Abstract
Proline‐, glutamic acid‐, and leucine‐rich protein 1 (PELP1) is a proto‐oncogene that functions as coactivator of the estrogen receptor and is an independent prognostic predictor of shorter survival of breast cancer patients. The dysregulation of PELP1 in breast cancer has been implicated in oncogenesis, metastasis, and therapy resistance. Although several aspects of PELP1 have been studied, a complete list of PELP1 target genes remains unknown, and the molecular mechanisms of PELP1 mediated oncogenesis remain elusive. In this study, we have performed a whole genome analysis to profile the PELP1 transcriptome by RNA‐sequencing and identified 318 genes as PELP1 regulated genes. Pathway analysis revealed that PELP1 modulates several pathways including the molecular mechanisms of cancer, estrogen signaling, and breast cancer progression. Interestingly, RNA‐seq analysis also revealed that PELP1 regulates the expression of several genes involved in alternative splicing. Accordingly, the PELP1 regulated genome includes several uniquely spliced isoforms. Mechanistic studies show that PELP1 binds RNA with a preference to poly‐C, co‐localizes with the splicing factor SC35 at nuclear speckles, and participates in alternative splicing. Further, PELP1 interacts with the arginine methyltransferase PRMT6 and modifies PRMT6 functions. Inhibition of PRMT6 reduced PELP1‐mediated estrogen receptor activation, cellular proliferation, and colony formation. PELP1 and PRMT6 are co‐recruited to estrogen receptor target genes, PELP1 knockdown affects the enrichment of histone H3R2 di‐methylation, and PELP1 and PRMT6 coordinate to regulate the alternative splicing of genes involved in cancer. Collectively, our data suggest that PELP1 oncogenic functions involve alternative splicing leading to the activation of unique pathways that support tumor progression and that the PELP1–PRMT6 axis may be a potential target for breast cancer therapy.
Keywords: Alternative splicing, Breast cancer, Epigenetics, PELP1, PRMT6
Highlights
Genomic analysis using RNA‐sequencing identified proto‐oncogene PELP1 target genes.
Alternative splicing was identified as a novel PELP1 regulated pathway.
PRMT6 was identified as PELP1 novel interacting protein.
PRMT6 is needed for PELP1 oncogenic and splicing functions.
PELP1‐PRMT6 axis may be a potential target for breast cancer therapy.
Abbreviations
- BCAS2
breast cancer amplified sequence2
- ChIP
chromatin immunoprecipitation
- CDK
cell cycle dependent kinases
- E2
Estrogen
- ERα
estrogen receptor alpha
- HER2
human epidermal growth factor receptor 2
- IHC
immuno histochemistry
- MAPK
mitogen activated protein kinase
- NR
nuclear receptor
- PR
progesterone receptor
- PELP1
proline, glutamic acid, and leucine-rich protein-1
- PRMT6
protein arginine methyltransferase
- PI3K
phosphotidyl inositol 3-kinase
- qRTPCR
quantitative real time PCR
- RNA
ribonucleic Acid
- siRNA
small interfering RNA
- shRNA
short hairpin RNA
- hnRNPs
heterogeneous nuclear ribonucleoproteins
- SRs
serine/arginine-rich proteins
1. Introduction
Proline, glutamic acid, leucine‐rich protein 1 (PELP1) is a proto‐oncogene that functions as an estrogen receptor alpha (ERα) coregulator that provides breast cancer cells with a distinct growth and survival advantage (Nair et al., 2010a; Rajhans et al., 2007; Vadlamudi and Kumar, 2007; Vadlamudi et al., 2001). PELP1 interacts with several proteins including pRB, histone H1, and BCAS3 and acts as a scaffolding molecule that regulates cellular signaling (Balasenthil and Vadlamudi, 2003; Gururaj et al., 2007; Nair et al., 2004). PELP1 is overexpressed in hormonal cancers, promotes estrogen‐mediated cell proliferation, and is linked to shorter breast cancer specific survival, therapy resistance, and metastasis (Chakravarty et al., 2010; Habashy et al., 2010; Kumar et al., 2009; Rajhans et al., 2007; Vadlamudi and Kumar, 2007; Vallabhaneni et al., 2011; Yang et al., 2012). Overexpression of PELP1 results in cellular transformation, increased migratory potential, anchorage‐independent growth, and rapid tumor growth in xenograft studies (Rajhans et al., 2007). Inhibition of PELP1 by siRNA‐liposomes reduces tumorigenic potential in breast and ovarian cancer xenograft models (Chakravarty et al., 2011; Cortez et al., 2012). PELP1 regulates epigenetic modifications of estrogen receptor target genes via its interactions with epigenetic modifiers KDM1 and CARM1 (Mann et al., 2013; Nair et al., 2010b). Although several aspects of PELP1 signaling have been studied, a complete list of PELP1 target genes remains unknown.
Alternative splicing (AS) is essential in regulating gene expression and results in protein diversity; however, dysregulation leads to cancer development with tumor‐specific splice variants (Xu and Lee, 2003). A genome‐wide analysis found cancer specific splice variants in 316 human genes, with a majority having a functional role in cancer, that act by disrupting a tumor suppressor function (Xu and Lee, 2003). Splicing and transcription are coupled for the majority of intron‐containing genes through terminal exon pausing that allows for a rapid response to changes in growth conditions to coordinate levels of RNA processing factors with mRNA levels (Carrillo Oesterreich et al., 2010; Ip et al., 2011). Steroid receptors such as ERα play a role in this process; however, the molecular mechanisms and the role of ERα coregulators in this process remain elusive. PELP1 localizes to the nucleolar compartment and plays a critical role in active ribosomal RNA transcription (Finkbeiner et al., 2011; Gonugunta et al., 2011). Further, PELP1 associates with the Rix1 complex and is needed for proper processing of the 32S rRNA intermediate (Castle et al., 2012). Even through these studies indicate a role of PELP1 in processing ribosomal RNA, it remains unknown whether PELP1 plays a role in the alternative splicing of genes that commonly occurs in tumors.
Recent studies suggest that PELP1 recognizes arginine methylation (Mann et al., 2013). Protein arginine methylation is a modification that is involved in signal transduction, pre‐RNA metabolism, and transcriptional activation (Friesen et al., 2001). Spliceosomal snRNPs have arginine‐ and glycine‐rich domains which are post‐translationally modified by di‐methylarginines that enhance and regulate protein–protein interactions (Miranda et al., 2005). Protein arginine methyltransferase 6 (PRMT6), which methylates Histone H3 Arginine 2, plays a key role in the coupling of transcription and AS by functioning as a transcriptional coactivator (Cheng et al., 2007). PRMT6 also coactivates ERα in a hormone‐dependent manner (Dowhan et al., 2012; Harrison et al., 2010). PRMT6 is dysregulated in cancer and increases proliferation by regulating the cell cycle, RNA processing, and DNA replication (Limm et al., 2013; Yoshimatsu et al., 2011). It remains unknown whether PELP1 has crosstalk with PRMT6 and whether PRMT6 plays a role in the oncogenic functions of PELP1.
In this study, we have performed a whole genome analysis to profile the PELP1 transcriptome by RNA‐sequencing and identified 318 genes as PELP1 regulated genes. Pathway analysis revealed that PELP1 modulates several pathways including the molecular mechanisms of cancer, estrogen signaling, and breast cancer progression. Functional analysis of PELP1 target genes revealed PELP1's role in RNA splicing. Mechanistic studies revealed that PELP1 oncogenic actions involve splicing that is mediated via PELP1 interactions with PRMT6. Collectively, our data suggest that PELP1 oncogenic functions involve alternative splicing leading to the activation of unique pathways that support tumor progression.
