Abstract
Basal-like breast cancer is a highly aggressive tumor subtype associated with poor prognosis. Aberrant activation of NF-κB signaling is frequently found in triple-negative basal-like breast cancer cells, but the cause of this activation has remained elusive. Here we report that α-catenin functions as a tumor suppressor in E-cadherin-negative basal-like breast cancer cells by inhibiting NF-κB signaling. Mechanistically, α-catenin interacts with IκBα protein and stabilizes IκBα by inhibiting its ubiquitination and its association with the proteasome. This stabilization in turn prevents nuclear localization of RelA and p50, leading to decreased expression of TNFα, IL-8 and RelB. In human breast cancer, CTNNA1 expression is specifically downregulated in the basal-like subtype, correlates with clinical outcome and inversely correlates with TNF and RELB expression. Taken together, these results uncover a previously undescribed mechanism by which the NF-κB pathway is activated in E-cadherin-negative basal-like breast cancer.
Breast cancer can be classified into three subtypes based on the membrane receptor status: estrogen receptor (ER)+, HER2+ and triple-negative – defined by lack of expression of ER, progesterone receptor (PR) and HER21. A molecular classification scheme, based on gene expression profiling, classifies breast cancer into the luminal A, luminal B, basal-like, normal-like and HER2+ subtypes2, 3. Basal-like breast cancer is associated with a worse prognosis than are other subtypes4, 5. Approximately 75% of triple-negative breast cancer (TNBC) cases are basal-like6. Patients with ER+ and HER2+ breast cancer are treated with tamoxifen and trastuzumab (or lapatinib), respectively7, 8, but so far there is no FDA-approved targeted therapy for TNBC4, 5.
Cell-cell adherens junctions (AJs) are intercellular junctions that are abundant in normal epithelia and reduced in cancers. The transmembrane core of AJs is composed of E-cadherin, whose cytoplasmic domain interacts with β-catenin, which in turn binds to α-catenin9. α-catenin integrates AJs with the actin cytoskeleton and promotes intercellular adhesion9. Alterations in AJ genes and their protein products are found in human cancer. The gene encoding E-cadherin, CDH1, is mutated in 7% of human breast cancer10. Loss of E-cadherin has been shown to induce breast cancer invasion and metastasis11, 12. β-catenin, which links AJs and the Wnt pathway, promotes both tumorigenesis and metastasis in multiple cancer types13. α-catenin is a putative tumor suppressor in myeloid leukemia14, glioblastoma15 and skin cancer16, 17. Loss of CTNNA1 (the gene encoding α-catenin) causes global loss of cell adhesion in E-cadherin-expressing human breast carcinoma cells18, which demonstrated the importance of α-catenin in maintaining the integrity of AJs.
Hyperactivation of NF-κB signaling is frequently found in triple-negative basal-like breast cancer cells19, but its cause is unclear. Genes regulated by the NF-κB pathway are implicated in various hallmarks of cancer, including proliferation, survival, cell death, invasion, angiogenesis and inflammation20, 21. The five subunits of the NF-κB transcription factor family, RelA (p65), RelB, cRel, NF-κB1 (p50 and its precursor p105) and NF-κB2 (p52 and its precursor p100), form homodimers or heterodimers22. NF-κB signaling consists of canonical and noncanonical pathways23. In the canonical pathway, IκB, the inhibitor of NF-κB, sequesters the RelA-p50 heterodimer in the cytoplasm under nonstimulated conditions. The key regulatory step in this pathway involves ligand-induced activation of an IκB kinase (IKK) complex. The activated IKK complex phosphorylates IκB, which is then ubiquitinated and degraded. Subsequently, the RelA-p50 dimer enters the nucleus, where it regulates gene transcription24. In the noncanonical pathway, activated IKKα phosphorylates p100, the main inhibitor of RelB. The resulting processing of p100 then leads to RelB-p50 and RelB-p52 nuclear translocation and DNA binding20. Extensive efforts have been made to develop agents targeting the NF-κB pathway20, 25.
In the study reported herein, we investigated E-cadherin-independent functions of α-catenin and identified α-catenin as a tumor-suppressing protein in E-cadherin-negative basal-like breast cancer cells. Mechanistically, α-catenin inhibits tumorigenesis by interacting with IκBα, and the resulting stabilization of IκBα leads to cytoplasmic retention of RelA and downregulation of TNFα, IL-8 and RelB. In human breast cancer, CTNNA1 expression is specifically downregulated in the basal-like subtype and is negatively associated with NF-κB signaling.
RESULTS
α-catenin suppresses proliferation and colony formation of E-cadherin-negative basal-like breast cancer cells
α-catenin is a putative tumor suppressor in epithelial cancer cells that express E-cadherin, presumably owing to its essential role in maintaining the integrity of AJs18. To investigate the role of α-catenin in E-cadherin-negative basal-like breast cancer cells, we first determined α-catenin protein levels in a panel of human mammary cell lines. Whereas immortalized human mammary epithelial (HMLE) cells and the MCF7 and T47D luminal breast cancer cells showed abundant α-catenin expression, this protein had moderate or negative expression in five of the six basal-like breast cancer cell lines tested: MDA-MB-231, SUM159, MDA-MB-436, MDA-MB-157 and MDA-MB-468. Only one basal-like cell line, BT549, exhibited α-catenin protein levels comparable to those in luminal-like cells (Fig. 1a). The MDA-MB-157 and MDA-MB-468 cell lines are α-catenin negative due to a frameshift mutation in CTNNA126.
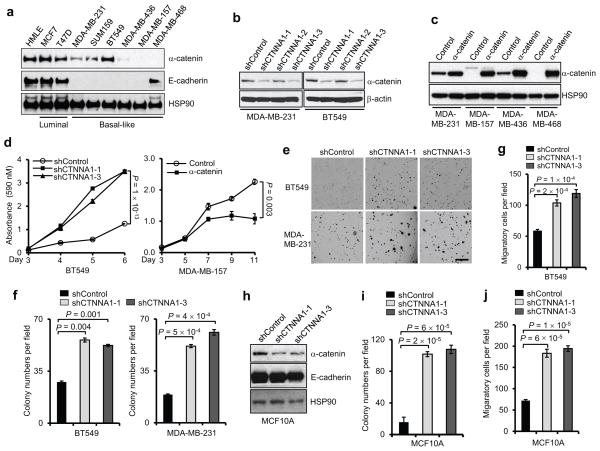
Figure 1. α-catenin inhibits proliferation and colony formation of basal-like breast cancer cells.
(a) Immunoblotting of α-catenin, E-cadherin and HSP90 in HMLE, luminal and basal-like breast cancer cell lines.
(b) Immunoblotting of α-catenin and β-actin in MDA-MB-231 and BT549 cells transduced with three independent α-catenin shRNAs.
(c) Immunoblotting of α-catenin and HSP90 in α-catenin-transduced MDA-MB-231, MDA-MB-157, MDA-MB-436 and MDA-MB-468 cells.
(d) Growth curves of BT549 cells with knockdown of α-catenin and MDA-MB-157 cells with ectopic expression of α-catenin. shControl: the pGIPZ-GFP lentiviral vector with a scrambled sequence that does not target any mRNA. Control: the pLOC lentiviral vector with an RFP open reading frame. n = 4 wells per group.
