity (Fig. 4c), and fast on- and off-kinetics (Fig. 1h, 1i, 4d), we examined whether these properties would translate into spiking at low blue irradiances. Chronos reliably drove 100% spiking at light powers as low as 0.05 mW/mm2, and maintained this fidelity over 2 orders of magnitude to 20 mW/mm2 (Fig. 4e–f). When we determined the minimum irradiance threshold to achieve 100% spiking (MIT100), Chronos had consistently lower MIT100 than ChR2 for similar GFP fluorescence levels (Fig. 4f and Supplementary Fig. 17a), suggesting that Chronos’ high effective light sensitivity is not due to higher expression levels, and that Chronos can robustly mediate light sensitive control of neural spiking across a range of expression levels (Fig. 4f, left) without altering neural excitability (Fig. 4g). These properties make Chronos an ideal blue candidate to be assessed against Chrimson.
Two-color slice validation
To determine acceptable blue and red irradiances to selectively drive Chronos and Chrimson without spiking crosstalk, we first examined the powers at which 470 nm and 625 nm LEDs could drive spiking in Chronos and Chrimson neurons, and the conditions at which spike-level crosstalk occurred (Fig. 5a–e). We expressed Chrimson or Chronos in mouse cortical (layer 2/3) neurons via in utero electroporation, and measured spike probabilities in opsin-expressing neurons: as expected, red light elicited spikes only in Chrimson-expressing cells (Fig. 5c) and blue light elicited spikes in Chronos-expressing cells at as low as 0.05 mW/mm2. Chrimson-expressing neurons began to spike in the blue only above 0.5 mW/mm2, suggesting Chronos-expressing neurons can be driven with high fidelity with zero Chimson spike probability over an operational blue irradiance range between 0.2–0.5 mW/mm2. Throughout this operational blue range, Chrimson-expressing cells showed blue subthreshold crosstalk levels (Fig. 5e) comparable to the culture data (Fig. 4b).
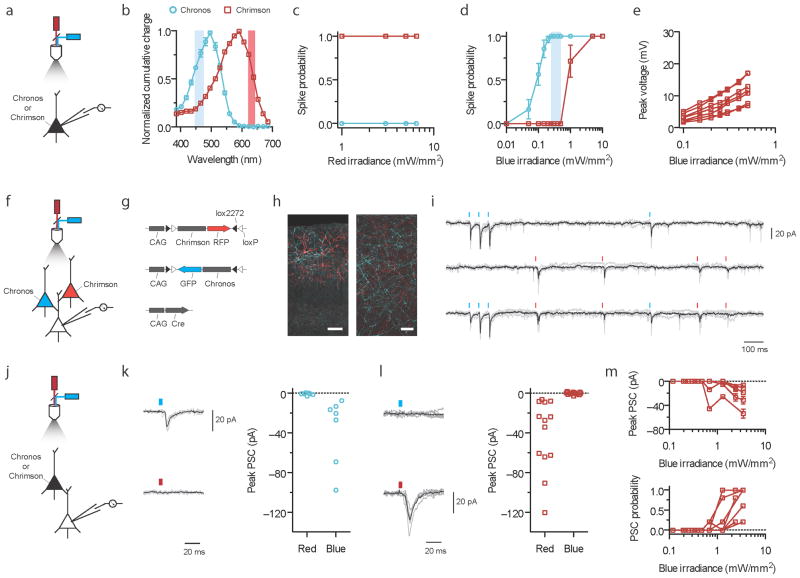
Figure 5. Independent optical excitation of neural populations in mouse cortical slice using Chrimson and Chronos.
(a–e) Spike and crosstalk characterization in opsin-expressing cells. a, f, and j indicate experimental optical configurations. (b) Chrimson and Chronos action spectra emphasizing (vertical shaded bars) the blue (470 nm) and red (625 nm) wavelengths used in this figure. (c–e) Current-clamp characterizations of Chrimson or Chronos expressing neurons in slice to determine optimal irradiance range for two-color excitation. Chrimson-GFP and Chronos-GFP were independently expressed in cortical layer 2/3 neurons in separate mice. 5 ms, 5 Hz light pulses were used; n = 7 cells from 3 animals for Chrimson; n = 11 cells from 4 animals for Chronos. (c) Red light spike probability vs. irradiance. (d) Blue light spike probability vs. irradiance. The blue vertical shaded bar represents the blue irradiance range where Chronos drove spikes at 100% probability and no crosstalk spike was ever observed for any Chrimson neurons. (e) Chrimson subthreshold crosstalk voltage in individual neurons vs. blue irradiances; compare to Fig. 4b. (f–i) Post-synaptic currents (PSC) in non-opsin-expressing neurons downstream of Chrimson and Chronos expressing neurons in brain slice with both opsins introduced into separate neural populations. 0.3 mW/mm2 for blue, 4 mW/mm2 for red, 5 ms pulses; 6 neurons from 3 animals. All synaptic transmission slice experiments were done using widefield illumination (Supplementary Fig. 18). (g) Triple plasmid electroporation scheme for mutually exclusive Chrimson and Chronos expression in different sets of layer 2/3 cortical pyramidal cells. (h) Histology of intermingled Chrimson- (red) and Chronos-expressing (blue) neurons in layer 2/3 (left) and their axons (right). Scale bar is 100 μm and 20 μm for the left and right images respectively. (i) PSCs in response to optical Poisson stimulation with blue and red light; shown are raw voltage traces (gray) with average trace (black) from a single neuron experiencing blue (top), red (middle) or both (bottom) light pulses. See Supplementary Fig. 18e for PSC traces from 5 different neurons downstream of mutually exclusive Chrimson and Chronos expressing neurons in response to blue or red light. (j–m) PSCs in non-opsin-expressing neurons downstream of Chronos- or Chrimson-expressing neurons. Same illumination conditions as in f–i, except pulses delivered at 0.2 Hz. n = 7 cells from 2 animals for Chronos; n = 12 cells from 4 animals for Chrimson. Black trace is the averaged response, grey traces are individual trials, throughout. (k) Chronos driven PSCs under blue or red light, obtained from a representative neuron (left), with population data (right). (l) Chrimson driven PSCs under blue or red light traces, obtained from a representative neuron (left), with population data (right). (m) Chrimson driven PSC amplitudes (top) and the probability of observing a PSC at all (bottom) vs. blue irradiances.