2. Materials and methods
2.1. Cell culture and chemicals
The ZR75 and MCF7 cells were obtained from the American Type Culture Collection (ATCC). ZRGST, ZRPELP1GST, MCF7GFP, and MCF7PELP1GFP stable cell lines were described earlier (Nair et al., 2010a). All of the model cells were passaged in the user's laboratory for fewer than 6 months after receipt or resuscitation. Antibodies used were purchased from various companies: H3R2me2a (cat# 07‐585) from Millipore; PRMT6 (cat# A310‐297A) from Bethyl Laboratories; glutathione‐S‐transferase (GST) from Cell Signaling Technology (cat# 2624) and PELP1 from Bethyl Laboratories. PRMT6 ON‐TARGETplus SMARTpool siRNA was purchased from Dharmacon (cat# L‐007773‐00‐0005) and transfected using oligofectamine as described previously [4].
2.2. Histone methyltransferase assay
Recombinant PRMT6 (cat# 11313‐H18H, Sino Biological Inc.) and Histone H3 (cat# 14‐411, Millipore) were used for the histone methyltransferase assay. The manufacturer's protocol was followed: Histone H3, S‐adenosyl‐l‐methionine, PRMT6, and purified bacterial full‐length PELP1 were added in methyltransferase buffer (100 mM Tris‐HCL, pH 8.0, 1 M NaCL, 2 mM ethylenediaminetetracetic acid) and incubated for 2 h at 30 °C. The reaction was stopped by the addition of Laemmli reducing sample buffer and run on 15% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis gel.
2.3. Cell proliferation, colony formation, reporter gene, and GST pull‐down assays
Proliferation assays were done as described previously (Nair et al., 2010a). The colony formation assays were performed as described previously (Rajhans et al., 2007). Reporter gene assays were performed as described previously with PRMT6 and ERE‐luciferase plasmids as described (Mann et al., 2013). Pull‐down assays were performed using bacterial PELP1‐GST deletions described previously (Nair et al., 2010b).
2.4. Western analysis and immunoflourescence
Cell lysates for western blot and immunoprecipitation were prepared as described previously (Nair et al., 2010b). For confocal analysis, MCF7 breast cancer cells cultured on glass coverslips were then fixed and co‐stained with antibodies against PELP1 (green) and SC35 (red) and analyzed by confocal microscopy (Nair et al., 2010b).
2.5. RNA polymer binding assay
RNA binding assays were done as described (Chen et al., 2002). Briefly, PELP1 fragments of various lengths were labeled with 35S‐methionine and incubated with various RNA polymers immobilized on Agarose beads for 4 h at 40 °C. The RNA polymers polyadenylic [poly(A)], polycytidylic acid [poly(C)], polyguanylic acid [poly(G)], and polyuridylic [poly(U)] were purchased from Sigma and are prepared by the catalytic action of the enzyme polynucleotide phosphorylase and the appropriate ribonucleotide as substrate. The bound PELP1 fragments were pulled down and washed six times with NP40 lysis buffer, eluted with 2XSDS buffer, separated by SDS‐PAGE, and developed by autoradiography.
2.6. Minigene splicing assay
HeLa cells were cotransfected with ERα and ERE‐CD44 mini gene along with vector or PELP1 expressing plasmid and treated with or without estradiol (Auboeuf et al., 2002). Total RNA was extracted and splicing pattern was analyzed by RT‐PCR using CD44 specific radiolabeled primers.
2.7. RNA‐sequencing
Total RNA was isolated from ZR75shControl and ZR75shPELP1 cells treated with estradiol for 5 h. Illumina TruSeq RNA Sample Preparation was performed following manufacturer's protocol. Samples were run on an Illumina HiSeq 2000 in duplicate. The combined raw reads were aligned to UCSC hg19 and genes were annotated by Tophat (Trapnell et al., 2009). Genes were annotated and quantified by HTSeq‐DESeq pipeline (Anders; Anders and Huber, 2010). Differential expression analysis was performed by DEseq and significant genes were chosen with abs (log2FoldChange) > 2.
2.8. Real time PCR‐isoform specific primers
Total RNA was extracted from cells using Trizol (Invitrogen), according to manufacturer's protocol. RNA was normalized by measuring in Nanodrop spectrophotometer and cDNA was synthesized using SuperScript III First Strand (Invitrogen), according to manufacturer's protocol. Gene expression levels were analyzed using SyberGreen on an Illumina Real‐Time PCR system with the following primers. Primers designed to amplify the specific alternatively spliced isoforms, were NIN inclusion – forward: GAGTCACTATTTTAGTTAAGC, NIN inclusion – reverse: AGATTCATATCTCA‐GAAGTTC, NIN skipping – forward GAACTAGAGCAGTTTCACCAG, NIN skipping – reverse CAGATTCATATCTTCCCTCCT (Dowhan et al., 2012). In order to detect levels of total VEGF transcript (VEGFtotal) and three splice products VEGF121, VEGF165 and VEGF189, a common forward primer (5′‐CCCTGATGAGATCGAGTACATCTT‐3′) located on exon 3 was used (Harrison et al., 2010). Specific amplification of each spliced product was achieved by using reverse primers spanning exon boundaries specific for the relevant product. Reverse primers used were: VEGF121 (5′‐GCCTCGGCTTGTCACATTTT‐3′), VEGF165 (5′‐AGCAAGGCCCACAGGGATTT‐3′), VEGF189 (5′‐AACGCTCCAGGACTTATACCG‐3′) and VEGFtotal (5′‐ACCGCCTCGGCTTGTCAC‐3′). Relative changes in expression were calculated with the ΔΔCt method. Each sample was measured in triplicate and normalized to actin.
2.9. Immunohistochemistry
Tissues from previously established xenografts were used for immunohistochemical analysis (Cortez et al., 2012). In brief, in this study nu/nu mice were inoculated with MCF7‐PELP1 cells and after establishment of tumors, they were treated with either control siRNA or PELP1 siRNA liposomes. Immunohistochemistry (IHC) was done according to previously established protocol with anti‐PELP1 (1:500), anti‐PRMT6 (1:50) and anti‐H3R2me2a (1:50) antibodies (Cortez et al., 2012).
2.10. Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) analysis was performed as described previously (Nair et al., 2010b). Sequential ChIP was performed using the Re‐ChIP‐IT Magnetic Chromatin Re‐Immunoprecipitation Kit (cat# 53016, Active Motif) according to the manufacturer's protocol with anti‐PELP1 and anti‐immunoglobulin G (IgG) antibodies. Primers for GREB1C: forward: TTGTTGTAGCTCTGGGAGCA; reverse: CAACCAGCCAAGAGGCTAAG.