(e, f) Representative images (e) and data quantification (f) of soft agar colony formation by BT549 and MDA-MB-231 cells transduced with two independent α-catenin shRNAs. Scale bar: 100 μm. n = 3 wells per group.
(g) Transwell migration assays of BT459 cells transduced with two independent α-catenin shRNAs. n = 3 wells per group.
(h) Immunoblotting of α-catenin, E-cadherin and HSP90 in MCF10A cells transduced with two independent α-catenin shRNAs.
(i, j) Soft agar colony formation (i) and Transwell migration assays (j) of MCF10A cells transduced with two independent α-catenin shRNAs. n = 3 wells per group.
Data in (d), (f), (g), (i) and (j) are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Student’s t-test. The experiments were repeated three times. The source data can be found in Supplementary Table 4. Uncropped images of blots are shown in Supplementary Fig. 7.
Next, we performed both loss-of-function and gain-of-function analyses of α-catenin in basal-like breast cancer cell lines (Fig. 1b, c). Two independent α-catenin shRNAs both increased the proliferation of BT549 cells (Fig. 1d). In contrast, restoring α-catenin expression in MDA-MB-157 cells reduced their proliferation (Fig. 1d).
We also examined the effect of α-catenin loss on anchorage-independent growth and cell motility. In both BT549 and MDA-MB-231 cells, depletion of α-catenin significantly increased the cells’ colony-forming ability in soft agar (Fig. 1e, f) and migratory ability (Fig. 1g). Knockdown of α-catenin in a luminal mammary cell line, MCF10A, also promoted oncogenic transformation and cell motility (Fig. 1h–j). Taken together, these results indicate that α-catenin may function as a tumor suppressor in both luminal and basal-like cells.
α-catenin inhibits NF-κB signaling in E-cadherin-negative basal-like breast cancer cells
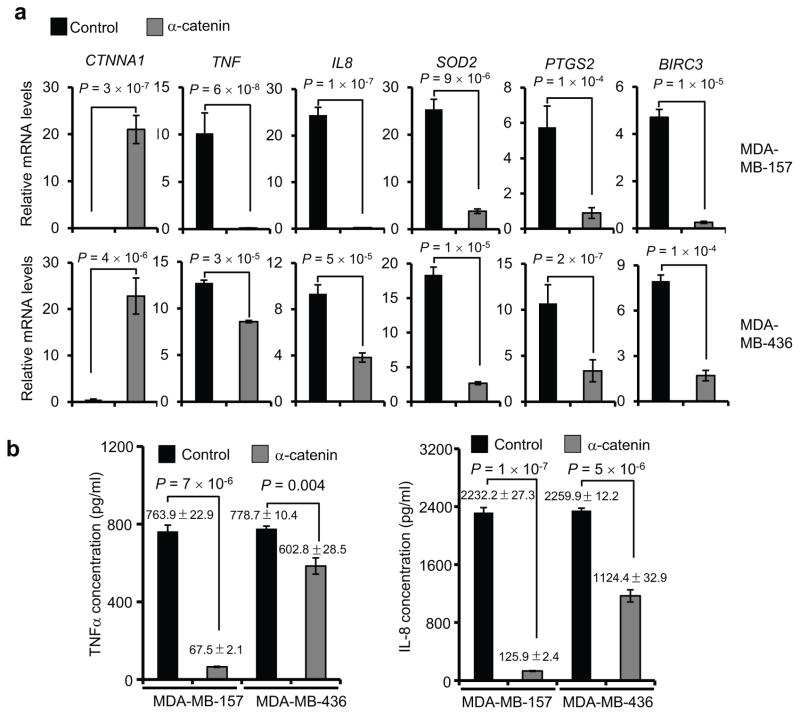
The association with AJs cannot explain the effect of α-catenin in E-cadherin-negative basal-like cells. To identify the signaling pathways regulated by α-catenin in basal-like cells, we performed a Human Cancer PathwayFinder qPCR array analysis. Interestingly, all three most significantly downregulated genes in α-catenin-overexpressing MDA-MB-157 cells—TNF, IL8 and MMP1—are NF-κB target genes27–29 (Supplementary Table 1). To confirm that α-catenin downregulates NF-κB response genes, we performed quantitative real-time PCR analysis of two α-catenin-overexpressing E-cadherin-negative basal-like breast cancer cell lines, MDA-MB-157 and MDA-MB-436. Among several NF-κB target genes downregulated by α-catenin, the most dramatically downregulated ones were TNF and IL8 in MDA-MB-157 cells (Fig. 2a), which was consistent with the array results. Moreover, MDA-MB-157 and MDA-MB-436 cells with overexpression of α-catenin also exhibited a significant reduction in the levels of secreted TNFα and IL-8 (Fig. 2b). In contrast, in the E-cadherin-positive, α-catenin-negative basal-like cell line MDA-MB-468, the levels of TNFα, IL-8 and other components of the NF-κB pathway were either unaffected or upregulated upon ectopic expression of α-catenin (Supplementary Fig. 1a–c). These findings indicate that α-catenin inhibits NF-κB signaling specifically in E-cadherin-negative basal-like breast cancer cells.
Figure 2. α-catenin inhibits NF-κB signaling in basal-like breast cancer cells.
(a) qPCR of NF-κB response genes in α-catenin-transduced MDA-MB-157 and MDA-MB-436 cells. n = 3 samples per group.
(b) ELISA of TNFα and IL-8 secreted by α-catenin-transduced MDA-MB-157 and MDA-MB-436 cells. n = 3 wells per group.
Data in (a) and (b) are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Student’s t-test. The experiments were repeated three times. The source data can be found in Supplementary Table 4.
α-catenin stabilizes IκBα protein by inhibiting IκBα ubiquitination
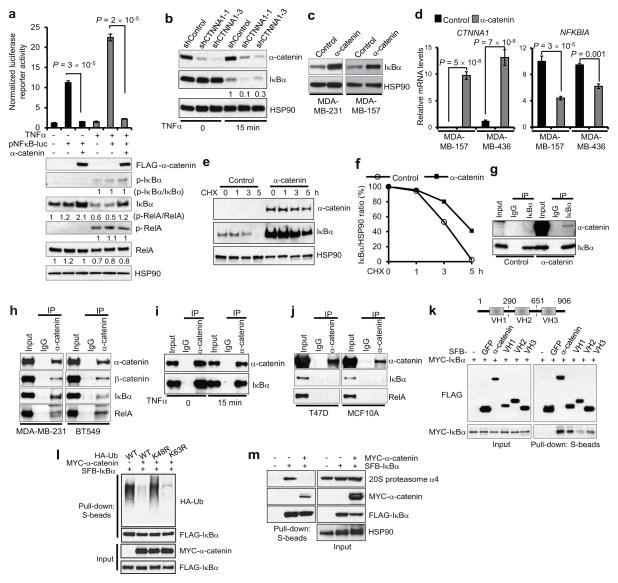
To further confirm that α-catenin inhibits NF-κB activity, we used a pNFκB luciferase reporter gene with specific NF-κB binding sites in the promoter region30. As anticipated, overexpression of α-catenin in MDA-MB-157 cells markedly suppressed the pNFκB luciferase activity and upregulated IκBα protein in the presence or absence of TNFα stimulation, whereas neither RelA total protein level nor its phosphorylation was altered (Fig. 3a). Moreover, expression of α-catenin did not affect IKK phosphorylation in MDA-MB-157 cells (Supplementary Fig. 2), suggesting that the observed upregulation of IκBα by α-catenin was not due to the alteration in IKK activity.