We expressed Chrimson and Chronos in independent sets of neurons within the same cortical microcircuit in layer 2/3 neurons by electroporating Cre-on and Cre-off vectors38, and patched post-synaptic non-opsin-expressing neurons (Fig. 5f–h). For all of these experiments, we chose a blue irradiance of 0.3 mW/mm2 to elicit 100% Chronos spiking without Chrimson activation, and used red irradiances of 1–4 mW/mm2 to reliably recruit Chrimson. We observed distinct post-synaptic current amplitudes for different wavelengths (Fig. 5i and Supplementary Fig. 18e). To examine whether these post-synaptic responses are crosstalk free at the synaptic transmission level, just as spiking events were at the action potential level, we singly expressed each opsin in distinct mouse brains, and patched cells post-synaptic to opsin-expressing cells (Fig. 5j–m). As expected, Chronos reliably drove synaptic events in response to blue light, and never under red light (Fig. 5k; P = 0.1, paired t-test of peak current 30 ms before vs. 30 ms after red light delivery). Chrimson-expressing neurons reliably drove synaptic events upon red illumination, and never upon blue illumination (Fig. 5l; P = 0.43, paired t-test of current before vs. after blue light delivery). Blue light-induced EPSCs by Chrimson-expressing neurons began only outside this range, at 0.65 mW/mm2 (Fig. 5m). We thus conclude it is possible to independently drive Chronos and Chrimson at the neural synaptic transmission level, using light powers determined by the operational blue irradiance range. We here have defined, and also demonstrated a strategy for end users to implement independent two-color excitation of synaptic transmission with zero post-synaptic crosstalk. As a final demonstration of Chronos’s experimental utility, we found pure axonal Chronos stimulation of retinal ganglion cell axons in the superior colliculus reliably elicited EPSCs in downstream neurons (Supplementary Fig. 19).
DISCUSSION
We here present the results from a broad systematic screen of 61 algal opsins, sequenced de novo as part of a massive plant transcriptome endeavor21 (www.onekp.com). Just as genomic diversity has yielded red vs. green fluorescent proteins from different organisms, here we find that a broad sweep of nature is capable of yielding molecules with diverse biophysical properties. We discovered Chronos, an ultra light-sensitive blue channelrhodopsin with faster kinetics than any previously described, and which might represent an excellent general use channelrhodopsin.
We also discovered and characterized Chrimson, a red-light drivable channelrhodopsin 45 nm more red-shifted than any previous channelrhodopsin, and which might be useful in scenarios where red light stimulation is essential. All previous channelrhodopsins that can be driven off-peak in the red (630 nm) have on/off kinetics exceeding 100 ms9,19: perhaps a fundamental limitation of their original VChR1 scaffold. Chrimson and Chronos are novel molecules which represent significant forward jumps in the field of opsin engineering: their spectral and kinetic properties enable fundamentally new types of experiments.
We explored the utility of Chrimson to mediate processes requiring red light, using Drosophila melanogaster. Chrimson was able to mediate responses in larval and adult flies with extremely low light powers across all wavelengths tested, most likely due to the robust high expression achieved with the expression cassette used (see methods). Further, light induced startle responses of adult flies were significantly reduced at longer wavelength (720 nm, but not at 470nm or 617nm) as long as visual distractors of shorter wavelength were provided. Thus, Chrimson may be useful for temporally precise neuronal stimulation in a variety of Drosophila behavioral experiments. Curiously, our results suggest that the photosensitivity of the fly eye extends well beyond the wavelength of typical room light for fly experiments (~650 nm), highlighting the utility of the 720 nm stimulation protocol we introduce for a wider range of behavioral experiments. Wavelengths lower than 720 nm also be useful in situations in which startle responses do not affect measurements of the parameters under study, such as stimulating neurons in the brain through intact cuticle to induce non-visually-driven behaviors as here shown with 617 nm light and CO2-responding neurons, something that has not yet been reliably possible using optogenetic techniques.
Using both Chronos and Chrimson, we found an ample blue light irradiance range that evokes reliable Chronos-induced spikes with zero Chrimson-induced spikes in mouse cortical slice, and also allowed Chronos-induced synaptic transmission with zero Chrimson-induced synaptic transmission. As most two-color experimental setups will not be identical to our slice demonstration, several constraints exist upon the use of effective blue light sensitivity to achieve two-color separation. Whenever possible, Chronos should be expressed under the stronger promoter and the more excitable neuron type than Chrimson to minimize Chrimson-induced depolarization. The experiments in this paper characterized two-color excitation with symmetric promoters and cell types to stringently test whether Chronos and Chrimson can, without exploiting differences in cellular excitability, present a clear separation due to biophysical properties. Neuroscientists seeking to manipulate two different cell populations may find expressing Chronos at high levels in the more excitable cell type may further increase the reliable blue dynamic range.
Although Chronos has the fastest kinetic properties of all reported channelrhodopsins, blue light pulses delivered at very high frequencies as part of a two-color experiment could in principle lead to charge integration and thus Chrimson blue spiking crosstalk (Fig. 4a and Supplementary Fig. 17c). The temporal precision of Chronos vs. Chrimson mediated synaptic events may also depend on light power, potentially limiting usage in scenarios requiring sub-millisecond timing of synaptic release, although ~1 ms jitter is achievable (Supplementary Fig. 20). We have additionally observed in post-synaptic experiments that the stimulation frequency for both red and blue pulses is fundamentally limited by the wild-type Chrimson, the slowest of the opsin pair (Supplementary Fig. 21). Accordingly, we engineered ChrimsonR, a faster Chrimson kinetic mutant, which has similar blue light sensitivity to the wildtype (Supplementary Fig. 17d), but allows modulation at >20Hz range (Fig. 2e–f and Supplementary Fig. 13b). However, it would still be desirable to further improve Chrimson’s kinetics and decrease its overall light sensitivity to enable high frequency modulation in both the red and blue channels and to increase the usable blue irradiance range.