3. Results
3.1. PELP1 genomic regulation involves activation of genes that participate in multiple pathways
In this study, we performed a transcriptome profile of the breast cancer cell line ZR75 with PELP1 knockdown by RNA‐sequencing (RNA‐seq). PELP1 knockdown resulted in an upregulation of 99 genes and downregulation of 219 genes by at least two‐fold (Table S1, Figure 1A). Ingenuity pathway analysis of the differentially expressed genes revealed the top pathways regulated by PELP1. The pathway of the molecular mechanisms of cancer emerged as a top pathway regulated by PELP1 (Figure 1B). Additional identified pathways include breast cancer regulation and estrogen receptor signaling, which validates the importance of PELP1 in breast cancer, and novel pathways such as oxidative stress response and protein ubiquitination. We further validated the expression of some of the top differentially expressed genes involved in the breast cancer and estrogen signaling pathways by qRTPCR in ZR75 cells with stable PELP1 knockdown (Figure 1C, D). The differential expression was also validated in MCF7 cells with stable PELP1 knockdown (Figure S1A) and in ZR75 cells with transient siRNA PELP1 knockdown (Figure 1E). The specificity of the results was further confirmed by the reintroduction of a siRNA‐resistant PELP1 cDNA into PELP1 knockdown cells (Figure S1B). IPA analysis of gene function revealed that PELP1 regulated genes are involved in cell cycle progression, DNA replication and repair, as well as cellular growth and proliferation (Figure 1F). Intriguingly, biological function analysis also revealed a novel function regulated by PELP1 as alternative splicing (AS). We confirmed the differential expression of several genes involved in AS including BCAS2, PRMT6, SRSF12, and AFF2. Accordingly, the PELP1 regulated genome contains several genes that are alternatively spliced (Table S2).
Figure 1.
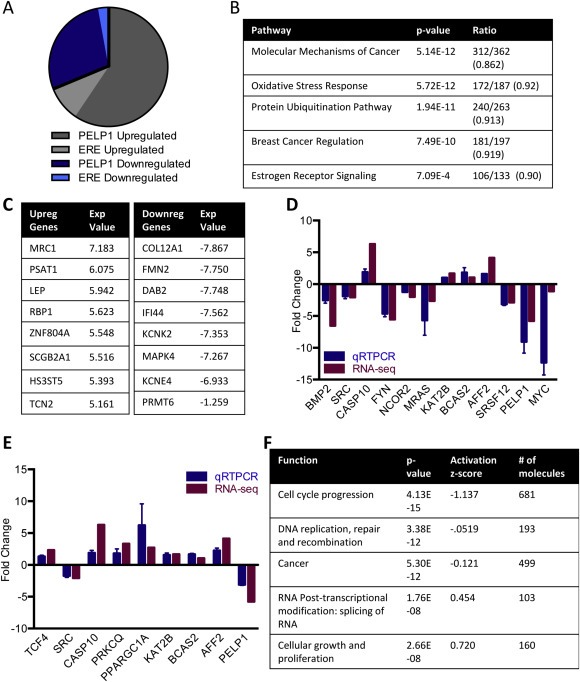
PELP1 genome wide analysis. A) A list of classical estrogen responsive genes was compared to the differentially expressed genes shown to be regulated by PELP1 from the RNA‐sequencing analysis (Tang et al., 2004). Of the 219 genes downregulated by PELP1, 30 are estrogen responsive and of the 99 genes upregulated by PELP1, 9 are estrogen responsive. B) ZR75shControl and ZR75shPELP1 cells were estrogen starved for 72 h in 5% Dextran charcoal stripped media and treated with estradiol 10−8 M for 5 h. Bioinformatics analysis of RNA sequencing results revealed genes with differential expression and genes with greater than two fold change in expression were further analyzed by Ingenuity Pathway Analysis. The top pathways from IPA are shown with the ratio of differentially expressed genes over total known genes in pathway. C) RNA‐sequencing of top differentially expressed genes with expression value given as fold change of ZR75shPELP1/ZR75conshRNA. D) Differential gene expression from RNA‐sequencing was validated with ZR75 cells stably transfected with either shRNA control or shRNA PELP1, starved of estrogen for 72 h in 5% Dextran charcoal stripped media, and treated with estradiol 10−8 M for 5 h using qRTPCR in triplicate. E) Differential gene expression from RNA‐sequencing was validated with ZR75 cells transfected with either siControl or siPELP1 and treated with estradiol for 5 h using qRTPCR in triplicate. F) IPA analysis of differentially expressed genes for PELP1 regulated biological functions.
3.2. PELP1 signaling regulates alternative splicing
Since RNA sequencing data revealed PELP1's potential to regulate the splicing of genes, we examined whether PELP1 plays an important role in AS. SC35 or SFRS2 is an arginine/serine‐rich splicing factor that is required for the early steps of spliceosome assembly and influences splice‐site selection (Fu, 1993). Immunoflourescence studies showed that PELP1 colocalizes with the spliceosome marker SC35 at nuclear speckles suggesting that PELP1 localizes to the spliceosome (Figure 2A) (Lin et al., 2008). Accordingly, immunoprecipitation studies confirmed that PELP1 and SC35 form a complex (Figure S1C). An in vitro RNA polymer binding assay in which PELP1 deletions were labeled with 35S‐methionine, incubated with synthetic ribonucleotide homopolymers, poly(A), poly(C), poly(U), and poly(G), and immobilized on agarose beads revealed that PELP1 interacts with RNA with a preference to poly‐C (Figure 2B). To examine whether PELP1 regulates splicing, we used a CD44 minigene assay with exons 4 and 5. The results from these assays revealed that PELP1 promotes the skipping of both exons 4 and 5 with estrogen stimulation (Figure 2C) (Konig et al., 1998). One of the genes we found to be alternatively spliced by Cufflink analysis of the RNA‐sequencing data was vascular endothelial growth factor (VEGF), which is a mediator of angiogenesis and its expression positively correlates with malignant progression (Wellmann et al., 2001). Since PELP1 is proto‐oncogene and VEGF isoforms are implicated in tumor progression, we determined whether PELP1 affects the exon skipping of endogenous VEGF. Alternative splicing of VEGF produces three isoforms including VEGF 121, VEGF 165, and VEGF 189 (Tischer et al., 1991). Each of the isoforms appears to play a role in cancer; increased expression of VEGF 165 and VEGF 189 correlates with tumor progression and poor prognosis while others have shown there is an angiogenic switch to VEGF 121 and VEGF 165 during malignant progression (Wellmann et al., 2001). Using isoform specific qRTPCR, we found that siRNA knockdown of PELP1 results in a decrease of VEGF transcripts with a significantly higher expression of the VEGF 189 isoform as compared to the other isoforms when normalized to total VEGF expression (Figure 2D).
Figure 2.
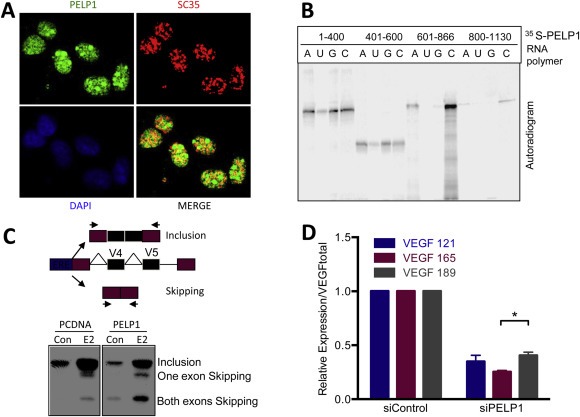
PELP1 regulation of alternative splicing. A) Immunoflourescence of MCF7 cells was performed for PELP1 (green) or SC35 (red) and analyzed by confocal microscopy. Colocalization of PELP1 and SC35 is visualized as yellow. Dapi (blue) was used to localize the nucleus. B) PELP1 fragments were labeled with 35S‐methionine and incubated with various RNA polymers including poly(A), poly(C), poly(G) and poly(U) immobilized on agarose beads. The bound PELP1 fragments were pulled down and washed six times with NP40 lysis buffer, eluted with 2XSDS buffer, separated by SDS‐PAGE, and developed by autoradiography. C) Top: Schematic of ERE‐CD44 minigene with exons 4 and 5. Bottom: HeLa cells were cotransfected with ERα, ERE‐CD44 mini gene and vector or PELP1 expressing plasmid, and treated with or without estradiol. The splicing pattern was analyzed by qRTPCR using CD44 specific radiolabeled primers. D) qRTPCR of VEGF isoforms 121, 165 and 189 normalized over total VEGF from ZR75 cells transfected with either siControl or siPELP1. Relative expression is from triplicates normalized to actin.