Figure 3. α-catenin stabilizes IκBα protein by inhibiting IκBα ubiquitination and abrogating IκBα interaction with the proteasome.
(a) Upper panel: luciferase assays of NF-κB activity in MDA-MB-157 cells transfected with a pNFκB luciferase reporter alone or in combination with FLAG-α-catenin, with or without TNFα treatment. Lower panel: immunoblotting of FLAG, p-IκBα, IκBα, p-RelA, RelA and HSP90. n = 3 wells per group.
(b) Immunoblotting of α-catenin, IκBα and HSP90 in α-catenin shRNA-transduced BT549 cells, with or without TNFα treatment.
(c) Immunoblotting of IκBα and HSP90 in α-catenin-transduced MDA-MB-231 and MDA-MB-157 cells.
(d) qPCR of CTNNA1 and NFKBIA in α-catenin-transduced MDA-MB-157 and MDA-MB-436 cells. n = 3 samples per group.
(e) α-catenin-transduced MDA-MB-157 cells were treated with 100 μg/ml of cycloheximide (CHX), harvested at different time points and immunoblotted with antibodies to α-catenin, IκBα and HSP90.
(f) Quantification of IκBα protein levels in (e).
(g) α-catenin was immunoprecipitated from α-catenin-transduced MDA-MB-157 cells and immunoblotted with antibodies to α-catenin and IκBα.
(h) α-catenin was immunoprecipitated from MDA-MB-231 and BT549 cells and immunoblotted with antibodies to α-catenin, β-catenin, IκBα and RelA.
(i) α-catenin was immunoprecipitated from untreated or TNFα-treated BT549 cells and immunoblotted with antibodies to α-catenin and IκBα.
(j) α-catenin was immunoprecipitated from T47D or MCF10A cells and immunoblotted with antibodies to α-catenin, IκBα and RelA.
(k) Upper panel: schematic representation of three vinculin domains of α-catenin. Lower panel: 293T cells were cotransfected with MYC-IκBα and SFB-tagged full-length α-catenin or fragment VH1, VH2 or VH3. α-catenin and the three fragments were purified with S-protein beads and immunoblotted with antibodies to FLAG and MYC.
(l) HA-tagged wild-type ubiquitin (Ub) or the K48R or K63R mutant was cotransfected with MYC-α-catenin and SFB-IκBα into 293T cells. Cells were treated with 10 μM MG132 and 20 ng/ml of TNFα for 30 minutes. IκBα was purified with S-protein beads and immunoblotted with antibodies to HA and FLAG.
(m) 293T cells were cotransfected with MYC-α-catenin and SFB-IκBα and then treated with 10 μM MG132 and 20 ng/ml of TNFα for 30 minutes. IκBα was purified with S-protein beads and immunoblotted with antibodies to the 20S proteasome subunit α4, MYC and FLAG.
Data in (a) and (d) are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Student’s t-test. The experiments were repeated three times. The source data can be found in Supplementary Table 4. Uncropped images of blots are shown in Supplementary Fig. 7.
In nonstimulated cells, RelA and NF-κB are sequestered in the cytoplasm by IκB inhibitory proteins, whereas TNFα treatment induces degradation of IκB, leading to RelA and NF-κB nuclear translocation20, 24. shRNA-mediated silencing of α-catenin decreased IκBα protein in TNFα-treated BT549 cells (Fig. 3b), suggesting that α-catenin suppresses the response to TNFα in these cells. In contrast, in luminal cell lines T47D and MCF10A, IκBα protein levels were not altered by knockdown of α-catenin, either with or without TNFα treatment (Supplementary Fig. 3a–c).
In both MDA-MB-231 and MDA-MB-157 breast cancer cells, overexpression of α-catenin increased IκBα protein levels (Fig. 3c). In contrast to IκBα protein, NFKBIA mRNA (encoding IκBα) was downregulated in α-catenin-expressing cells (Fig. 3d), which suggested that α-catenin-mediated upregulation of IκBα might be due to increased protein stability instead of upregulation of its mRNA. We therefore examined IκBα protein levels in the presence of cycloheximide, an inhibitor of protein translation. As expected, overexpression of α-catenin in MDA-MB-157 cells significantly increased the stability of endogenous IκBα protein (Fig. 3e,f).
We next investigated whether α-catenin-mediated stabilization of IκBα involves the physical interaction of α-catenin with IκBα. Coimmunoprecipitation assays showed that overexpressed α-catenin could be detected in endogenous IκBα immunoprecipitates from MDA-MB-157 cells (Fig. 3g), and that endogenous IκBα was present in endogenous α-catenin immunoprecipitates from MDA-MB-231 and BT549 cells (Fig. 3h). This interaction did not require TNFα stimulation (Fig. 3i). In contrast, α-catenin did not interact with IκBα in E-cadherin-positive luminal-like cell lines, T47D and MCF10A (Fig. 3j). In agreement with cytoplasmic sequestration of RelA by IκBα20, 24, we observed existence of endogenous IκBα and RelA in endogenous α-catenin-containing protein complexes in both MDA-MB-231 and BT549 cells (Fig. 3h). β-catenin was also detectable in α-catenin immunoprecipitates (Fig. 3h). However, knockdown or overexpression of α-catenin did not affect β-catenin protein level, localization or activity in basal-like breast cancer cells (Supplementary Fig. 4). It has been reported that α-catenin contains three vinculin homology (VH) domains31: VH1, VH2 and VH3 (Fig. 3k). Whereas all three VH domains exhibited association with IκBα, the VH1 fragment showed the strongest interaction (Fig. 3k).
To address how α-catenin increases IκBα protein stability, we examined ubiquitination of IκBα. Expression of α-catenin dramatically reduced the polyubiquitination level of IκBα, and this effect was abrogated when wild-type ubiquitin was substituted with a K48R mutant, but not a K63R mutant (Fig. 3l), suggesting that α-catenin inhibits K48-linked ubiquitination, which is known to be involved in protein degradation32. Moreover, the interaction of IκBα with the proteasome α4 subunit was abolished upon α-catenin overexpression (Fig. 3m). We therefore conclude that α-catenin interacts with IκBα, inhibits IκBα polyubiquitination, abrogates IκBα interaction with the proteasome and, as a result, stabilizes IκBα.