In vivo, it would be important to illuminate the circuit region of interest with powers that fall within the windows here defined, which may require alternative illumination methods such as 3D optical waveguides39 or wireless LED implants40. While these constraints add complexity to experiments, they may also enable in vivo two-color experiments not previously possible.
METHODS
Molecular Cloning
All opsin genes were synthesized (Genscript) with mammalian codon optimization and subcloned as previously described3. C1V1TT and ReaChR were synthesized with the same codon usage as previously described9,19. For cultured neuron studies, all genes were subcloned into lentiviral backbone under CaMKII promoter and with a C-terminal GFP fusion. No trafficking sequences were added to any opsin genes. The tdTomato plasmid used for co-transfection in cultured neuron is also subcloned into lentivral backbone, but with a ubiquitin promoter instead. For in utero electroporation experiments, all genes were subcloned into pCAGIG vector (Addgene 11159) with C-terminal GFP, tdTomato, or mOrange2 fusion. The Cre-dependent vectors were generated using the same pCAGIG vector, but with lox sites flanking protein coding region as done previously38.
We performed RACE on some of the sequences (Supplementary Fig. 1) by using 5′ RNA ligation to enrich for full length transcripts (Supplementary Table 2). Protocol is the same as Ambion RLM-RACE kit and PCR primers were designed based on partial transcriptome sequencing results. RACE products were blunt cloned (Invitrogen Zero Blunt kit) into vector for Sanger sequencing.
Characterization of channel kinetics and ion selectivity in HEK293FT cells
Whole-cell patch clamp recordings were performed in isolated HEK293FT cells to avoid space clamp issues. All recordings were performed using an Axopatch 200B amplifier and Digidata 1440 digitizer (Molecular Devices) at room temperature. In order to allow isolated cell recording, cells were plated at a density of 15,000 cells per well in 24-well plates that contained round glass coverslips (0.15 mm thick, 25 mm in diameter, coated with 2 % Growth Factor Reduced Matrigel in DMEM for 1 h at 37 °C). To ensure accurate measurements, cells with access resistance less than 25 MΩ, holding current less than ± 50 pA were used. Typical membrane resistance was between 500 MΩ – 2 GΩ and pipette resistance was between 3 – 6 MΩ. Photostimulation of patch clamped cells was conducted by a 470 nm LED (Thorlabs) at irradiance of 10 mW/mm2 for ChR2, ChR2 E123A, and Chronos, and by a 590 nm LED (Thorlabs) at irradiance of 4.6 mW/mm2 for Chrimson.
For characterizing channel kinetics, the extracellular solution (Tyrode’s solution) consisted of (in mM) 125 NaCl, 2 KCl, 3 CaCl2, 1 MgCl2, 10 HEPES, 30 Glucose, pH 7.3 NaOH adjusted, 305 mOsm and the intracellular solution consisted of (in mM) 125 K-Gluconate, 8 NaCl, 0.1 CaCl2, 0.6 MgCl2, 1 EGTA, 10 HEPES, 4 MgATP, 0.4 NaGTP, pH 7.3 (KOH adjusted), with 295–300 mOsm (sucrose adjusted). Channel closing rates (τoff; e.g. as in Fig. 3a) were measured by fitting the decay of photocurrent after 2 ms light pulses to monoexponential curves, and time to peak (time to reach 90% of peak photocurrent after the beginning of illumination, as in Fig. 3b) were measured from 1s pulses.
For characterizing ion selectivity in channelrhodopsins, the intracellular solution’s ion compositions were (in mM) 140 K, 5.1×10−5 H, 5 EGTA, 2 MgCl2, 10 HEPES. The 145 mM NaCl extracellular solution’s ion compositions were (in mM) 145 Na, 5 K, 1 Ca, 5.1×10−5 H, 10 HEPES, 5 glucose, 2 MgCl2. The 145 mM KCl extracellular solution’s ion compositions were (in mM) 145 K, 1 Ca, 5.1×10−5 H, 10 HEPES, 5 glucose, 2 MgCl2. The 90 mM CaCl2 extracellular solution’s ion compositions were (in mM) 5 K, 91 Ca, 5.1×10−5 H, 10 HEPES, 5 glucose, 2 MgCl2. The 5 mM NaCl extracellular solution’s ion compositions were (in mM) 5 Na, 5 K, 1 Ca, 5.1×10−4 H, 135 NMDG, 10 HEPES, 5 glucose, 2 MgCl2. All solutions were balanced to pH 7.4, except for the 5 mM NaCl solution that was balanced to pH 7.3. The liquid junction potentials were 5.8 mV, and 4.9 mV for 90mM CaCl2 and 5 mM NaCl, respectively, and were corrected during recording; the others were < 1 mV in liquid junction potential. Peak component of the photocurrent during a 1s illumination was used to calculate photocurrent density ratios.
HEK 293FT cell culture and transfection
HEK 293FT cells (Invitrogen) were maintained between 10–70% confluence in D10 medium (Cellgro) supplemented with 10% fetal bovine serum (Invitrogen), 1% penicillin/streptomycin (Cellgro), and 1% sodium pyruvate (Biowhittaker)). For recording, cells were plated at 5–20% confluence on glass coverslips coated with Matrigel (BD Biosciences). Adherent cells were transfected approximately 24 hours post-plating either with TransLT 293 lipofectamine transfection kits (Mirus) or with calcium phosphate transfection kits (Invitrogen), and recorded via whole-cell patch clamp between 36–72 hours post-transfection. 1.25 μg of DNA was delivered. 1 uM of retinal was supplemented to the culture media for an hour before patch clamp experiments.