One of the alternative splicing genes that we found to be regulated by PELP1 from the RNA sequencing data is PRMT6; specifically, PRMT6 is downregulated in the PELP1 knockdown cells (Figure 1C). PRMT6 is an arginine methyltransferase that was recently reported to regulate the splicing of several genes including VEGF and NIN (Dowhan et al., 2012). Since PELP1 has been shown to affect the functions of another arginine methyltransferase CARM1 (Mann et al., 2013), we examined whether PELP1 status affects PRMT6's ability to alternatively splice these genes. By performing qRTPCR with isoform specific primers for NIN and VEGF, we found that PELP1 status significantly affected the ability of PRMT6 to promote exon skipping (Figure 3A, B). PRMT6 knockdown produced a knockdown of VEGF 165 with no significant difference in VEGF 121 or VEGF 189. However, transient knockdown of PRMT6 in stable PELP1 knockdown cells resulted in an increase in VEGF 165 and significant decreases in VEGF 121 and VEGF 189 (Figure 3B). Knockdown of PRMT6 results in inclusion of the NIN exon 19 as shown previously (Dowhan et al., 2012); however, knockdown of PRMT6 in cells without PELP1 expression causes no significant difference in the inclusion or skipping of exon 19 (Figure 3C, D). These results suggest that PELP1 and PRMT6 work together to regulate the alternative splicing of selective genes.
Figure 3.
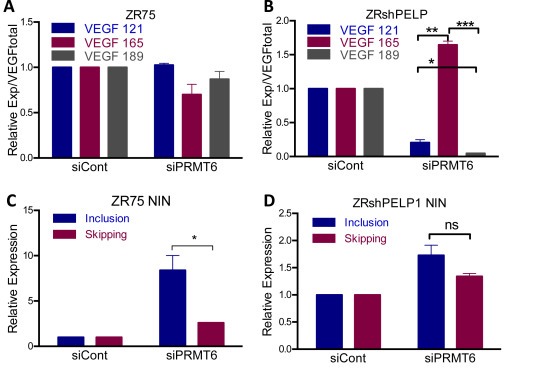
Coordination of PELP1 and PRMT6 in alternative splicing. A,B) qRTPCR of VEGF isoforms 121, 165 and 189 normalized over total VEGF from ZR75shControl or ZR75shPELP1 cells transfected with either siControl or siPRMT6. Relative expression is from triplicates normalized to actin. C,D) Isoform specific qRTPCR of inclusion or skipping of exon 19 of NIN from ZR75shControl or ZR75shPELP1 cells transfected with siControl or siPRMT6. Relative expression is from triplicates normalized to actin.
3.3. Role of PRMT6 in PELP1 oncogenic functions
PRMT6 is implicated in cell death, cell cycle and gene expression and is dysregulated in ERα positive breast cancer (Harrison et al., 2010). Since the RNA‐sequencing data suggested that PELP1 may regulate PRMT6 and PELP1 affects PRMT6‐mediated alternative splicing, we tested whether PELP1 and PRMT6 pathways crosstalk via protein–protein interactions. We performed co‐immunoprecipitation of PELP1 or PRMT6 from ZR75 cells treated with estrogen and found PRMT6 and PELP1 to co‐immunoprecipitate in both assays (Figure 4A). A GST‐binding assay revealed that PRMT6 binds to the 600‐866 amino acid region of PELP1 (Figure 4B). We then tested the effect of PRMT6 knockdown in ERE reporter gene assays. As expected, PELP1 overexpressing cells (ZRPELP1 or MCF7PELP1) showed increased ERE reporter activity compared to ZR75 or MCF7 cells; however, knockdown of PRMT6 decreased PELP1's activation of the ERE reporter (Figure 4C, S1D). Transient knockdown of PRMT6 by siRNA resulted in a decrease in the PELP1‐mediated proliferation of both ZR75 and MCF7 breast cancer cells (Figure 4D, S1E). Further, PRMT6 knockdown also decreased the clonogenic ability of PELP1‐overexpressing ZR75 and MCF7 cells (Figure 4E, S1F). Importantly, knockdown of PRMT6 in cells with stable knockdown of PELP1 had no effect of the clonogenicity, suggesting that observed clonogenic potential of PRMT6 is dependent upon PELP1 (Figure 4F). Collectively, these results suggest that PRMT6 plays an important role on PELP1 mediated oncogenic functions.
Figure 4.
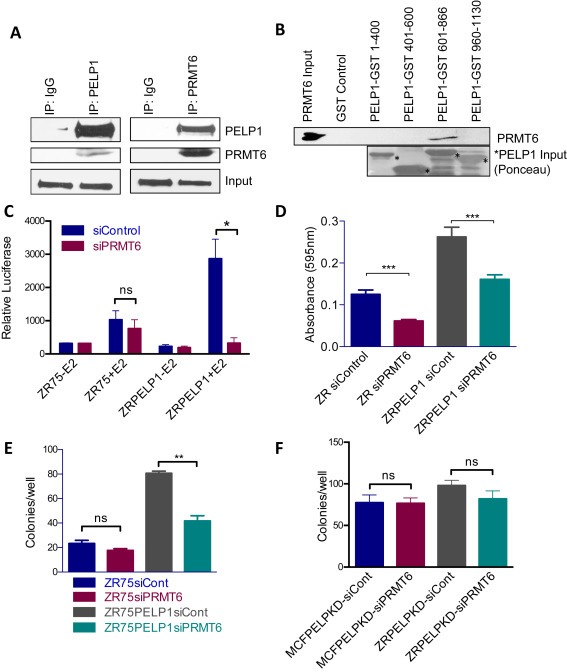
Effect of PRMT6 on PELP oncogenic functions. A) Immunoprecipitation of PELP1 or PRMT6 was performed from nuclear lysates of ZR75 cells grown in 8% FBS‐RPMI. Western blot analysis was performed for co‐immunoprecipitation of PRMT6 or PELP1 with IgG and Input as the controls. B) GST binding assay with bacterial purified PELP1‐GST deletions and recombinant PRMT6 with GST as the control. Western blot is shown for PRMT6 expression with PRMT6 input as the control and ponceau stain shows PELP1 input protein denoted by asterisk. C) An ERE‐luciferase gene reporter assay was performed with ZR75vector and ZR75PELP1 cells transfected with siControl or siPRMT6 and ERE‐luciferase in triplicate. Cells were starved of estrogen in 5% DCC media for 48 h and treated with estradiol 10−8 M for 12 h. Cells were lysed with passive lysis buffer and relative luciferase activity was measured by a luminometer. D) An MTT proliferation assay was performed with ZR75vector and ZR75PELP1 cells transfected with either control siRNA or PRMT6 siRNA. E) ZR75 and ZR75PELP1 cells were transfected with either control siRNA or PRMT6 siRNA and plated in triplicate in 6‐well plates for a colony formation assay. Colonies were stained by crystal violet and counted on day 14. F) MCF7PELP1shRNA and ZR75PELP1shRNA cells were transfected with either control siRNA or PRMT6 siRNA and plated in triplicate in 6‐well plates for a colony formation assay. Colonies were stained by crystal violet and counted on day 14.