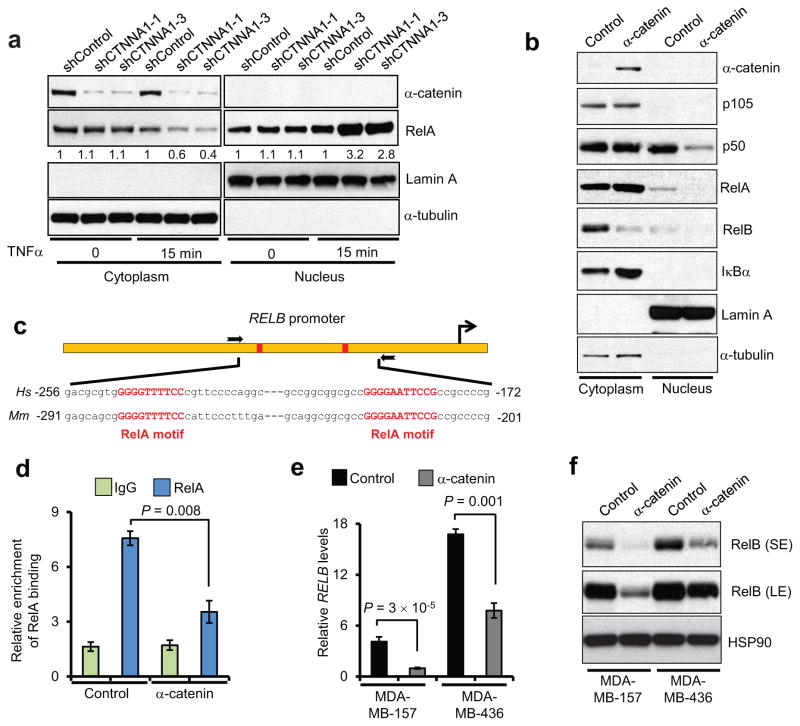
α-catenin inhibits RelA-p50 nuclear localization and downregulates RelB
Activation of the canonical NF-κB pathway leads to nuclear translocation of the RelA-p50 dimer, which functions as a transcriptional activator20, 24. Compared with control shRNA-infected BT549 cells, two independent α-catenin shRNAs both induced nuclear translocation of RelA in response to TNFα treatment (Fig. 4a), consistent with the effect on IκBα (Fig. 3b). Moreover, compared with mock-infected MDA-MB-157 cells, α-catenin-overexpressing MDA-MB-157 cells had increased cytoplasmic RelA and reduced nuclear RelA and p50, as gauged by fractionation assays (Fig. 4b).
Figure 4. α-catenin inhibits RelA-p50 nuclear localization and downregulates RelB.
(a) Immunoblotting of α-catenin and RelA in cytoplasmic and nuclear fractions of BT549 cells transduced with two independent α-catenin shRNAs, with or without TNFα treatment.
(b) Immunoblotting of α-catenin, p105, p50, RelA, RelB and IκBα in cytoplasmic and nuclear fractions of α-catenin-transduced MDA-MB-157 cells. α-tubulin and Lamin A were used as cytoplasmic and nuclear markers, respectively, in (a) and (b).
(c) Schematic representation of the RELB promoter containing two RelA binding sites (red rectangles). The two boxed arrows indicate the primers used for ChIP-qPCR. Hs: Homo sapiens, Mm: Mus musculus.
(d) ChIP-qPCR analysis of RelA binding to the RELB promoter in α-catenin-transduced MDA-MB-157 cells. qPCR was performed with primers specific to the RelA binding motifs. Data were normalized to the input. n = 3 samples per group.
(e) qPCR of RELB in α-catenin-transduced MDA-MB-157 and MDA-MB-436 cells. n = 3 samples per group.
(f) Immunoblotting of RelB and HSP90 in α-catenin-transduced MDA-MB-157 and MDA-MB-436 cells. SE: short exposure; LE: long exposure.
Data in (d) and (e) are the mean of biological replicates from a representative experiment,, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Student’s t-test. The experiments were repeated three times. The source data can be found in Supplementary Table 4. Uncropped images of blots are shown in Supplementary Fig. 7.
Besides activating the transcription of TNF and IL8, RelA can also induce RELB expression through direct binding of the RELB promoter33. We therefore reasoned that α-catenin-mediated cytoplasmic retention of RelA may lead to reduced binding of the RELB promoter by RelA. To investigate this possibility, we performed chromatin immunoprecipitation (ChIP) assays of α-catenin-overexpressing MDA-MB-157 cells, followed by quantitative PCR of the conserved RELB promoter region encompassing two consensus RelA-binding motifs (Fig. 4c). Chromatin immunoprecipitates with the RelA antibody were enriched for the RELB promoter region compared with those precipitated with the IgG control, confirming that this region does contain the RelA binding site (Fig. 4d). Expression of α-catenin resulted in an approximately 50% reduction in binding of the RELB promoter by RelA (Fig. 4d). In agreement with these findings, overexpression of α-catenin in MDA-MB-157 and MDA-MB-436 cells downregulated both mRNA (Fig. 4e) and protein (Fig. 4b,f) levels of RelB. These findings confirm that α-catenin suppresses RelA-p50 nuclear localization, leading to inhibition of RELB transcription.
α-catenin functions as a tumor suppressor in basal-like breast cancer cells by inhibiting NF-κB signaling
To determine whether NF-κB mediates the growth-promoting effect of α-catenin shRNA, we expressed two independent RelA shRNAs in α-catenin-depleted MDA-MB-231 and BT549 cells (Fig. 5a). In both cell lines, knockdown of RelA reversed the effect of α-catenin shRNA on promoting anchorage-independent growth (Fig. 5b).
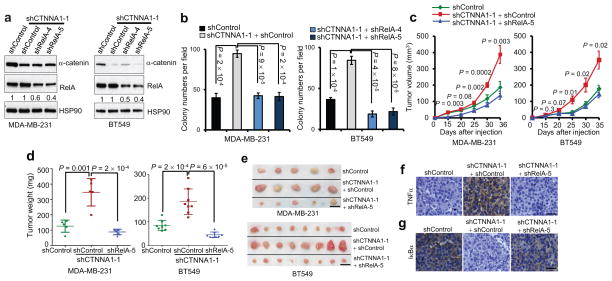
Figure 5. Loss of α-catenin promotes tumor growth in basal-like breast cancer cells by activating NF-κB signaling.
(a) Immunoblotting of α-catenin, RelA and HSP90 in MDA-MB-231 and BT549 cells transduced with α-catenin shRNA alone or in combination with RelA shRNA.
(b) Soft agar colony formation by MDA-MB-231 and BT549 cells transduced with α-catenin shRNA alone or in combination with RelA shRNA. n = 4 wells per group.
(c) Tumor growth by subcutaneously implanted MDA-MB-231 (3 × 106 cells injected) or BT549 (4 × 106 cells injected) cells infected with α-catenin shRNA alone or in combination with RelA shRNA. P values correspond to comparisons between α-catenin shRNA alone and α-catenin shRNA in combination with RelA shRNA.
(d, e) Tumor weight (d) and tumor images (e) 5 weeks after mice were injected subcutaneously with MDA-MB-231 or BT549 cells transduced with α-catenin shRNA alone or in combination with RelA shRNA. Scale bar: 1 cm. n = 5 (for MDA-MB-231 cells) or 8 (for BT549 cells) mice per group in (c) and (d).
(f, g) TNFα (f) and human-specific IκBα (g) immunohistochemical staining of subcutaneous tumors formed by MDA-MB-231 cells transduced with α-catenin shRNA alone or in combination with RelA shRNA, at 5 weeks after implantation. Scale bar: 50 μm.
Data in (b) – (d) are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Student’s t-test. The experiments were repeated three times. The source data can be found in Supplementary Table 4. Uncropped images of blots are shown in Supplementary Fig. 7.