Primary neuron culture and transfection
All procedures involving animals were in accordance with the National Institutes of Health Guide for the care and use of laboratory animals and approved by the Massachusetts Institute of Technology Animal Care and Use Committee. Hippocampal neuron culture was prepared from postnatal day 0 or day 1 Swiss Webster (Taconic or Charles River) mice as previously described3, but with the following modifications: dissected hippocampal tissues were digested with 50 units of papain (Worthington Biochem) for 5 minutes and the digestion was stopped with ovomucoid trypsin inhibitor (Worthington Biochem). Cells were plated at a density of 16,000–20,000 per glass coverslip coated with Matrigel (BD Biosciences).
Cultured neurons were transfected at 4 days in vitro (DIV) with commercial calcium phosphate kit (Invitrogen). We added an additional washing with acidic MEM buffer (pH 6.8 – 6.9) after calcium phosphate precipitate incubation to completely re-suspend residual precipitates41. tdTomato was used as a co-transfectant DNA reagent to assist with unbiased selection of opsin-expressing neurons (see main text); in this condition, we would deliver 2 μg of opsin DNA and 0.2 μg tdTomato. When no tdTomato was used, we used 2 μg of opsin DNA alone.
Whole-cell electrophysiology in vitro and in slice
Whole cell patch clamp recordings were made using Axopatch 200B or Multiclamp 700B amplifier, a Digidata 1440 digitizer, and a PC running pClamp (Molecular Devices). For in vitro current-clamp recordings, neurons were patched 14–18 DIV (10–14 days post-transfection) to allow for sodium channel maturation. Neurons were bathed in room temperature Tyrode containing 125 mM NaCl, 2 mM KCl, 3 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 30 mM glucose, 0.01 mM NBQX and 0.01 mM GABAzine. The Tyrode pH was adjusted to 7.3 with NaOH and the osmolarity was adjusted to 300 mOsm with sucrose. For in vitro voltage-clamp recordings, neurons were patched 11–14 DIV (7–10 days post-transfection) and were done under similar conditions as current-clamp recordings, except tyrode also contains 1 μM of tetrodotoxin (TTX, Tocris Bioscience). No retinal was supplemented for any cultured neuron recordings.
For slice recordings, room temperature artificial cerebrospinal fluid (ACSF) was continuously perfused over slices and no blockers were used. ACSF contained: 127 mM NaCl, 2.5 mM KCl, 25 mM NaHCO3, 1.25 mM NaH2PO4, 12 mM D-glucose, 0.4 mM sodium ascorbate, 2 mM CaCl2, 1 mM MgCl2, and was bubbled continuously with carbogen.
For both in vitro and slice recordings, borosilicate glass pipettes (Warner Instruments) with an outer diameter of 1.2 mm and a wall thickness of 0.255 mm were pulled to a resistance of 3–7 MΩ with a P-97 Flaming/Brown micropipette puller (Sutter Instruments) and filled with a solution containing 125 mM K-gluconate, 8 mM NaCl, 0.1 mM CaCl2, 0.6 mM MgCl2, 1 mM EGTA, 10 mM HEPES, 4 mM Mg-ATP, and 0.4 mM Na-GTP. The pipette solution pH was adjusted to 7.3 with KOH and the osmolarity was adjusted to 298 mOsm with sucrose. For voltage clamp experiments, cells were clamped at −65 mV for in vitro (HEK293, cultured neuron) recordings and between −65 to −80 mV for slice recordings. For current clamp experiments, <50 pA constant current injection was used for in vitro recordings and no current injection was used for slice recordings. To ensure accurate measurements, cells with access resistance between 5–35 MΩ, holding current less than ± 100 pA (at −65 mV, in voltage clamp) were used. Access resistance was monitored throughout recording. Data was analyzed using Clampfit (Molecular Devices) and custom MATLAB scripts (Mathworks, Inc.)
Light delivery and imaging
All neuron culture and slice experiments were done with the LEDs mounted on microscope for wide-field illumination, with nominal wavelength at 470 nm, 530 nm, 625 nm, 660 nm and 735 nm (Thorlabs, M470L2, M530L2, M625L3, M660L3 and M735L3 respectively). We additionally filtered LEDs spectrum with the following bandpass filters (Semrock): 530 nm LED with 543 nm ± 11 nm filter, 625 nm LED with 632 nm ± 11 nm, and 660 nm LED with 661 nm ± 10 nm. The 735 nm LED was not additionally filtered. Light power was controlled through LED driver using analog voltage modulation (LEDD1B or DC4100, Thorlabs), and voltage pulse width was adjusted to obtain the desired illumination duration as measured with photometer (S120VC, Thorlabs). Illumination spot size with different objectives were measured by focusing on a microscope slide coated with appropriate wavelength Alexa dye and photobleaching it for 10 minutes under full intensity illumination. The photobleached slide was then imaged with a micrometer calibration slide to determine the photobleached radius.
Action spectrum data was taken with a monochromator (Till-Photonics Polychrome IV, bandwidth 15 nm centered around each value) and a separate (Uniblitz) shutter was used to gate the light pulses to 10 ms. Equal photon fluxes of ~2.5 × 1021 photons/s/m2 (~1.1 mW/mm2 at 450 nm) were used across wavelengths by using the monochromator’s builtin intensity adjustment. We additionally corrected for any photon dosage difference (12% at the most) between wavelengths by dividing the recorded opsin response by the measured photon dose. A (QuickMacros) script was used to automate the wavelength and intensity selections and to synchronize with electrophysiology recording. For each cell, wavelengths were swept from blue-to-red and red-to-blue, and the responses were averaged.
Photocurrent measurements were single trial and input resistance was monitored throughout. For color ratio comparisons, the order of illumination wavelength was shuffled between different cells. For photocurrent recovery kinetics measurements, an additional two minutes wait in the dark preceded the protocol to allow for recovery before beginning the measurement. In protocols where irradiances (or stimulation frequencies) were varied, the irradiances (or stimulation frequencies) were either shuffled or measured both from low-to-high and high-to-low. In the acute slice post-synaptic experiments, at least 5 sweeps were taken for each condition.
Unless otherwise specified, all experiments included 20–30 seconds wait in the dark between sweeps to allow for opsin recovery.