3.4. PELP1 regulates PRMT6 activity and its recruitment to target genes
Earlier studies revealed that PELP1 is a scaffolding protein, interacts with histones, and regulates the activity of chromatin‐modifying enzymes (Cortez et al., 2012; Mann et al., 2013; Vadlamudi and Kumar, 2007). PRMT6 is an arginine methyltransferase that di‐methylates Histone H3R2. To determine if PELP1 affects the ability of PRMT6 to methylate histones, we performed an in vitro methylation assay. When purified PELP1 was added, there was an increase in H3R2me2a suggesting that PELP1 enhances PRMT6's methyltransferase activity (Figure 5A). To determine if the enrichment of the arginine di‐methylation modification is affected by PELP1 status at ERα target genes, we performed ChIP of H3R2me2 in ZR75 control and PELP1 stable knockdown cells with or without estrogen. When the cells are treated with estrogen in the presence of PELP1, there is a significant increase in H3R2me2 enrichment at the promoter of GREB1C; however, when PELP1 is knocked down there is almost no enrichment of the histone modification (Figure 5B). Therefore, PELP1 is required for the optimal enrichment of the H3R2me2 modification by PRMT6. The recruitment of the enzyme itself is also affected by the status of PELP1. There is strong PRMT6 recruitment to GREB1C when PELP1 is present, but when PELP1 is knocked down there is no recruitment of PRMT6 suggesting that PELP1 affects not only the methyltransferase activity of PRMT6 but also its recruitment to certain genes (Figure 5C). In a sequential ChIP, we observed that PELP1 and PRMT6 are co‐recruited to the estrogen responsive gene GREB1C (Figure 5D). In a previous xenograft experiment, PELP1 siRNA liposome treatment resulted in a significant decrease in tumor growth (Cortez et al., 2012). We performed immunohistochemistry on the tissues from the xenograft assay to probe for expression of H3R2me2a and PRMT6 between the control siRNA and PELP1 siRNA‐treated groups. The results revealed a significant decrease in both histone modification and enzyme expression in the tumors treated with PELP1 siRNA, confirming our in vitro results that PELP1 affects the arginine di‐methylation catalyzed by PRMT6 as well as the expression of PRMT6 itself (Figure 5E).
Figure 5.
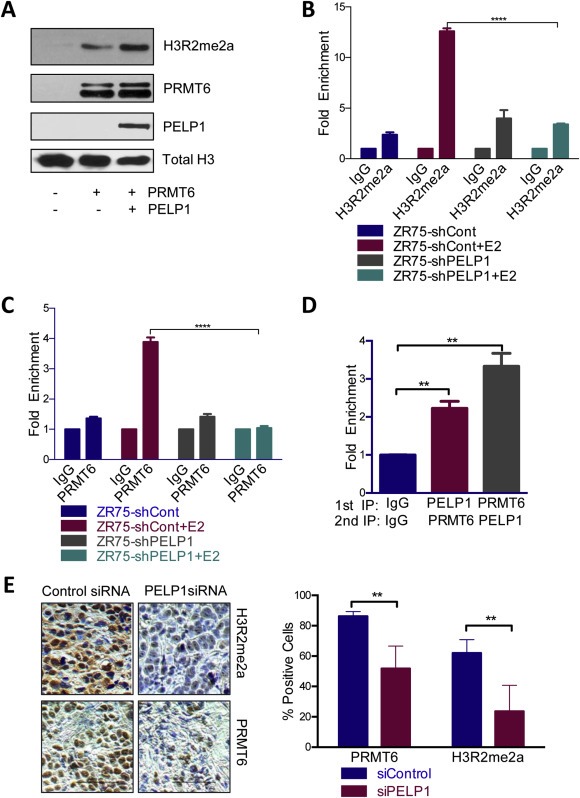
Regulation of PRMT6 activity by PELP1. A) Histone methyltransferase assay was performed with recombinant Histone H3, PRMT6 enzyme, purified PELP1 in buffer with S‐adenosyl methionine, estradiol and purified estrogen receptor alpha. Western blot analysis shows expression of H3R2me2a, PRMT6 and PELP1 with Total Histone H3 as the control. B) ChIP assay with ZR75shControl and ZR75shPELP1 cells starved of estrogen for 72 h and treated with estradiol for 30 min was performed with H3R2me2a or IgG. qRTPCR was done in triplicate for GREB1C. C) ChIP assay with ZR75shControl and ZR75shPELP1 cells starved of estrogen for 72 h and treated with estradiol for 30 min was performed with PRMT6 or IgG. qRTPCR was done in triplicate for GREB1C. D) Sequential ChIP of ZR75PELP1 cells grown in 2.5% DCC for 72 h and treated with estradiol 10−7 M for 30 min was done with PELP1, PRMT6 or IgG. qRTPCR was performed in triplicate for GREB1C. E) Immunohistochemistry of tissues of xenografts treated with siControl or siPELP1 liposomes probed for PRMT6 or H3R2me2a expression. Quantification of ten sections per group as percentage of positive cells over total cells is shown on right.
4. Discussion
PELP1 is an ERα coactivator that has been shown to play a role in breast cancer tumorigenesis. Classical estrogen genes are characterized as having estrogen response elements. As expected from earlier studies, we found that 12% of the PELP1 regulated genes are known classical estrogen responsive genes. Further, the results of this study revealed that, in addition to its established role in ERα signaling, PELP1 has the potential to regulate multiple signaling pathways that promote breast cancer progression. Gene network analysis suggested that PELP1 oncogenic functions involve genes regulated by Cyclin D1, Rb/E2F1, AKT/Myc and p53 pathways, which is in agreement with previous studies suggesting that PELP1 regulates various cell cycle genes and opens the avenue to elucidate the role of PELP1 in the p53 network and DNA damage pathway (Figures S2–6) (Nair et al., 2010a). PELP1 was also shown to be involved in the pathways of oxidative stress response and protein ubiquitination that can be studied further for a better understanding of PELP1's functions. PELP1 also functions as a coregulator for other transcription factors including AP1, SP1, PR, GR, ERR, and STAT3 (Girard et al., 2013), and this may explain PELP1's ability to regulate non‐ERE containing genes probably via non‐classical mechanisms. With the novel list of PELP1 target genes we have compiled, these alternate mechanisms can be explored to further elucidate the role of PELP1 in breast cancer by identifying which downstream target genes are necessary for its oncogenic functions (Figure S7).
Intriguingly, our results revealed an unexpected role of PELP1 in the regulation of alternative splicing. Splicing involves the removal of introns and selection of exons that are included or skipped to join together and form mRNA that is translated; it is regulated by the complex spliceosome composed of hundreds of proteins (Kramer, 1996). Alternative splicing controls multiple regulatory processes, including chromatin modification and signal transduction (Kornblihtt et al., 2013). The molecular diversity achieved through alternative splicing is suggested to play a critical role in diseases such as cancer. The selection of splice sites can depend on trans‐acting factors that function through the binding of splicing enhancers and silencers including serine/arginine‐rich proteins (SRs) and heterogeneous nuclear ribonucleoproteins (hnRNPs) (Kornblihtt et al., 2013). One of the genes we found to be regulated by PELP1, SRSF12 (serine/arginine‐rich splicing factor), is an essential splicing regulator and oncoprotein that influences the selection of alternative splice sites and assists in the formation of the spliceosome (Wu and Maniatis, 1993).
