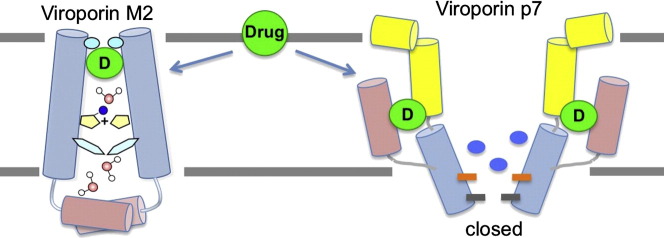
Abstract
Many viral genomes encode small, integral membrane proteins that form homo-oligomeric channels in membrane, and they transport protons, cations, and other molecules across the membrane barrier to aid various steps of viral entry and maturation. These viral proteins, collectively named viroporins, are crucial for viral pathogenicity. In the past five years, structures obtained by nuclear magnetic resonance (NMR), X-ray crystallography, and electron microscopy (EM) showed that viroporins often adopt minimalist architectures to achieve their functions. A number of small molecules have been identified to interfere with their channel activities and thereby inhibit viral infection, making viroporins potential drug targets for therapeutic intervention. The known architectures and inhibition mechanisms of viroporins differ significantly from each other, but some common principles are shared between them. This review article summarizes the recent developments in the structural investigation of viroporins and their inhibition by antiviral compounds. This article is part of a Special Issue entitled: Viral Membrane Proteins—Channels for Cellular Networking.
Keywords: Viroporin, Structure, M2, p7, Influenza, Hepatitis C
Graphical abstract
Highlights
-
•
Provide an overview of the known architectures of viroporins
-
•
Review of the minimalist structural solutions adopted by influenza H + channels and HCV p7 channel
-
•
Discuss how structures explain the molecular mechanism of ion selectivity and transport
-
•
Discuss how adamantane derivatives block M2 and p7 viroporins and mechanism of drug resistance
1. Introduction
The past few decades of virus research have witnessed the emergence and growth of a special family of membrane-embedded channel proteins encoded by the genome of viruses [1], [2], [3]. This family of proteins, collectively termed viroporins, generally has very low sequence homology with the ion channels from prokaryotes and eukaryotes, though certain transmembrane (TM) segments can be similar [4]. Viroporin proteins share some common features; they are small (50–120 amino acids), hydrophobic, and often dynamic, and due to the small size, these membrane proteins must oligomerize in order to form channels. Moreover, these viral channels appear to adopt minimalist but functional structural solutions that are drastically different from the known channel structures from other organisms, making structural and functional exploration of viroporins both interesting and rewarding.
Early investigations of viroporins have focused on a few systems including 2B of poliovirus, 6K of togavirus, and M2 from influenza A virus (IAV). The 2B is a well-known viroporin implicated in membrane permeabilization to ions and small molecules during later stages of viral infection [5], [6], [7]. The 6K has been shown to form cation-selective channels when inserted into lipid bilayers [8], but the role of 6K in the viral life cycle is still poorly characterized [9], [10], [11]. The M2 proton channel has by far received the biggest spotlight because 1) it is the target of the once successful anti-flu drug (amantadine or rimantadine) and 2) it is a pH-dependent proton conducting channel (our knowledge of proton channel has for a long time been very limited) [12], [13], [14]. Since the discovery of above proteins, the viroporin family has grown substantially (Table 1 ). The human immunodeficiency virus type 1 (HIV-1) Vpu and the hepatitis C virus (HCV) p7 have been shown to conduct cations [15], [16], [17], [18], [19]. The influenza B viral genome encodes the BM2 protein that is a functional homolog of M2 but is not sensitive to the adamantane family of drugs [20]. There is the viral K+ channel from Paramecium bursaria chlorella virus 1 (PBCV-1) that contains the selectivity filter of the prokaryotic and eukaryotic K+ channels [21] and was proposed to adopt similar structure as KcsA [22]. Apart from the ion channels, poliovirus developed a VP4 channel that permeates single-stranded RNA across the membrane [23], further expanding the list of substrates that viroporins transport.
Table 1.