Quantitative fluorescence comparison of opsin-GFP and cytosolic tdTomato expressed in cultured neurons was imaged with Leica HCX APO L 20x objective (air, NA=0.5) using Hamamatsu Orca Flash 2.8 under identical illumination conditions throughout: 445 nm LED (Thorlabs) at 4.28 mW/mm2 using EN GFP filter cube (Leica) for GFP fluorescence; 530 nm LED (Thorlabs) with 543 nm ± 11 nm filter (Semrock) at 2.14 mW/mm2 using Custom R filter cube (Leica) for tdTomato fluorescence.
In utero electroporation and virus delivery for acute slice experiments
All procedures were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Massachusetts Institute of Technology Committee on Animal Care. C57BL/6J E16-timed pregnant mice were used for electroporation. Surgery was done under ketamine-xylazine anesthesia and buprenorphine analgesia. For cortical experiments, DNA solution containing plasmids of interest were injected into lateral ventricle of each embryo using a pulled capillary tube. Five square pulses (50ms width, 1Hz, 35V) were applied using tweezer electrode for electroporation (Harvard Apparatus, ECM 830). Direct opsin-expressing experimental mice were electroporated with pCAG-opsin-GFP plasmid. Post-synaptic experimental mice were electroporated with pCAG-FLEX-rc[Chronos-GFP] and/or pCAG-FLEX-Chrimson-mOrange2, and pCAG-Cre plasmids. pCAG-Chrimson-tdTomato was additionally used in half of the single post-synaptic experiments.
For the retinal ganglion cell-superior colliculus experiment, intravitreal virus injection was performed on P0 C57BL/6 mice with Nanoject II (Drummond) under cold anesthesia. 100 nL of rAAV2/8-Synapsin-Chronos-GFP (titer 1.4×1013 particles/mL) was injected into the eye. AAV particles were produced by the University of North Carolina Chapel Hill Vector Core.
Data analysis
Phylogenetic trees were generated using Neighbor-joining method with p-distance model (MEGA5).
Opsin-GFP and tdTomato fluorescence was measured with custom MATLAB script using masks around neuron soma region. Masks were generated (based on either GFP or tdTomato images) using CellProfiler (www.cellprofiler.org) and manually edited in ImageJ to remove neuronal processes. The same mask was used to quantify both GFP and tdTomato fluorescence.
Action spectra were computed by integrating charge from light-onset to the half-max time. Half-max time refers to the time that the highest amplitude response wavelength has half of it’s peak current. This is to ensure the measured response is the transition from ground state, as oppose to secondary photoproducts. Given the integration window is the same across wavelengths for a given cell (typically 1–3 ms), this is effectively measuring the initial slope as done previously by other labs13.
EC50 in Fig. 4c was calculated by fitting 5 ms blue irradiance photocurrents to the one-site specific binding model (Y = Bmax*x/(Kd + x) using Prism (GraphPad Software), where the Kd corresponds to EC50. This is the same convention as previously published paper18.
Spikes were defined as depolarizing above 0 mV and then repolarizing below –30 mV within each stimulation interval. Spike threshold was measured with a 500 ms current ramp (400–800 pA) current injection protocol and determined with custom MATLAB script. Threshold was defined as the voltage at which the slope (dV/dt) has exceeded 0.3 mV/s.
Slice preparation
P20–P40 mice were used for slice preparation unless otherwise specified. Mice were anesthetized with isofluorane and transcardialy perfused with ice-cold cutting solution containing 110 mM choline chloride, 25 mM NaHCO3, 25 mM D-glucose, 11.6 mM sodium ascorbate, 7 mM MgCl2, 3.1 mM sodium pyruvate, 2.5 mM KCl, 1.25 mM NaH2PO4 and 0.5 mM CaCl2. The brain was then carefully removed and mounted in vibrating blade microtome (Leica VT1000S). 300-mm thick coronal slices of the cortex (varying regions of brain were sectioned, depending on where the electroporation resulted in expressing neurons; they were as a rule found in layer 2/3 of the cortical area obtained) were cut with a vibrating metal blade at 90 Hz and 0.1 mm/s cutting speed. Sectioned slices were incubated in 37 °C cutting solution for 30–45 minutes before transfer to room temperature oxygenated artificial cerebrospinal fluid (ACSF) for recording. ACSF contained 127 mM NaCl, 2.5 mM KCl, 25 mM NaHCO3, 1.25 mM NaH2PO4, 12 mM D- glucose, 0.4 mM sodium ascorbate, 2 mM CaCl2, and 1 mM MgCl2. For spiking characterization (Fig. 5c–e), we used a Nikon CFI Apo 60x NIR objective (water immersion, NA=1); for synaptic characterizations (Fig. f–m), we used a Nikon CFI Super Plan Fluor ELWD 20xc objective (air, NA=0.45) in order to recruit as many synapses as possible to insure stringency of the zero crosstalk synaptic control. For slice experiments, electrical artifacts were apparent in the recorded traces as a result of LED on/off coupling to fluids flowing by the slice, since LED was in the Faraday cage; we include raw traces with artifacts interpolated in Fig. 5, and without artifact interpolation in Supplementary Fig. 22.
Fly stocks
Chrimson constructs were prepared with additional Golgi and ER export motifs42–45 in the pJFRC7–20XUAS-IVS vector46 and inserted in attP18/+;+/+;+/+. To express Chrimson in larval motor neurons, we crossed UAS-Chrimson-mVenus flies to flies containing a GAL4 driver (OK371-GAL4) that drives expresses in all glutamatergic neurons47. We used the same GAL4 line to express ChR233 in larval motorneurons. Gr64f-gal4 flies were obtained from J.R. Carlson34, UAS-ChR2 flies from W.D. Tracey Jr.33, and pBDP-Gal4 from G.M. Rubin48. Control flies for adult fly experiments were obtained by crossing Chrimson virgin female flies to wild type Berlin (WTB) flies. VT031497-Gal4 flies from B. Dickson (personal communication).