HnRNPs are known to be methylated at specific arginine residues by arginine methyltransferases to regulate transcription and RNA, arginine methylation of RNA helicase A is required for it to translocate to the nucleus, and RNA binding proteins are also substrates of arginine methyltransferases (Cote et al., 2003; Herrmann et al., 2004; Smith et al., 2004). One of these arginine methyltransferases, PRMT6, was identified to be up‐regulated by PELP1 in our RNA‐sequencing analysis. PRMT6 has been shown to require its enzymatic activity to regulate splicing by methylating RNA‐binding splicing proteins that contain glycine‐arginine‐rich domains including RDA288 and hnRNP D (Cheng et al., 2007; Harrison et al., 2010). Interestingly, PRMT6's dual role in regulating gene expression and alternative splicing can occur independently of each other (Harrison et al., 2010). We have shown that PELP1 is an RNA‐binding protein with preference to poly‐C and that PELP1 enhances the arginine methyltransferase activity of PRMT6; therefore, PELP1 may regulate PRMT6‐mediated alternative splicing by altering the methylation of the splicing proteins.
PELP1 is a transcriptional regulator that functions by recruiting other regulator proteins and remodeling chromatin to facilitate access to the promoter (Nair et al., 2004). Several nuclear receptor coregulators have been shown to be a link between transcriptional machinery and the recognition and processing of the transcript in the spliceosome (Auboeuf et al., 2005). Published studies showed that poly‐C binding proteins act at multiple levels during gene expression including transcriptional activation and regulation of RNA splicing (Choi et al., 2009). We have shown that PELP1 regulates the expression of several genes involved with splicing including BCAS2, PRMT6, SRSF12, and AFF2, colocalizes with the splicing marker SC35 at nuclear speckles, promotes the exon skipping of the CD44 minigene with estrogen activation, and regulates the PRMT6‐mediated alternative splicing of VEGF and NIN. Taken together, these results suggest that PELP1 may be another nuclear receptor coregulator that plays a role in coupling transcription and alternative splicing by functioning as a poly‐C binding scaffolding molecule.
Recent studies suggest that histone marks affect splicing outcome by influencing the recruitment of splicing regulators via chromatin‐binding proteins, and that histone modification patterns regulate alternative splicing patterns (Venables et al., 2009). PELP1 is a unique coregulator that interacts with multiple chromatin modifying enzymes and specifically recognizes dimethyl‐modified histones (Mann et al., 2013; Nair et al., 2010b). PELP1's ability to bind several chromatin modifiers and its ability to recognize dimethyl arginine modifications may play a role in splicing either by facilitating arginine modifications of key enzymes involved in splicing or by facilitating the recruitment of PRMT6 to the splicing machinery. Since PELP1's association with the Rix complex is implicated in the proper splicing of ribosomal RNA (Castle et al., 2012), it is possible that PELP1‐mediated alternative splicing of other cellular genes may also involve the Rix complex and future studies are needed to explore this hypothesis.
Several recent studies have shown the potential of targeting PELP1 oncogenic functions as a promising cancer therapeutic strategy (Chakravarty et al., 2011; Cortez et al., 2012; Mann et al., 2013; Nair et al., 2011; Vallabhaneni et al., 2011). In this study, we have shown that PELP1 and PRMT6 form a complex and that PRMT6 binds to PELP1 near its histone‐binding region. We have also shown that inhibition of PRMT6 can inhibit PELP1‐mediated estrogen activation, cellular proliferation, and colony formation. Also, PELP1 and PRMT6 are co‐recruited to estrogen responsive genes, PELP1 knockdown affects the enrichment of histone H3R2 di‐methylation, and PELP1 and PRMT6 coordinate to regulate the alternative splicing of genes involved in cancer including VEGF and NIN. Since both PELP1 and PRMT6 are deregulated in breast cancer, our findings suggest a novel role of the PELP1–PRMT6 axis in ERα‐mediated RNA splicing, and their deregulation may have implications in ERα‐driven breast cancer.
5. Conclusion
Our results suggest that PELP1 regulates the expression of both classical and non‐classical estrogen regulated genes that are involved in cell cycle progression, DNA replication/repair, cellular growth and proliferation. Further, PELP1 oncogenic functions involve alternative splicing leading to the activation of unique pathways that support breast tumor progression. Our mechanistic studies demonstrated that PELP1 interacts with and modifies the functions of PRMT6, both PELP1 and PRMT6 are needed for optimal splicing, and the PELP1–PRMT6 axis represents a potential target of breast cancer therapy. Further analysis of the transcriptional regulation by PELP1 elucidated from the RNA‐sequencing may aide in the understanding of the molecular pathogenic mechanisms underlying PELP1‐mediated oncogenesis, as well as lead to the development of novel therapeutic targets.
Competing interests
The authors declare that they have no conflict of interest.
Author contributions
M.M, D.B and R.V. designed the experiments. M.M. and R.V. performed the experiments. Y.Z. and Y.C. performed the bioinformatics analysis for the RNA‐sequencing. M.M. and R.V. wrote the manuscript with input from all authors.
Supporting information
The following is the supplementary data related to this article:
Figure S1: A) Differential gene expression from RNA‐sequencing was validated with MCF7 and MCF7PELP1KD cells treated with estradiol for 5 h using qRTPCR in triplicate. B) Differential gene expression from RNA‐sequencing was validated with ZR75 cells transfected with siControl or siPELP1 and either control vector or siRNA‐resistant‐PELP1 plasmid. After transfection, cells were starved of estrogen for 72 h and treated with estradiol 10−8 M for 5 h. Gene expression is shown as siControl/siPELP1 and (siControl + PELP1)/(siPELP + PELP1) normalized to Actin. C) Co‐immunoprecipitation of nuclear lysates of ZR75 cells was performed with PELP1, SC35, and IgG antibodies. Input of PELP1 and SC35 is shown. D) An ERE‐luciferase gene reporter assay was performed with MCF7 and MCF7PELP1 cells transfected with siControl or siPRMT6 and ERE‐luciferase in triplicate. Cells were starved of estrogen in 5% DCC media for 48 h and treated with estradiol 10−8 M for 12 h. Cells were lysed with passive lysis buffer and relative luciferase activity was measured by a luminometer. E) Proliferation assay was performed with MCF7 and MCF7PELP1 cells transfected with either control siRNA or PRMT6 siRNA. F) MCF7 and MCF7PELP1 cells were transfected with either control siRNA or PRMT6 siRNA and plated in triplicate in 6‐well plates for a colony formation assay. Colonies were stained by crystal violet and counted on day 14. Figure S2‐6: Molecular networks from IPA based on gene/molecule connectivity with other genes/molecules. Genes that are downregulated by PELP1 knockdown are shown in green and upregulated by PELP1 knockdown are in red. Figure S7: Schematic representation of PELP1 signaling from RNA‐sequencing analysis. Supplementary Table 1: RNA‐sequencing genes regulated by PELP1 with significance of p < 0.05 and log2(Fold Change) > 2 as analyzed by DESeq. Supplementary Table 2: Genes found to be significantly alternatively spliced by RNA‐sequencing using Cufflink analysis.
Acknowledgments
We thank Dr. Sujit Nair for the confocal assay and the CD44 minigene assay. We thank Dr. Rakesh Kumar for the ERE‐CD44 minigene plasmid. We thank Lai Zhao and Dawn Garcia at the Next Generation Sequencing Core for the RNA sequencing. The confocal images were generated in the Core Optical Imaging Facility which is supported by UTHSCSA, NIH‐NCI P30 CA54174 (CTRC at UTHSCSA) and NIH‐NIA P01AG19316. This research was funded by the NIH CA095681 and NIH 1 F31 CA173909‐01A1.
Supplementary data 1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2013.12.012.