To explore the function of α-catenin in basal-like breast cancer cells in vivo, we subcutaneously implanted α-catenin-depleted BT549 and MDA-MB-231 cells with or without expression of RelA shRNA into nude mice. Hosts of α-catenin shRNA-expressing cancer cells had larger tumor volumes throughout the experiment than mice implanted with control shRNA-infected cells (Fig. 5c). At 5 weeks after tumor cell implantation, we observed a 2.8-fold increase and a 2.2-fold increase in the weight of the tumors formed by α-catenin-depleted MDA-MB-231 and BT549 cells, respectively (Fig. 5d,e). Notably, the tumor growth of α-catenin-depleted cells that also expressed RelA shRNA was similar to the tumor growth of control shRNA-infected cells (Fig. 5c–e). Compared with the tumors formed by control MDA-MB-231 cells or MDA-MB-231 cells with simultaneous knockdown of α-catenin and RelA, α-catenin-depleted MDA-MB-231 tumors exhibited upregulation of TNFα (Fig. 5f) and downregulation of IκBα (Fig. 5g), suggesting that loss of α-catenin can lead to activation of NF-κB signaling in vivo.
To further confirm that α-catenin suppresses tumorigenesis by inhibiting NF-κB signaling, we used two independent shRNAs to silence IκBα in α-catenin-overexpressing MDA-MB-157 cells. Knockdown of IκBα rescued RelB expression (Fig. 6a), in vitro cell proliferation (Fig. 6b,c) and colony formation (Fig. 6d,e). We then implanted these cells subcutaneously into mice. Notably, mice implanted with either the control MDA-MB-157 cells or MDA-MB-157 cells with simultaneous α-catenin overexpression and IκBα knockdown showed similar tumor growth rates, whereas mice bearing α-catenin-overexpressing MDA-MB-157 cells showed markedly inhibited tumorigenesis (Fig. 6f–h). Taken together, these results suggest that stabilization of IκBα mediates, at least in part, the tumor-suppressing effect of α-catenin in E-cadherin-negative basal-like breast cancer cells.
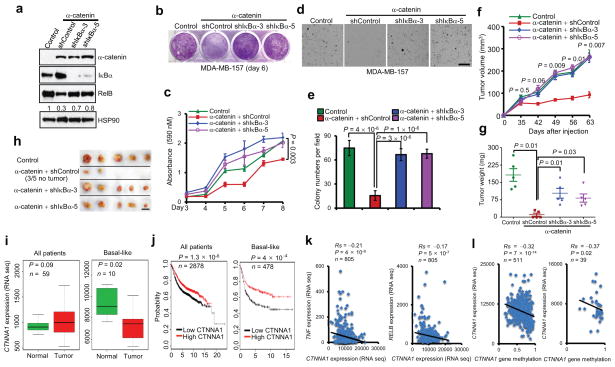
Figure 6. α-catenin inhibits tumorigenesis and is downregulated in human basal-like breast cancer.
(a) Immunoblotting of α-catenin, IκBα, RelB and HSP90 in MDA-MB-157 cells transduced with α-catenin alone or in combination with IκBα shRNA.
(b, c) Images (b) and quantification (c) of growth curves of cells described in (a). n = 3 wells per group.
(d, e) Images (d) and quantification (e) of soft agar colony formation by cells described in (a). Scale bar: 100 μm. n = 5 wells per group.
(f) Tumor growth by 3 × 106 subcutaneously injected cells described in (a). P values correspond to comparisons between α-catenin alone and α-catenin in combination with IκBα shRNA (shIκBα-3) in (c) and (f).
(g, h) Tumor weight (g) and tumor images (h) of mice described in (f). Scale bar: 1cm. n = 5 mice per group in (f) and (g).
(i) Box plots comparing CTNNA1 expression in normal breast tissues and in total (n = 59) and basal-like (n = 10) breast tumors. Statistical significance was determined by the Wilcoxon test. The boxes show the median and the interquartile range. The whiskers show the minimum and maximum.
(j) Kaplan-Meier curves of relapse-free survival times of total breast cancer patients (n = 2878) and patients with basal-like breast cancer (n = 478), stratified by CTNNA1 expression levels. Data were obtained from http://kmplot.com/analysis/34. Statistical significance was determined by the log-rank test.
(k) Scatterplots showing the inverse correlation of CTNNA1 with TNF (left panel) or RELB (right panel) expression in human breast tumors (n = 805).
(l) Scatterplots showing the inverse correlation between methylation of the CTNNA1 gene and CTNNA1 expression in total (left panel, n = 511) and basal-like (right panel, n = 39) breast tumors. Statistical significance in (k) and (l) was determined by Spearman rank correlation test. Rs = Spearman rank correlation coefficient.
Data in (c) and (e) – (g) are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Student’s t-test. The experiments were repeated three times. The source data can be found in Supplementary Table 4. Uncropped images of blots are shown in Supplementary Fig. 7.
α-catenin expression is downregulated in human basal-like breast tumors and negatively correlates with the activity of the NF-κB pathway
To investigate the relevance of our findings to human breast cancer, we analyzed gene expression data from The Cancer Genome Atlas (TCGA)10. We found that CTNNA1 expression was significantly downregulated in the basal-like subtype of breast cancer compared with normal breast tissues, whereas other breast cancer subtypes exhibited either upregulation of CTNNA1 or no significant difference (Fig. 6i and Supplementary Fig. 5a). We also evaluated the prognostic value of CTNNA1 in a microarray dataset of breast tumors from 2,878 patients34. Of 478 patients with basal-like breast cancer, those with low levels of CTNNA1 (the median value was used as the cutoff for low and high expression) had much shorter relapse-free survival (P = 4 × 10−4) than did patients with high levels of CTNNA1, whereas the difference in patients with other subtypes of breast cancer was either not significant or less significant (Fig. 6j and Supplementary Fig. 5b).
Next, we investigated whether CTNNA1 expression is negatively associated with the activity of NF-κB signaling. Both RNA-sequencing and microarray data from TCGA10 revealed a significant inverse correlation of CTNNA1 with TNF and RELB expression levels in human breast tumors (Fig. 6k and Supplementary Fig. 5c).
We asked how α-catenin expression is downregulated or lost in breast cancer. Although CTNNA1 gene deletion was found in tumors from one basal-like breast cancer patient35, analysis of breast cancer data from TCGA10 revealed that deletion (1 of 482 patients) or mutation (4 of 482 patients) of CTNNA1 was rare. On the other hand, we observed a significant inverse correlation between CTNNA1 mRNA level and CTNNA1 gene methylation in both total and basal-like breast tumors (Fig. 6l), which indicated that CTNNA1 gene hypermethylation, but not genetic alteration, is likely to be a main cause of α-catenin downregulation or loss in human breast cancer.
DISCUSSION
We conclude that α-catenin acts as a tumor-suppressing protein in multiple cancer types and subtypes, but the mechanism of action varies depending on the tissue type. In epidermal cells, α-catenin inhibits tumorigenesis by inducing phosphorylation, cytoplasmic retention and functional inactivation of the oncoprotein YAP16, 17 (Supplementary Fig. 6a, left panel). However, we did not observe this effect on YAP phosphorylation in the basal-like breast cancer cell lines MDA-MB-157 and MDA-MB-436 (Supplementary Fig. 6b). Moreover, α-catenin is a structural link between the E-cadherin-catenin complex and the actin cytoskeleton and is involved in epithelial tumor suppression9, 18 (Supplementary Fig. 6a, left panel), but this mechanism is irrelevant in basal-like cells that have lost E-cadherin expression. In the present study, we identified α-catenin as a tumor-suppressing protein in E-cadherin-negative basal-like breast cancer cells and found that α-catenin inhibits NF-κB signaling by stabilizing IκBα and sequestering RelA-p50 in the cytoplasm (Supplementary Fig. 6a, right panel). In contrast, α-catenin does not inhibit the NF-κB pathway in luminal cells or E-cadherin-positive basal-like cells, indicating that when localized at the E-cadherin-catenin complex, α-catenin may not be capable of regulating IκBα ubiquitination and RelA-p50 localization, at least in mammary cells. It should be noted that Kobielak and Fuchs have demonstrated deregulation of NF-κB signaling components in α-catenin-deficient mouse skin cells36, but the functional relevance and the mechanistic link have not been determined in the skin.