List of identified viroporin proteins.a
| Name of virus | Name of viroporin | # of amino acid | Oligomeric state | Ion or substrate | References |
|---|---|---|---|---|---|
| IAV | M2 | 97 | Tetramer | H+ | Ref. [37], [38], [74] |
| IBV | BM2 | 115 | Tetramer | H+ | Ref. [39] |
| HCV | p7 | 63 | Hexamer | Cation | Ref. [18], [62] |
| HIV-1 | Vpu | 81 | Pentamer | Cation | Ref. [70], [71], [90] |
| SARS-CoV | 3a | 274 | Tetramer | K+ | Ref. [72] |
| E | 76 | Pentamer | Na+, K+ | Ref. [91] | |
| Alphavirus | 6 K | 61 | N/A b | Na+, K+, Ca2 + | Ref. [8] |
| Poliovirus | VP4 | 68 | N/A b | RNA | Ref. [23] |
| 2B | 97 | N/A b | Ca2 + | Ref. [7], [92] | |
| HPV | E5 | 83 | N/A b | N/A | Ref. [93], [94] |
| PBCV-1 | Kcv | 94 | Tetramer | K+ | Ref. [21], [95] |
The viruses listed are influenza A virus (IAV), influenza B virus (IBV), hepatitis C virus (HCV), human immunodeficiency virus type 1 (HIV-1), severe acute respiratory syndromes-associated coronavirus (SARS-CoV), alphavirus, poliovirus, human papillomavirus (HPV), and Paramecium bursaria chlorella virus 1 (PBCV-1).
Pore formation indicated but a defined oligomeric state either does not exist or is not characterized.
The exact functions of viroporins in many viruses are not yet known, though their roles have been implicated in entry, viral assembly and viral release. In particular, the roles of the M2 proton channel during influenza replication have more or less been accepted. One role of the M2 channel is to equilibrate pH across the viral membrane during cell entry, which acidifies the virion interior after endocytosis and facilitates RNA release [24], [25]. Another role of M2 is to equilibrate pH across the trans-Golgi membrane of infected cells during viral maturation, which preserves the pre-fusion form of hemagglutinin through de-acidification of the Golgi lumen [26]. The roles of other cation-selective viroporins are however not so clear. In the case of HCV, the p7-mediated channel activity has been reported to facilitate virus release [27], [28]. In fact, one of the general roles proposed for viroporin is to depolarize either the endoplasmic reticulum (ER) or plasma membrane to facilitate membrane curvature during budding [3], [29]. Another general role for viroporin is to simply cause cation leakage and cellular stress, which would induce programmed cell death [30]. In addition to ion permeation, several viroporins are known to participate in viral assembly through interaction with other proteins. For example, the IAV M2 and IBV BM2 proteins both have significant cytoplasmic domains that are involved in recruiting matrix proteins M1 during virus assembly [31], [32], [33]. The HCV p7 protein has also been reported to interact with other non-structural proteins such as NS2, and this interaction appears to be crucial for the production of infectious HCV particles [34], [35].
While the precise functions of ion conduction activity of many viroporins remain to be discovered, advances in structural characterization of influenza M2, BM2 and HCV p7 proteins have provided compelling evidences that viroporins are not simple porins but can adopt interesting architectures for regulated transport activities such as pH dependent proton transport and selective cation conductance. Furthermore, structural characterizations of interactions between viroporins and small molecule inhibitors have revealed conceptually very different mechanisms by which these channel proteins can be inhibited, and thus alluded to new opportunities of using viroporins as antiviral targets. The current review describes the recently characterized viroporin architectures, their intriguing and minimalist modes of assembly, and how the new structural information explains the observed functional properties of the viroporins. This review also describes examples of viroporins being inhibited by the known antiviral drugs and explains the mechanism of drug inhibition and drug resistance.
2. The architectures of viroporins
Structural studies of viroporins have not been easy because these small membrane proteins are typically dynamic and very sensitive to the hydrophobic surroundings. In the last five years, multiple biophysical techniques including solution NMR, X-ray crystallography, solid-state NMR, and EM have been used to gradually fill the knowledge gap. For example, there are now structural information of the channel domain of M2 in crystal and solution states, in lipid bilayer, under different pHs, and bound to different small molecules. These complementary structural data allow elucidation of the functional mechanism from different view angles. In addition to the influenza proton channels, the structure of the full-length p7 channel of HCV has recently been determined. In both cases, structure determination revealed unexpected structural features that are important for understanding the functional mechanism.