Mann Monica, Zou Yi, Chen Yidong, Brann Darrell and Vadlamudi Ratna, (2014), PELP1 oncogenic functions involve alternative splicing via PRMT6, Molecular Oncology, 8, doi: 10.1016/j.molonc.2013.12.012.
Contributor Information
Monica Mann, Mannm3@livemail.uthscsa.edu.
Yi Zou, Email: zou@uthscsa.edu.
Yidong Chen, Email: cheny8@uthscsa.edu.
Darrell Brann, Email: dbrann@gru.edu.
Ratna Vadlamudi, Email: vadlamudi@uthscsa.edu.
References
- Anders, S., HTSeq: Analysing High-throughput Sequencing Data with Python. [DOI] [PMC free article] [PubMed]
- Anders, S. , Huber, W. , 2010. Differential expression analysis for sequence count data. Genome Biol.. 11, R106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf, D. , Honig, A. , Berget, S.M. , O'Malley, B.W. , 2002. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science (New York, N.Y.). 298, 416–419. [DOI] [PubMed] [Google Scholar]
- Auboeuf, D. , Dowhan, D.H. , Dutertre, M. , Martin, N. , Berget, S.M. , O'Malley, B.W. , 2005. A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol. Cell. Biol.. 25, 5307–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasenthil, S. , Vadlamudi, R.K. , 2003. Functional interactions between the estrogen receptor coactivator PELP1/MNAR and retinoblastoma protein. J. Biol. Chem.. 278, 22119–22127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich, F. , Preibisch, S. , Neugebauer, K.M. , 2010. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell. 40, 571–581. [DOI] [PubMed] [Google Scholar]
- Castle, C.D. , Cassimere, E.K. , Denicourt, C. , 2012. LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol. Biol. Cell. 23, 716–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty, D. , Nair, S.S. , Santhamma, B. , Nair, B.C. , Wang, L. , Bandyopadhyay, A. , Agyin, J.K. , Brann, D. , Sun, L.Z. , Yeh, I.T. , Lee, F.Y. , Tekmal, R.R. , Kumar, R. , Vadlamudi, R.K. , 2010. Extranuclear functions of ER impact invasive migration and metastasis by breast cancer cells. Cancer Res.. 70, 4092–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty, D. , Roy, S.S. , Babu, C.R. , Dandamudi, R. , Curiel, T.J. , Vivas-Mejia, P. , Lopez-Berestein, G. , Sood, A.K. , Vadlamudi, R.K. , 2011. Therapeutic targeting of PELP1 prevents ovarian cancer growth and metastasis. Clin. Cancer Res.: An Off. J. Am. Assoc. Cancer Res.. 17, 2250–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.C. , Lin, W.C. , Tsay, Y.G. , Lee, S.C. , Chang, C.J. , 2002. An RNA helicase, DDX1, interacting with poly(A) RNA and heterogeneous nuclear ribonucleoprotein K. J. Biol. Chem.. 277, 40403–40409. [DOI] [PubMed] [Google Scholar]
- Cheng, D. , Cote, J. , Shaaban, S. , Bedford, M.T. , 2007. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell. 25, 71–83. [DOI] [PubMed] [Google Scholar]
- Choi, H.S. , Hwang, C.K. , Song, K.Y. , Law, P.Y. , Wei, L.N. , Loh, H.H. , 2009. Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem. Biophys. Res. Commun.. 380, 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez, V. , Mann, M. , Tekmal, S. , Suzuki, T. , Miyata, N. , Rodriguez-Aguayo, C. , Lopez-Berestein, G. , Sood, A.K. , Vadlamudi, R.K. , 2012. Targeting the PELP1-KDM1 axis as a potential therapeutic strategy for breast cancer. Breast Cancer Res.: BCR. 14, R108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote, J. , Boisvert, F.M. , Boulanger, M.C. , Bedford, M.T. , Richard, S. , 2003. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol. Biol. Cell. 14, 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan, D.H. , Harrison, M.J. , Eriksson, N.A. , Bailey, P. , Pearen, M.A. , Fuller, P.J. , Funder, J.W. , Simpson, E.R. , Leedman, P.J. , Tilley, W.D. , Brown, M.A. , Clarke, C.L. , Muscat, G.E. , 2012. Protein arginine methyltransferase 6-dependent gene expression and splicing: association with breast cancer outcomes. Endocr. Relat. Cancer. 19, 509–526. [DOI] [PubMed] [Google Scholar]
- Finkbeiner, E. , Haindl, M. , Muller, S. , 2011. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J.. 30, 1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen, W.J. , Massenet, S. , Paushkin, S. , Wyce, A. , Dreyfuss, G. , 2001. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell. 7, 1111–1117. [DOI] [PubMed] [Google Scholar]
- Fu, X.D. , 1993. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 365, 82–85. [DOI] [PubMed] [Google Scholar]
- Girard, B.J. , Daniel, A.R. , Lange, C.A. , Ostrander, J.H. , 2013. PELP1: a review of PELP1 interactions, signaling, and biology. Mol. Cell Endocrinol.. 382, 642–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonugunta, V.K. , Nair, B.C. , Rajhans, R. , Sareddy, G.R. , Nair, S.S. , Vadlamudi, R.K. , 2011. Regulation of rDNA transcription by proto-oncogene PELP1. PloS One. 6, e21095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururaj, A.E. , Peng, S. , Vadlamudi, R.K. , Kumar, R. , 2007. Estrogen induces expression of BCAS3, a novel estrogen receptor-alpha coactivator, through proline-, glutamic acid-, and leucine-rich protein-1 (PELP1). Mol. Endocrinol. (Baltimore, Md.). 21, 1847–1860. [DOI] [PubMed] [Google Scholar]
- Habashy, H.O. , Powe, D.G. , Rakha, E.A. , Ball, G. , Macmillan, R.D. , Green, A.R. , Ellis, I.O. , 2010. The prognostic significance of PELP1 expression in invasive breast cancer with emphasis on the ER-positive luminal-like subtype. Breast Cancer Res. Treat.. 120, 603–612. [DOI] [PubMed] [Google Scholar]
- Harrison, M.J. , Tang, Y.H. , Dowhan, D.H. , 2010. Protein arginine methyltransferase 6 regulates multiple aspects of gene expression. Nucl. Acids Res.. 38, 2201–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, F. , Bossert, M. , Schwander, A. , Akgun, E. , Fackelmayer, F.O. , 2004. Arginine methylation of scaffold attachment factor A by heterogeneous nuclear ribonucleoprotein particle-associated PRMT1. J. Biol. Chem.. 279, 48774–48779. [DOI] [PubMed] [Google Scholar]
- Ip, J.Y. , Schmidt, D. , Pan, Q. , Ramani, A.K. , Fraser, A.G. , Odom, D.T. , Blencowe, B.J. , 2011. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res.. 21, 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig, H. , Ponta, H. , Herrlich, P. , 1998. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J.. 17, 2904–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt, A.R. , Schor, I.E. , Allo, M. , Dujardin, G. , Petrillo, E. , Munoz, M.J. , 2013. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol.. 14, 153–165. [DOI] [PubMed] [Google Scholar]
- Kramer, A. , 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem.. 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Zhang, H. , Holm, C. , Vadlamudi, R.K. , Landberg, G. , Rayala, S.K. , 2009. Extranuclear coactivator signaling confers insensitivity to tamoxifen. Clin. Cancer Res. An Off. J. Am. Assoc. Cancer Res.. 15, 4123–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limm, K. , Ott, C. , Wallner, S. , Mueller, D.W. , Oefner, P. , Hellerbrand, C. , Bosserhoff, A.K. , 2013. Deregulation of protein methylation in melanoma. Eur. J. Cancer (Oxford, Engl.: 1990). 49, 1305–1313. [DOI] [PubMed] [Google Scholar]