The effect of α-catenin shRNA on IκBα stability (Fig. 3b) and RelA nuclear translocation (Fig. 4a) was only observed upon stimulation with TNF-α. Similarly, it has been shown that deletion of either IKKα or IKKβ stabilized IκBα protein in TNFα-treated mouse embryonic fibroblast cells (MEFs), but had no effect on IκBα stability in nonstimulated MEFs37, 38. This loss-of-function effect of α-catenin is biologically relevant, because cancer cells within a tumor are usually exposed to TNF-α secreted by infiltrated macrophages or by tumor cells themselves.
The NF-κB pathway can be activated by a variety of stimuli21. Activated NF-κB regulates approximately 200 target genes encoding cytokines, chemokines, growth factors and transcription factors21, 39, 40. Aberrant activation of NF-κB signaling is associated with various human cancers including breast cancer41. Although HER2 can trigger NF-κB activation through the PI3K-AKT pathway42, the cause of the elevated NF-κB activity frequently observed in triple-negative basal-like breast cancer cells19 remains elusive. Our findings identify loss of α-catenin as a mechanism by which the NF-κB pathway is activated in the basal-like subtype of breast cancer. This is highly relevant in human tumors, as α-catenin is specifically downregulated in human basal-like breast cancer, correlates with recurrence-free survival and negatively correlates with the activity of NF-κB signaling. Targeting the NF-κB pathway may provide therapeutic benefits to patients with basal-like, triple-negative breast cancer.
METHODS
Cell culture
The HMLE immortalized human mammary epithelial cells were described previously43. The SUM159 cell line, provided by Stephen Ethier, was cultured as described (http://www.asterand.com/asterand/human_tissues/159PT.htm). Cell lines MCF10A, MCF7, T47D, BT549, MDA-MB-231, MDA-MB-157, MDA-MB-436, MDA-MB-468 and 293T were purchased from the American Type Culture Collection and cultured under conditions specified by the provider.
RNA isolation and real-time RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) and was then reverse transcribed with an iScript cDNA Synthesis Kit (Bio-Rad). The resulting cDNA was analysed by qPCR using the SYBR Green Gene Expression Assays (Bio-Rad), and data were normalized to an endogenous control, GAPDH. Real-time PCR and data collection were performed on a CFX96 instrument (Bio-Rad). The primers used in this study are listed in Supplementary Table 2.
Plasmids and shRNA
The following shRNA and ORF clones were obtained from Open Biosystems through the shRNA and ORFeome Core facility of The University of Texas MD Anderson Cancer Center: human CTNNA1 shRNA, V3LMM-492867 (5′-AATTTGTAGACATCTGCCA-3′, designated shCTNNA1-1) and V3LMM-492872 (5′-TGCTTGTTCAACAGATGCA-3′, designated shCTNNA1-3); human IκBα shRNA, V3LHS_375160 (5′-CGTCCTCTGTGAACTCCGT-3′, designated shIκBα-3) and V3LHS_410688 (5′-TTTTTGATAACCTTCTCCA-3′, designated shIκBα-5); human RelA shRNA, V3LHS_633762 (5′-ATCTGTGCTCCTCTCGCCT-3′, designated shRelA-4) and V3LHS_633764 (5′-TCTATAGGAACTTGGAAGG-3′, designated shRelA-5); human α-catenin ORF, PLOH-10000388. The human IκBα ORF was cloned from the cDNA of HMLE cells. The α-catenin and IκBα ORFs were subcloned into an SFB (S-protein, FLAG tag and streptavidin-binding peptide)-tagged expression vector (provided by Junjie Chen). The pNFκB luciferase reporter construct was purchased from Stratagene (219078-51). The vectors used in this study are listed in Supplementary Table 3.
Human cancer pathway PCR array analysis
The Cancer PathwayFinder RT2 Profiler PCR Array, consisting of 84 genes representative of six tumor signaling pathways, was used to profile α-catenin-expressing MDA-MB-157 cells according to the manufacturer’s instructions (http://www.sabiosciences.com/rt_pcr_product/HTML/PAHS-033A.html). Briefly, total RNA was extracted and reverse transcribed into cDNA using an RT2 First Strand Kit (Qiagen). The cDNA was combined with an RT2 SYBR Green qPCR Master Mix (Qiagen), and then equal aliquots of this mixture (25 μl) were added to each well of the same PCR Array plate that contained the predispensed gene-specific primer sets. Real-time PCR and data collection were performed on a CFX96 instrument (Bio-Rad).
Lentiviral transduction
Lentivirus was produced and target cells were infected as described previously44. Virus-containing supernatant was collected 48 hours and 72 hours after co-transfection of pCMV-VSV-G, pCMVΔ8.2 and the shRNA- or ORF-containing vector into 293T cells, and then added to the target cells. 24 hours later, the infected cells were selected with 10 μg/ml blasticidin (for pLOC vector) or 2 μg/ml puromycin (for pGIPZ vector).
ELISA
Cells were grown to near confluence in serum-containing medium, washed three times with PBS and then incubated with serum-free medium (DMEM/F12 containing penicillin and streptomycin). After a 48-h (for IL-8) or 96-h (for TNFα) incubation, conditioned medium was collected for ELISA using the Quantikine Kit (IL8: D8000C; TNFα: DTA00C, R&D Systems) according to the manufacturer’s protocol.
Cell proliferation assay
To determine growth curves, we plated equal numbers of cells in 12-well plates in triplicate. Beginning on day 3, cells were fixed with 10% methanol and stained with 0.1% crystal violet (dissolved in 10% methanol) every day. After staining, wells were washed three times with PBS and destained with acetic acid, and the absorbance of the crystal violet solution was measured at 590 nm.
Anchorage-independent growth assay
Cells were suspended in 1 ml of 0.35% low-melting-point agarose (Invitrogen) in DMEM supplemented with 10% FBS and plated in triplicate in 6-well plates on 1 ml of presolidified 0.65% agarose in the same medium, with 1 ml of medium covering the cells. After incubation at 37°C in 5% CO2 for 3–6 weeks, plates were stained with crystal violet and scanned on a GelCount system (Oxford Optronix). Colonies were counted by using the ImageJ program (http://rsbweb.nih.gov/ij/download.html).
Transwell migration assay
Cells (1–2 × 105) were plated in the top chamber with the noncoated membrane (24-well insert; pore size, 8 μm; BD Biosciences) in medium without serum or growth factors. Medium supplemented with growth factors (for MCF10A) or serum (for BT549) was used as a chemoattractant in the lower chamber. The cells were incubated for 20 h, and cells that did not migrate through the pores were removed with a cotton swab. Cells on the lower surface of the membrane were stained with the Diff-Quick Staining Set (Dade) and counted.