Larval NMJ: fly handling and preparation
Flies were raised on standard cornmeal based media with 0.2 mM all-trans-retinal at 25°C. Wandering 3rd instar larvae were dissected in HL3.1 physiological saline containing (in mM) 70 NaCl, 5 KCl, 0.4 CaCl2, 4 MgCl2, 10 NaHCO3, 5 trehalose, 115 sucrose, 5 HEPES, pH 7.15. Animals were filleted as described previously 32 and the central nervous system was removed. Larval fillets were then mounted on a standard intracellular electrophysiology rig. Temperature was maintained at ~22°C.
Larval NMJ: intracellular recording and stimulation parameters
We used 10–15 MΩ sharp intracellular electrodes filled with 3M KCl to record light evoked excitatory junctional potentials in larval muscle 6 (m6). The electrode was maneuvered using a MP-285 micromanipulator (Sutter Instruments). Recordings were amplified with an Axopatch 200B (Molecular Devices) and collected using a Powerlab 16/30 and Chart 7.1 software (both from AD Instruments, Colorado Springs, Co.). We delivered 1, 2, 4, 8, and 16 ms light pulses at either 470 nm (0.14 mW/mm2) and 617 nm (0.06 mW/mm2), or 10, 20, 40, 80, and 160 ms at 720 nm (1 mW/mm2). Light pulses were generated with LED light sources (470 nm, 617 nm: OptoLED, Cairn Instruments, Kent, UK; 720 nm: Thorlabs LED controller), triggered by the Powerlab 16/30 data acquisition system. For each genotype tested, we recorded from 2 muscles in each of 3 separate animals. Electrophysiological data were analyzed with custom scripts in Spike2 (Cambridge Electronic Designs).
Adult flies: fly handling
Flies were raised from the egg stage49 on standard cornmeal and soybean based media with 0.2 mM retinal. Vials were wrapped in aluminum foil to protect retinal from light and kept in a 23°C, 60% humidity incubator. Flies were transferred to fresh retinal food vials on the first day of eclosion. We used 2- to 3-day-old female flies in PER characterization experiments for 470nm and 617nm, and 4- to 5-day-old female flies for 720nm. All flies used to measure behavioral artifacts and the corresponding PER were crossed on the same day, raised side-by-side in two vials of the same batch of media and collected on the same day.
Adult flies: setup for behavioral experiments
The fly preparation and LED display have been published elsewhere50. Briefly, female flies were cold anesthetized and placed in a sarcophagus under a dissection microscope. Then, they were tethered with a wire placed between the head and the thorax with UV-activated glue. They were centered in a modular display system50 (IO Rodeo), which consists of 5 rows and 7 columns of 8×8 LED panels (Bright LED Electronics Corp., BM-10B88MD) covering the upper visual field from −105° to +105°. The LED emission surface was covered with a layer of conductive film to minimize electrical noise (Clear Shield, Transparent Conductive Film), two layers of bandpass filter gel (ROSCO, roscolux #59 indigo), another layer of conductive film, and a layer of stencil paper to prevent reflection. Behavior was recorded using a camera (camera: PointGrey, FFMV-03M2M, lens: Computar, MLM3x-MP, software: MATLAB Image Acquisition Toolbox, MathWorks) with an IR illumination source (Oslon, SFH4715S), longpassed at 850nm (Thorlabs, FGL850) to prevent Chrimson activation. The same filter was used in front of the imaging camera to prevent image corruption from LEDs used for Chrimson activation. We used three high power LEDs for Chrimson activation (Thorlabs, M470L2, M617L2, M735L3 respectively) after collimation (Thorlabs, ACL2520-DG6-A for 470nm and 617nm, ACL2520-DG6-B for 735nm, additional bandpass filtering for 735nm: Semrock, FF01-720/12-25). Light intensity was measured at the position of the fly (Thorlabs, S130C). The intensity of the random dot pattern was 40 nW/mm2 and that of IR illumination was 240 μW/mm2. The overall configuration is shown in Supplementary Video 1. PER to 470 nm and 617 nm light sources by control flies are shown in Supplementary Figure 15.
Adult flies: design and analysis of PER experiments
Pulse width (ms) and LED light intensity (mW/mm2) were varied in PER experiments. The number of pulses (25) and the frequency (40 Hz) were fixed. For each fly, a total of 10 parameter sets (5 per parameter) were tested in each lighting condition with 5 repetitions. Inter-stimulus-interval (ISI) was 30 seconds. To minimize desensitization, trials were ordered from low to higher light intensity and low to higher pulse widths. The LED was controlled using a data acquisition device (Measurement Computing, USB-1208FS) and the Data Acquisition Toolbox (MathWorks). Fly behavior was recorded using a camera from 1 second before to 2 seconds after the initiation of each trial. The positions of the neck connectives and the root of the antennae were manually determined. The position of the tip of the proboscis was determined by finding the maximum horizontal coordinate of non-zero pixels after the image was thresholded. The PER score was scaled by the size of the head capsule (Supplementary Fig. 15a). All trials were manually proofread to correct image corruption by leg movement. Five flies were used for each combination of wavelength and fly group.
Adult flies: measurement of light-induced behavioral artifacts
Setup used to measure light-induced behavioral artifacts was similar to that used for PER characterization with the following exceptions: Nine Chrimson flies (Gr64f x Chrimson) and nine control flies (WTB x Chrimson) were tested for all conditions in a single day. Flies were given 3 hours to settle after tethering. For each wavelength, we used the minimum LED intensity that reliably evoked PER (Fig. 3f). We manually triggered each trial only when flies stopped moving (minimum ISI: 6.5 seconds). Flow speed for the blue random dot pattern presented on the LED display was 20 pixels/s. For the dark condition, the LED arena was removed from the setup. The startle score was manually assessed by counting the number of legs moving within 2 seconds of initiation of each trial (scores thus ranged from 0–6 with flight assigned a score of 7). Trials were discarded if flies moved before stimulation. No pair of dark and lit arena conditions showed significant difference (paired t-test or Wilcoxon signed rank test after Jarque-Bera normality test; p-level 0.05) except the 720 nm condition for WTB x Chrimson (Wilcoxon signed rank test; P = 0.004). To test for differences between the random dot condition at 720nm and all other conditions (WTB x Chrimson), data in all 6 conditions were randomly reassigned into each condition to obtain a null distribution of differences between the conditions (10,000 repetitions). No sample showed greater difference than the original one (thus P < 1/10,000). MATLAB and the Statistics Toolbox (MathWorks) were used for all analyses.