- Lin, S. , Coutinho-Mansfield, G. , Wang, D. , Pandit, S. , Fu, X.D. , 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol.. 15, 819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, M. , Cortez, V. , Vadlamudi, R. , 2004. PELP1 oncogenic functions involve CARM1 regulation. Carcinogenesis. 34, 1468–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, T.B. , Webb, K.J. , Edberg, D.D. , Reeves, R. , Clarke, S. , 2005. Protein arginine methyltransferase 6 specifically methylates the nonhistone chromatin protein HMGA1a. Biochem. Biophys. Res. Commun.. 336, 831–835. [DOI] [PubMed] [Google Scholar]
- Nair, S.S. , Mishra, S.K. , Yang, Z. , Balasenthil, S. , Kumar, R. , Vadlamudi, R.K. , 2004. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res.. 64, 6416–6423. [DOI] [PubMed] [Google Scholar]
- Nair, B.C. , Nair, S.S. , Chakravarty, D. , Challa, R. , Manavathi, B. , Yew, P.R. , Kumar, R. , Tekmal, R.R. , Vadlamudi, R.K. , 2010. Cyclin-dependent kinase-mediated phosphorylation plays a critical role in the oncogenic functions of PELP1. Cancer Res.. 70, 7166–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, S.S. , Nair, B.C. , Cortez, V. , Chakravarty, D. , Metzger, E. , Schule, R. , Brann, D.W. , Tekmal, R.R. , Vadlamudi, R.K. , 2010. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep.. 11, 438–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, B.C. , Vallabhaneni, S. , Tekmal, R.R. , Vadlamudi, R.K. , 2011. Roscovitine confers tumor suppressive effect on therapy-resistant breast tumor cells. Breast Cancer Res.: BCR. 13, R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajhans, R. , Nair, S. , Holden, A.H. , Kumar, R. , Tekmal, R.R. , Vadlamudi, R.K. , 2007. Oncogenic potential of the nuclear receptor coregulator proline-, glutamic acid-, leucine-rich protein 1/modulator of the nongenomic actions of the estrogen receptor. Cancer Res.. 67, 5505–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, W.A. , Schurter, B.T. , Wong-Staal, F. , David, M. , 2004. Arginine methylation of RNA helicase a determines its subcellular localization. J. Biol. Chem.. 279, 22795–22798. [DOI] [PubMed] [Google Scholar]
- Tang, S. , Han, H. , Bajic, V.B. , 2004. ERGDB: estrogen responsive genes database. Nucl. Acids Res.. 32, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer, E. , Mitchell, R. , Hartman, T. , Silva, M. , Gospodarowicz, D. , Fiddes, J.C. , Abraham, J.A. , 1991. The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J. Biol. Chem.. 266, 11947–11954. [PubMed] [Google Scholar]
- Trapnell, C. , Pachter, L. , Salzberg, S.L. , 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinform. (Oxford, England). 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi, R.K. , Kumar, R. , 2007. Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl. Recep. Signal.. 5, e004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi, R.K. , Wang, R.A. , Mazumdar, A. , Kim, Y. , Shin, J. , Sahin, A. , Kumar, R. , 2001. Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor alpha. J. Biol. Chem.. 276, 38272–38279. [DOI] [PubMed] [Google Scholar]
- Vallabhaneni, S. , Nair, B.C. , Cortez, V. , Challa, R. , Chakravarty, D. , Tekmal, R.R. , Vadlamudi, R.K. , 2011. Significance of ER-Src axis in hormonal therapy resistance. Breast Cancer Res. Treat.. 130, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables, J.P. , Klinck, R. , Koh, C. , Gervais-Bird, J. , Bramard, A. , Inkel, L. , Durand, M. , Couture, S. , Froehlich, U. , Lapointe, E. , Lucier, J.F. , Thibault, P. , Rancourt, C. , Tremblay, K. , Prinos, P. , Chabot, B. , Elela, S.A. , 2009. Cancer-associated regulation of alternative splicing. Nature Struct. Mol. Biol.. 16, 670–676. [DOI] [PubMed] [Google Scholar]
- Wellmann, S. , Taube, T. , Paal, K. , Graf, V.E.H. , Geilen, W. , Seifert, G. , Eckert, C. , Henze, G. , Seeger, K. , 2001. Specific reverse transcription-PCR quantification of vascular endothelial growth factor (VEGF) splice variants by LightCycler technology. Clin. Chem.. 47, 654–660. [PubMed] [Google Scholar]
- Wu, J.Y. , Maniatis, T. , 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 75, 1061–1070. [DOI] [PubMed] [Google Scholar]
- Xu, Q. , Lee, C. , 2003. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucl. Acid Res.. 31, 5635–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Ravindranathan, P. , Ramanan, M. , Kapur, P. , Hammes, S.R. , Hsieh, J.T. , Raj, G.V. , 2012. Central role for PELP1 in nonandrogenic activation of the androgen receptor in prostate cancer. Mol. Endocrinol. (Baltimore, Md.). 26, 550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu, M. , Toyokawa, G. , Hayami, S. , Unoki, M. , Tsunoda, T. , Field, H.I. , Kelly, J.D. , Neal, D.E. , Maehara, Y. , Ponder, B.A. , Nakamura, Y. , Hamamoto, R. , 2011. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer (Journal international du cancer). 128, 562–573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Figure S1: A) Differential gene expression from RNA‐sequencing was validated with MCF7 and MCF7PELP1KD cells treated with estradiol for 5 h using qRTPCR in triplicate. B) Differential gene expression from RNA‐sequencing was validated with ZR75 cells transfected with siControl or siPELP1 and either control vector or siRNA‐resistant‐PELP1 plasmid. After transfection, cells were starved of estrogen for 72 h and treated with estradiol 10−8 M for 5 h. Gene expression is shown as siControl/siPELP1 and (siControl + PELP1)/(siPELP + PELP1) normalized to Actin. C) Co‐immunoprecipitation of nuclear lysates of ZR75 cells was performed with PELP1, SC35, and IgG antibodies. Input of PELP1 and SC35 is shown. D) An ERE‐luciferase gene reporter assay was performed with MCF7 and MCF7PELP1 cells transfected with siControl or siPRMT6 and ERE‐luciferase in triplicate. Cells were starved of estrogen in 5% DCC media for 48 h and treated with estradiol 10−8 M for 12 h. Cells were lysed with passive lysis buffer and relative luciferase activity was measured by a luminometer. E) Proliferation assay was performed with MCF7 and MCF7PELP1 cells transfected with either control siRNA or PRMT6 siRNA. F) MCF7 and MCF7PELP1 cells were transfected with either control siRNA or PRMT6 siRNA and plated in triplicate in 6‐well plates for a colony formation assay. Colonies were stained by crystal violet and counted on day 14. Figure S2‐6: Molecular networks from IPA based on gene/molecule connectivity with other genes/molecules. Genes that are downregulated by PELP1 knockdown are shown in green and upregulated by PELP1 knockdown are in red. Figure S7: Schematic representation of PELP1 signaling from RNA‐sequencing analysis. Supplementary Table 1: RNA‐sequencing genes regulated by PELP1 with significance of p < 0.05 and log2(Fold Change) > 2 as analyzed by DESeq. Supplementary Table 2: Genes found to be significantly alternatively spliced by RNA‐sequencing using Cufflink analysis.