Luciferase reporter assay
Cells of 70% confluence in 6-well plates were transfected using X-tremeGene9 (Roche). The firefly luciferase reporter gene construct (0.5 μg) and the pRL-SV40 Renilla luciferase construct (10 ng, for normalization) were used for cotransfection. Cell extracts were prepared 24–48 h after transfection, and the luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega).
Immunoblotting
Western blot analyses were performed with precast gradient gels (Bio-Rad) using standard methods. Briefly, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (Roche) and phosphatase inhibitors (Sigma). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto a PVDF membrane (Bio-Rad). Membranes were probed with the specific primary antibodies and then with peroxidase-conjugated secondary antibodies. The bands were visualized by chemiluminescence (Denville Scientific). The following antibodies were used: antibodies to α-catenin (1:1,000, Sigma, SAB1402163; 1:1,000, Cell Signaling Technology, 3240, Clone 23B2), α-tubulin (1:3,000, Sigma, T5168, Clone B-5-1-2), β-catenin (1:2,000, BD Transduction Laboratories, 610154, Clone 14/Beta-catenin), E-cadherin (1:1,000, BD Transduction Laboratories, 610182, Clone 36/E-Cadherin), p-RelA (Ser536; 1:1,000, Cell Signaling Technology, 3033, Clone 93H1), RelA (1:1,000, Cell Signaling Technology, 6956; 1:1,000, Cell Signaling Technology, 8242, Clone L8F6), RelB (1:1,000, Cell Signaling Technology, 4922, Clone C1E4), p-IκBα (Ser32/36; 1:1,000, Cell Signaling Technology, 9246, Clone 5A5), IκBα (1:1,000, Cell Signaling Technology, 9242), IKKβ (1:1,000, Cell Signaling Technology, 2678, Clone 2C8), p-IKKα/β (; Ser176/180; 1:1,000, Cell Signaling Technology, 2697, Clone 16A6), FLAG (1:2,000, Sigma, F3165; 1:2,000, Sigma, F7425, Clone M2), lamin A (1:2,000, Abcam, ab8980, Clone 133A2), 20S proteasome subunit α4 (1:1,000, Enzo Life Sciences, PW8120, Clone MCP34), HA (; 1:1,000, Roche, 11583816001, Clone 12CA5), MYC (1:2,000, Cell Signaling Technology, 2276, Clone# 9B11), YAP (1:1,000, Cell Signaling Technology, 4912), p-YAP (1:1,000, Cell Signaling Technology, 4911), β-actin (1:5,000, Sigma, A5441, Clone AC-15) and HSP90 (1:2,000 BD Transduction Laboratories, 610419, Clone 68/Hsp90). The ImageJ program (http://rsbweb.nih.gov/ij/download.html) was used for densitometric analysis of Western blots, and the quantification results were normalized to the loading control.
Cytoplasmic and nuclear fractionation
Nuclear and cytoplasmic proteins were fractionated using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo) according to the manufacturer’s protocol. After fractionation, 30 μg of protein was used for Western blot analysis of α-catenin, RelA, RelB and IκBα in the cytoplasm and nucleus. α-Tubulin and lamin A were used as cytoplasmic and nuclear markers, respectively.
Coimmunoprecipitation and pull-down assays
Coimmunoprecipitation was performed as described previously45. Cells were lysed in E1A lysis buffer (250 mM NaCl, 50 mM HEPES [pH 7.5], 0.1% NP-40, 5 mM EDTA, protease inhibitor cocktail [Sigma]). The antibodies to α-catenin (1:500, Sigma, C2081) and IκBα (1:500, Cell Signaling Technology, 9242) were used for immunoprecipitation. The SFB pull-down experiment was done as described previously46. Briefly, 293T cells were transfected with SFB-tagged protein and lysed in NETN buffer (200 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.05% NP-40, 1 mM EDTA, protease inhibitor cocktail [Sigma]) for 20 min at 4°C. Crude lysates were subjected to centrifugation at 14,000 rpm for 15 min at 4°C. Supernatants were incubated with S-protein beads for 4 h (Novagen). The beads were washed three times with NETN buffer. Proteins were eluted by boiling in 1× SDS running buffer and subjected to SDS-PAGE for immunoblotting.
ChIP assay
Cells were grown to 80% confluence, and crosslinking was performed with 1% formaldehyde for 10 min. ChIP assays were performed using a Magna ChIP G Chromatin Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. After immunoprecipitation with an antibody to RelA (Cell Signaling Technology, 8242) or normal rabbit IgG, protein-DNA crosslinks were reversed. DNA was then purified to remove the chromatin proteins and analysed by quantitative real-time PCR using the Qiagen EpiTect ChIP qPCR Primers (Cat# GPH1006956(-)01A).
In vivo tumorigenesis study
All animal experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of MD Anderson Cancer Center. When used in a power calculation, our sample size predetermination experiments indicate that 5 mice per group can identify the expected effect of α-catenin on tumor size and weight (P < 0.05) with 100% power. Animals were randomly assigned to different groups. Six- to eight-week-old female nude mice were used for subcutaneous injection of human breast cancer cells. Tumor cells in 30 μl of growth medium (mixed with Matrigel at a 1:1 ratio) were injected subcutaneously using a 100-μl Hamilton microliter syringe. Tumor size was measured once a week using a caliper, and tumor volume was calculated using the standard formula 0.5 × L × W2, where L is the longest diameter and W is the shortest diameter. Mice were euthanized when they met the institutional euthanasia criteria for tumor size and overall health condition. The tumours were removed, photographed and weighed. The freshly dissected tumor tissues were fixed in 10% buffered formalin overnight, washed with PBS, transferred to 70% ethanol, embedded in paraffin, sectioned and stained with H&E. A laboratory technician (Min Wang) who provided animal care and measured tumor growth was blinded to the group allocation during all animal experiments and outcome assessment.
Immunohistochemistry
Samples were deparaffinized and rehydrated. Antigen was retrieved using 0.01 M sodium-citrate buffer (pH 6.0) at a sub-boiling temperature for 10 min after boiling in a microwave oven. To block endogenous peroxidase activity, the sections were incubated with 3% hydrogen peroxide for 10 min. After 1 h of preincubation in 5% normal goat serum to prevent nonspecific staining, the samples were incubated with the antibody to IκBα (1:50, Cell Signaling Technology, 4814) or TNFα (1:50, Novus Biologicals, NBP1-19532) at 4°C overnight. The sections were incubated with a biotinylated secondary antibody (1:500, biotinylated anti-rabbit IgG(H+L), Vector Laboratories, BA-1000 for TNFα; 1:500, biotinylated anti-mouse IgG(H+L), Vector Laboratories, BA-9200 for IκBα) and then incubated with an avidin-biotin peroxidase complex solution (Vector Laboratories, PK-6100) for 30 min at room temperature. Color was developed using the DAB (diaminobenzidine) Substrate Kit (BD Biosciences, 550880). Counterstaining was carried out using Harris modified hematoxylin.