2.1. The influenza M2 channels: structural solutions for proton conduction
The M2 protein of IAV and BM2 protein of IBV are 97- and 109-residue single-pass membrane proteins, respectively, that form homotetramers in membrane [36], [37], [38], [39], [40]. The two proteins share almost no sequence homology except for the HxxxW sequence motif in the TM domain that is essential for channel activity. Their domain arrangements are also different. The TM region encompasses residues 24–46 in M2 and residues 4–33 in BM2. Hence the unstructured N-terminal segment preceding the TM domain is much longer in M2 and has been sought after as a possible target for vaccine development [41], [42], [43], [44], [45]. In addition to the membrane-embedded channel domain, M2 and BM2 both have relatively large C-terminal cytoplasmic regions. These regions have been suggested to play a role during viral assembly, by recruiting the matrix proteins to the cell surface during virus budding and/or by contributing to the coating of matrix proteins to the viral envelope [31], [32], [46], [47].
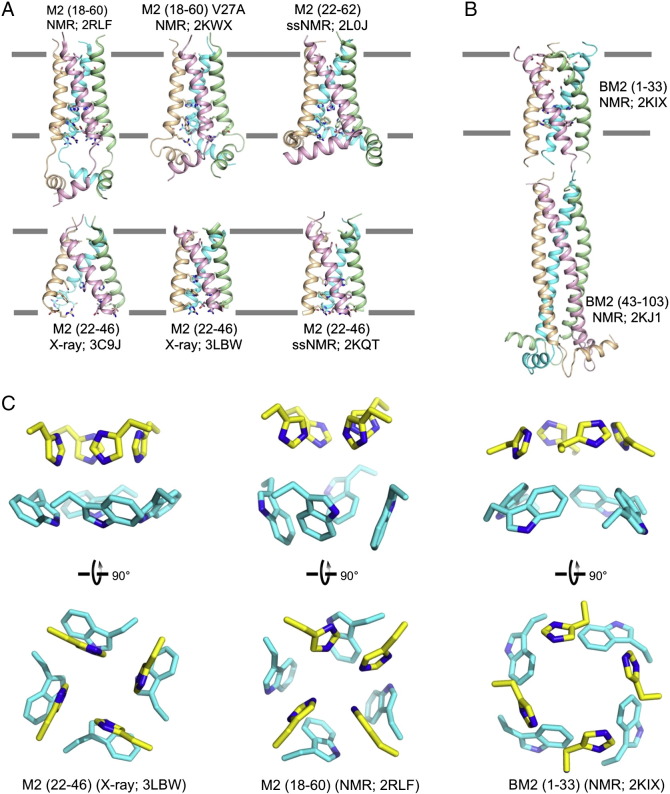
The first detailed structures of the M2 channel were determined concurrently by solution NMR spectroscopy and X-ray crystallography in 2008 [48], [49]. Subsequent X-ray and solution NMR studies determined the structures of the channel domain under different conditions [50] and with different drug resistance mutations [51], [52]. Moreover, structural characterization by solid-state NMR spectroscopy also generated working structural models for the M2 channel in the lipid bilayer environment [53], [54]. The above structures solved under different conditions show substantially different conformations (Fig. 1A), suggesting that the assembly of the M2 channel is quite dynamic and is sensitive to the reconstitution environment. This structural plasticity could have been one of the major factors that caused technical difficulties for structure determination.
Fig. 1.