Adult flies: optogenetics of freely behaving intact flies
An LED array (array of Luxeon Rebel, 700mA, 617 nm, Philips LXM2-PH01-0070) and its driver circuit were custom-designed to independently illuminate each quadrant of a circular light arena from the bottom, and controlled by a microcontroller (Arduino) using custom software (See Supplementary Fig. 16a for schematic of setup). A diffuser (Optically Colored Cast Acrylic Sheet, 1/8″ Thick, 12″ × 12″, White, McMaster 8505K11) was placed on top of an LED array to produce a homogeneous light field under a white opaque plastic panel with a circular hole, which served as the behavior arena (3D printed, Vero White, Objet Connex 350; hole dimension: 100mm diameter, 3mm depth). The arena was covered by a transparent plastic panel (Optically Clear Cast Acrylic Sheet, 1/8″ Thick, 12″ × 12″, McMaster 8560K239) to allow video recording. For fly movement imaging, the whole arena was illuminated from above by an IR light source (850 nm), and a thin IR absorption film (Laser-gard PVC film, YAG, Edmund Optics 53-738) was placed between the arena and the diffuser to prevent IR reflection. In a single session (see Supplementary Video 6), we illuminated two opposing quadrants of the arena with the LED array for 30s (0.015mW/mm2) and switched to the other two quadrants for the next 30s. The switching protocol was repeated 3 times for a total video recording of 120s. For each session (total 9 sessions for each genotype), we put 10~20 flies (2~5 day old males and females) in the arena. The behavior was recorded using a USB3 camera (Flea3, PointGrey, with long pass filter of 800nm) and custom software. Image analysis was done in MATLAB (MathWorks). The background image (an average image over all frames) was subtracted from each frame. Next, any pixels with less than 20% of the maximum pixel value of the frame were set to zero and others to one. This generates a white IR-reflected body image of flies with black background. This image was divided into four quadrants. The percentage of flies in quadrants 1 (northeast) and 3 (southwest) was approximated by the percentage of white pixels in those quadrants relative to the total number of white pixels. Finally, we picked a snapshot of each light condition just before switching (at 60s and 90s) and performed a paired t-test after Jarque-Bera normality test (VT031497-Gal4 x UAS-Chrimson attP18: df = 8, P = 0.007; WTB x UAS-Chrimson attP18: df = 8, P = 0.502).
Supplementary Material
Acknowledgments
We thank Alla Karpova (Janelia Farm) for technical advice, reagents and generous assistance with construct preparation for Drosophila, K. Hibbard and members of the Janelia Fly Core for fly husbandry and assistance with fly crosses, and Joanna Pulver for technical advice and assistance with data analysis software. We thank Aso, Y., Ming, W., and Rubin, G. (Janelia Farm) for kindly allowing us to use their circular light arena and for useful discussion. We also thank Negrashov, I., Sawtelle, S., and Liu, J. (Janelia Farm) for arena-related development and support.
SSK, SRP and VJ were supported by the Howard Hughes Medical Institute. The 1000 Plants (1KP) initiative, led by GKSW, is funded by the Alberta Ministry of Enterprise and Advanced Education, Alberta Innovates Technology Futures (AITF) Innovates Centre of Research Excellence (iCORE), Musea Ventures, and BGI-Shenzhen. BYC and ESB were funded by HR0011-12-C-0068. BYC was funded by National Science Foundation Biophotonics. MCP was funded by National Institutes of Health Grant 5R01EY014074-18. ESB was funded by MIT Media Lab, Office of the Assistant Secretary of Defense for Research and Engineering, Harvard/MIT Joint Grants in Basic Neurosci., National Science Foundation (especially CBET 1053233 and EFRI 0835878), National Institues of Health (especially 1DP2OD002002, 1R01NS067199, 1R01DA029639, 1R01GM104948, and 1R01NS075421), Wallace H. Coulter Foundation, Alfred P. Sloan Foundation, Human Frontiers Science Program, New York Stem Cell Foundation Robertson Neuroscience Investigator Award, Institution of Engineering and Technology A. F. Harvey Prize, and the Skolkovo Institute of Science and Technology.
Footnotes
Contributions
NCK, ESB, MCP, and VJ contributed to the study design and data analysis. GKSW and BYC oversaw transcriptomic sequencing. ESB and MCP supervised mammalian opto/electrophysiological parts of the project. NCK coordinated all experiments and data analysis. NCK, YKC, ASC, and TKM conducted and analyzed all in vitro electrophysiology. MM, BS, NCK, TKM, EC, ZT, JW, YX, ZY and YZ conducted algal RNA experiments or transcriptome sequencing and analysis. NCK, YM, and ABB performed and analyzed all slice electrophysiology. VJ prepared Chrimson for injection into Drosophila. SSK and VJ designed adult fly behavior experiments. SSK performed all fly behavior experiments and data analysis. SP designed, performed and analyzed all larval Drosophila experiments. All authors contributed to the discussions and writing of the manuscript.
Competing financial interests
Authors BYC, ESB, GKSW, NCK and YKC are inventors on pending patents covering the described work. ESB is an equity holder in Eos Neuroscience.