TCGA data analysis
We obtained level 3 data for mRNA expression and gene methylation of human breast tumors from Synapse (http://synapse.org) (syn1461151). mRNA expression was measured using the Agilent 244K Custom Gene Expression G4502A-07-3 (microarray) or the Illumina HiSeq 2000 RNA Sequencing version 2 analysis platform (RNA-Seq by Expectation Maximization, RSEM). Methylation was measured by using the Illumina Infinium Human DNA Methylation 450 platform. The breast cancer subtype information (luminal A, luminal B, basal-like and HER2 subtypes) was described previously10. The expression levels of CTNNA1 in normal and cancer samples for each subtype were compared using the Wilcoxon test. The association between CTNNA1 expression level and its gene methylation was assessed by the Spearman rank correlation test.
Statistical analysis
Each experiment was repeated three times or more. Unless otherwise noted, data are presented as mean ± s.e.m., and Student’s t-test (unpaired, two-tailed) was used to compare two groups of independent samples. The data analysed by t-test meet normal distribution; we used an F-test to compare variances, and the variances are not significantly different. Therefore, when using an unpaired t-test, we assumed equal variance, and no samples were excluded from the analysis. The log-rank test was used to compare Kaplan-Meier survival curves. Statistical methods used for TCGA data analysis are described above. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Stephen Ethier for the SUM159 cell line; Junjie Chen for plasmids; the shRNA and ORFeome Core and the Histology Core at The University of Texas MD Anderson Cancer Center for technical assistance; Jinsong Zhang and Juan Chen for assistance with graphics; Jeffrey M. Rosen, Xin Lin, Wenqi Wang and members of the Ma lab for discussion; and Hai-lan Piao, Caleb Chu and Arthur Gelmis for editing the manuscript. This work is supported by U.S. National Institutes of Health grants R00CA138572 (to L.M.) and R01CA166051 (to L.M.) and a Cancer Prevention and Research Institute of Texas Scholar Award R1004 (to L.M.).
Footnotes
AUTHOR CONTRIBUTIONS
L.M. conceived and supervised the project. H.-L.P. designed, performed and analyzed most of the experiments. Y.Y. and H.L. performed computational data analysis. M.W. performed immunohistochemical staining and provided animal care. Y.S. maintained shRNA and ORF clones and provided significant intellectual input. H.-L.P. and L.M. wrote the manuscript with input from all other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession number. The accession number of TCGA breast cancer data is syn1461151 (https://www.synapse.org/#!Synapse:syn1461151)
References
- 1.Di Cosimo S, Baselga J. Management of breast cancer with targeted agents: importance of heterogeneity. [corrected] Nat Rev Clin Oncol. 2010;7:139–147. doi: 10.1038/nrclinonc.2009.234. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakha EA, El-Sayed ME, Reis-Filho J, Ellis IO. Patho-biological aspects of basal-like breast cancer. Breast Cancer Res Treat. 2009;113:411–422. doi: 10.1007/s10549-008-9952-1. [DOI] [PubMed] [Google Scholar]
- 6.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16 (Suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 7.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 8.Slamon DJ, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 9.Kobielak A, Fuchs E. Alpha-catenin: at the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol. 2004;5:614–625. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Network TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onder TT, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 12.Derksen PW, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–449. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Liu TX, et al. Chromosome 5q deletion and epigenetic suppression of the gene encoding alpha-catenin (CTNNA1) in myeloid cell transformation. Nat Med. 2007;13:78–83. doi: 10.1038/nm1512. [DOI] [PubMed] [Google Scholar]
- 15.Ji H, Wang J, Fang B, Fang X, Lu Z. alpha-Catenin inhibits glioma cell migration, invasion, and proliferation by suppression of beta-catenin transactivation. J Neurooncol. 2011;103:445–451. doi: 10.1007/s11060-010-0413-4. [DOI] [PubMed] [Google Scholar]
- 16.Schlegelmilch K, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silvis MR, et al. alpha-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal. 2011;4:ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bajpai S, Feng Y, Krishnamurthy R, Longmore GD, Wirtz D. Loss of alpha-catenin decreases the strength of single E-cadherin bonds between human cancer cells. J Biol Chem. 2009;284:18252–18259. doi: 10.1074/jbc.M109.000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi N, et al. Constitutive activation of nuclear factor-kappaB is preferentially involved in the proliferation of basal-like subtype breast cancer cell lines. Cancer Sci. 2009;100:1668–1674. doi: 10.1111/j.1349-7006.2009.01228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 22.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 23.Baltimore D. Discovering NF-kappaB. Cold Spring Harb Perspect Biol. 2009;1:a000026. doi: 10.1101/cshperspect.a000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karin M, Yamamoto Y, Wang QM. The IKK NF-kappa B system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 25.Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817–6830. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 26.Hollestelle A, et al. Four human breast cancer cell lines with biallelic inactivating alpha-catenin gene mutations. Breast Cancer Res Treat. 2010;122:125–133. doi: 10.1007/s10549-009-0545-4. [DOI] [PubMed] [Google Scholar]
- 27.Collart MA, Baeuerle P, Vassalli P. Regulation of tumor necrosis factor alpha transcription in macrophages: involvement of four kappa B-like motifs and of constitutive and inducible forms of NF-kappa B. Mol Cell Biol. 1990;10:1498–1506. doi: 10.1128/mcb.10.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunsch C, Rosen CA. NF-kappa B subunit-specific regulation of the interleukin-8 promoter. Mol Cell Biol. 1993;13:6137–6146. doi: 10.1128/mcb.13.10.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincenti MP, Coon CI, Brinckerhoff CE. Nuclear factor kappaB/p50 activates an element in the distal matrix metalloproteinase 1 promoter in interleukin-1beta-stimulated synovial fibroblasts. Arthritis Rheum. 1998;41:1987–1994. doi: 10.1002/1529-0131(199811)41:11<1987::AID-ART14>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 30.Matluk N, Rochira JA, Karaczyn A, Adams T, Verdi JM. A role for NRAGE in NF-kappaB activation through the non-canonical BMP pathway. BMC Biol. 2010;8:7. doi: 10.1186/1741-7007-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lien WH, Gelfand VI, Vasioukhin V. Alpha-E-catenin binds to dynamitin and regulates dynactin-mediated intracellular traffic. J Cell Biol. 2008;183:989–997. doi: 10.1083/jcb.200805041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 33.Bren GD, et al. Transcription of the RelB gene is regulated by NF-kappaB. Oncogene. 2001;20:7722–7733. doi: 10.1038/sj.onc.1204868. [DOI] [PubMed] [Google Scholar]
- 34.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 35.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobielak A, Fuchs E. Links between alpha-catenin, NF-kappaB, and squamous cell carcinoma in skin. Proc Natl Acad Sci U S A. 2006;103:2322–2327. doi: 10.1073/pnas.0510422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y, et al. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 38.Yan J, et al. Inactivation of BAD by IKK inhibits TNFalpha-induced apoptosis independently of NF-kappaB activation. Cell. 2013;152:304–315. doi: 10.1016/j.cell.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 40.Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 41.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 42.Pianetti S, Arsura M, Romieu-Mourez R, Coffey RJ, Sonenshein GE. Her-2/neu overexpression induces NF-kappaB via a PI3-kinase/Akt pathway involving calpain-mediated degradation of IkappaB-alpha that can be inhibited by the tumor suppressor PTEN. Oncogene. 2001;20:1287–1299. doi: 10.1038/sj.onc.1204257. [DOI] [PubMed] [Google Scholar]
- 43.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang WL, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, et al. PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 2012;26:1959–1971. doi: 10.1101/gad.192955.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.