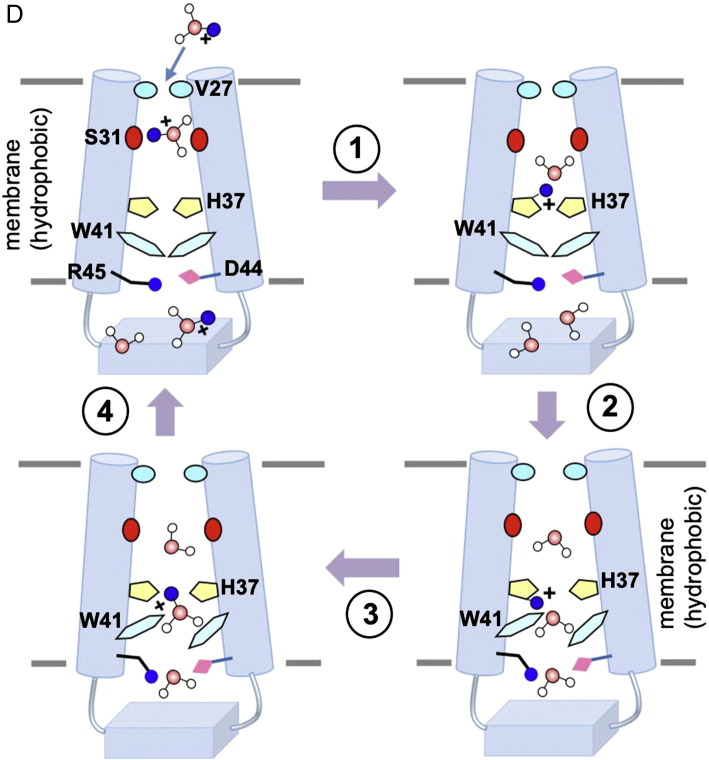
Structures of the influenza proton channels and mechanism of proton conduction. (A) The many structures of the influenza M2 channel. The PDB codes 2RLF and 2KWX2RLF2KWX represent the solution NMR structures of the wildtype and the V27A mutant determined using residues 18–60. The 3C9J and 3LBW3C9J3LBW are crystal structures of the TM domain (residues 22–46) determined at pH 7.3 and pH 6.5, respectively. The structures 2L0J and 2KQT2L0J2KQT were obtained using solid-state NMR using protein constructs that encompass residues 22–62 and residues 22-46, respectively. (B) Solution NMR structures of the BM2 protein. The PDB codes 2KIX and 2KJ12KIX2KJ1 represent the structures of the TM domain (residues 1–33) and the cytoplasmic domain (residues 43–103), respectively. (C) Isolated view of the pore-lining histidine and tryptophan sidechains in M2 and BM2 channels. Images are from the high resolution crystal structure (3LBW) and NMR structure (2RLF) of M2 and NMR structure of BM2 (2KIX). (D) Cartoon of the mechanism of proton conduction. (1) Channel breathing due to thermal energy can transiently enlarge the N-terminal opening, allowing protons to be relayed cross the Val27 barrier. (2) The protons bind to His37 imidazole, and protonation of His37 triggers opening of the Trp41 gate. (3) C-terminal water molecules accept protons from protonated His37. (4) Polar residues Asp44 and Arg45 facilitate proton exit.
Despite the conformational variability, the experimental structures converged to a common mode of channel assembly: a left-handed four-helix bundle forms the channel pore, and tetramerization of the four TM helices is further stabilized by intermolecular contacts between C-terminal amphipathic helices flanking the TM domain [48], [54]. This mode of TM helix assembly places the “H” and “W” of the HxxxW sequence motif inside the channel. Four imidazole rings of His37, which are pH trigger devices and are essential in transporting protons, are packed closely inside the pore. Moreover, packing of the Trp41 indoles creates a channel gate, which occludes the C-terminal end of the pore.
The detailed structure of the BM2 channel domain was solved using solution NMR methods [33]. The BM2 channel is also assembled through left-handed helical packing but unlike M2, the packing shows strong coiled-coil characteristics with regular heptad repeats [12] (Fig. 1B). In the BM2 coiled-coil tetramer, positions a and d of the heptad repeats are occupied mostly by serines and His19 to form the hydrophilic pore. Positions g and e are occupied by leucines and phenylalanines, respectively, to allow for hydrophobic packing between the peripheral leucines and phenylalanines. This arrangement for coiled-coil assembly in membrane is the opposite to that of water-soluble coiled-coil tetramer, in which positions a and d are typically hydrophobic residues and positions g and e are polar residues [55]. The inverse coiled-coil assembly in membrane places the histidine (His19) and tryptophan (Trp23) of the HxxxW motif inside the pore. The cytoplasmic domain of BM2 is also a left-handed coiled-coil tetramer, but it is water-soluble and has strong bipolar distribution of surface charged residues. This domain specifically interacts with the M1 matrix protein [33].
The structural arrangement of the histidine and tryptophan inside the pore in M2 and BM2 implies the roles of imidazoles as proton selection devices and indoles as channel gates (Fig. 1C). Indeed, functional assays [56] and NMR measurements [57] suggested that proton conduction across the M2 channel involves cycles of histidine protonation and deprotonation, and that the histidines serve as proton shuttling devices. A concern of histidine protonation is that when multiple histidines are protonated, the charge repulsion between them would be strong enough to destabilize the tetramer. This issue can however be resolved by the fact that not all histidines can be protonated at the same time because protonation of one histidine would increase the barrier for the protonation of another histidine [58]. Indeed, multiple pKa values have been observed in proton conduction assays of M2 [56]. Therefore, our current understanding of the proton conduction mechanism is that the His-Trp structural elements constitute the minimally required unit for pH-dependent proton transport (Fig. 1D). It was proposed in Ref. [58] that in the closed state two pairs of histidines in the tetramer each share one proton, which explains the high pKa ~ 8.2. Lowering pH results in the protonation of the third histidine from the N-terminal side that, in turn, results in disruption of the two histidine dimers and proton conduction. The remaining question to be addressed in the future is how does the third protonation result in conformational change of the tryptophan that would allow relaying the proton to the C-terminal side of the tryptophan gate [56]. Finally, the significant structural differences between the M2 and BM2 channels show strong ability of influenza virus to evolve different solutions to achieve the same goal.