References
- 1.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature neuroscience. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 2.Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PloS one. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 5.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyden ES. A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 biology reports. 2011;3:11. doi: 10.3410/B3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang F, et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nature neuroscience. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erbguth K, Prigge M, Schneider F, Hegemann P, Gottschalk A. Bimodal activation of different neuron classes with the spectrally red-shifted channelrhodopsin chimera C1V1 in Caenorhabditis elegans. PloS one. 2012;7:e46827. doi: 10.1371/journal.pone.0046827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prigge M, et al. Color-tuned channelrhodopsins for multiwavelength optogenetics. The Journal of biological chemistry. 2012;287:31804–31812. doi: 10.1074/jbc.M112.391185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, et al. Tuning the electronic absorption of protein-embedded all-trans-retinal. Science. 2012;338:1340–1343. doi: 10.1126/science.1226135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waddell WH, Schaffer AM, Becker RS. Visual pigments. 3. Determination and interpretation of the fluorescence quantum yields of retinals, Schiff bases, and protonated Schiff bases. Journal of the American Chemical Society. 1973;95:8223–8227. doi: 10.1021/ja00806a002. [DOI] [PubMed] [Google Scholar]
- 13.Govorunova EG, Spudich EN, Lane CE, Sineshchekov OA, Spudich JL. New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. mBio. 2011;2:e00115–00111. doi: 10.1128/mBio.00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nature neuroscience. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinlogel S, et al. Ultra light-sensitive and fast neuronal activation with the Ca(2)+-permeable channelrhodopsin CatCh. Nature neuroscience. 2011;14:513–518. doi: 10.1038/nn.2776. [DOI] [PubMed] [Google Scholar]
- 16.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nature neuroscience. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 17.Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 18.Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nature methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nature neuroscience. 2013 doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Govorunova EG, Sineshchekov OA, Li H, Janz R, Spudich JL. Characterization of a Highly Efficient Blue-shifted Channelrhodopsin from the Marine Alga Platymonas subcordiformis. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M113.505495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson MT, et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PloS one. 2012;7:e50226. doi: 10.1371/journal.pone.0050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin JY. A user’s guide to channelrhodopsin variants: features, limitations and future developments. Experimental physiology. 2011;96:19–25. doi: 10.1113/expphysiol.2009.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagel G, et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Current biology : CB. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 24.de Vries SE, Clandinin TR. Loom-sensitive neurons link computation to action in the Drosophila visual system. Curr Biol. 2012;22:353–362. doi: 10.1016/j.cub.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schroll C, et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 26.Gaudry Q, Hong EJ, Kain J, de Bivort BL, Wilson RI. Asymmetric neurotransmitter release enables rapid odour lateralization in Drosophila. Nature. 2013;493:424–428. doi: 10.1038/nature11747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honjo K, Hwang RY, Tracey WD., Jr Optogenetic manipulation of neural circuits and behavior in Drosophila larvae. Nat Protoc. 2012;7:1470–1478. doi: 10.1038/nprot.2012.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Ge W, Wang Z. A toolbox for light control of Drosophila behaviors through Channelrhodopsin 2-mediated photoactivation of targeted neurons. The European journal of neuroscience. 2007;26:2405–2416. doi: 10.1111/j.1460-9568.2007.05862.x. [DOI] [PubMed] [Google Scholar]
- 29.Xiang Y, et al. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature. 2010;468:921–926. doi: 10.1038/nature09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claridge-Chang A, et al. Writing memories with light-addressable reinforcement circuitry. Cell. 2009;139:405–415. doi: 10.1016/j.cell.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein JG, Garrity PA, Boyden ES. Optogenetics and thermogenetics: technologies for controlling the activity of targeted cells within intact neural circuits. Current opinion in neurobiology. 2012;22:61–71. doi: 10.1016/j.conb.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pulver SR, Pashkovski SL, Hornstein NJ, Garrity PA, Griffith LC. Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. Journal of neurophysiology. 2009;101:3075–3088. doi: 10.1152/jn.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang RY, et al. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahanukar A, Lei YT, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minke B, Kirschfeld K. The contribution of a sensitizing pigment to the photosensitivity spectra of fly rhodopsin and metarhodopsin. The Journal of general physiology. 1979;73:517–540. doi: 10.1085/jgp.73.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salcedo E, et al. Blue- and green-absorbing visual pigments of Drosophila: ectopic expression and physiological characterization of the R8 photoreceptor cell-specific Rh5 and Rh6 rhodopsins. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1999;19:10716–10726. doi: 10.1523/JNEUROSCI.19-24-10716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin HH, Chu LA, Fu TF, Dickson BJ, Chiang AS. Parallel neural pathways mediate CO2 avoidance responses in Drosophila. Science. 2013;340:1338–1341. doi: 10.1126/science.1236693. [DOI] [PubMed] [Google Scholar]
- 38.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zorzos AN, Scholvin J, Boyden ES, Fonstad CG. Three-dimensional multiwaveguide probe array for light delivery to distributed brain circuits. Optics letters. 2012;37:4841–4843. doi: 10.1364/OL.37.004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim TI, et al. Injectable, cellular-scale optoelectronics with applications for wireless optogenetics. Science. 2013;340:211–216. doi: 10.1126/science.1232437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nature protocols. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- 42.Stockklausner C, Klocker N. Surface expression of inward rectifier potassium channels is controlled by selective Golgi export. The Journal of biological chemistry. 2003;278:17000–17005. doi: 10.1074/jbc.M212243200. [DOI] [PubMed] [Google Scholar]
- 43.Stockklausner C, Ludwig J, Ruppersberg JP, Klocker N. A sequence motif responsible for ER export and surface expression of Kir2.0 inward rectifier K(+) channels. FEBS letters. 2001;493:129–133. doi: 10.1016/s0014-5793(01)02286-4. [DOI] [PubMed] [Google Scholar]
- 44.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain cell biology. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma D, et al. Role of ER export signals in controlling surface potassium channel numbers. Science. 2001;291:316–319. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- 46.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo A, et al. Conditioned visual flight orientation in Drosophila: dependence on age, practice, and diet. Learn Memory. 1996;3:49–59. doi: 10.1101/lm.3.1.49. [DOI] [PubMed] [Google Scholar]
- 50.Reiser MB, Dickinson MH. A modular display system for insect behavioral neuroscience. Journal of Neuroscience Methods. 2008;167:127–139. doi: 10.1016/j.jneumeth.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 51.Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. 2. Elsevier Academic Press; 2004. Compact. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.