2.2. The funnel architecture of the p7 channel
The viroporin protein p7 encoded by the HCV genome is a 63-residue protein that oligomerizes in membrane to form cation-selective channels [18], [19], with higher selectivity for Ca2 + than K+/Na+ [59], [60]. The channel activity of p7 is important for the assembly and release of infectious viruses, although the molecular mechanism of this function remains elusive [27], [28]. As in the case of M2, structural characterization of p7 was confronted with challenges of preparing concentrated and homogeneous sample of p7 oligomers. Earlier NMR studies found that the p7 monomer has three helical segments: two in the N-terminal half of the sequence and one near the C-terminus [60], [61]. These NMR studies were performed under conditions that are believed to support the monomeric state of p7. Although the monomeric state is unlikely to conduct ions, it could be involved in interacting with the NS2 protein during virus assembly [34], [35]. The first structural investigation of the assembled p7 oligomer was conducted using single-particle EM, which obtained a 16 Å resolution electron density map of the protein complex [62]. The map shows that the p7 from HCV genotype 2a (JFH-1 strain) forms a 42 kDa hexamer and adopts a flower-like shape that does not resemble any of the known ion channel structures in the database. While the above structural investigations validated p7 as a stable hexameric complex, the structural details required for understanding ion conduction and drug inhibition remained unknown. Recently, an effective solution NMR system of the p7 hexamer was established using p7 from genotype 5a (EUH1480 strain), which is one of the less hydrophobic sequences among the p7 variants. Using the NMR system, the detailed structure of the p7 hexamer was eventually determined [63].
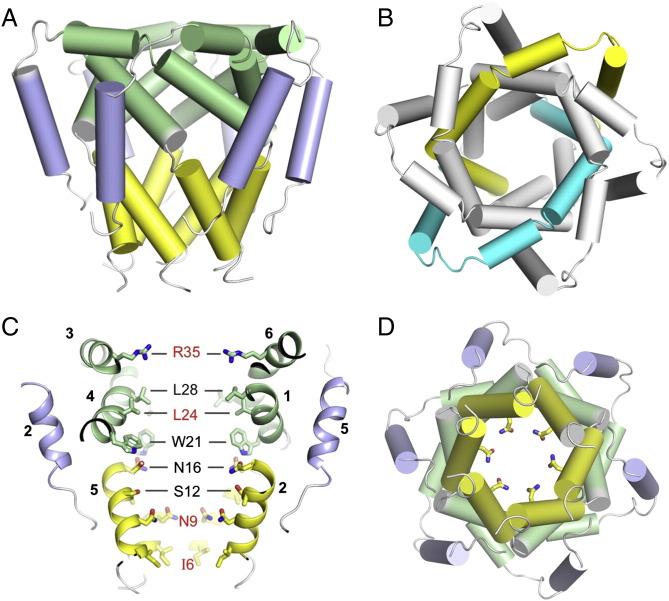
The NMR structure of the p7 channel shows that HCV has developed a solution to conduct cations that is substantially different from any of the known K+ or Ca2 + channels. The channel has a funnel-like architecture with six minimalist chains, each containing three helical segments, H1, H2 and H3. The H1 and H2 helices of the monomers form the narrow and wide regions of the funnel-shaped cavity, respectively, and the H3 helices wrap the channel peripheral by interacting with H2 of the i + 2 and H1 of the i + 3 monomers (Fig. 2A). There are no intramonomer contacts between the three helical segments in the context of the hexamer. The high order of oligomerization and the maximized interactions between the monomers allow a small hydrophobic peptide (63-residues) to form a 42 kDa channel complex.
Fig. 2.
The funnel architecture and pore elements of the p7 channel. (A) NMR structure of the p7 hexamer. Left panel: cartoon (cylinder) representation illustrating the funnel architecture of the channel. Right panel: global arrangement of H1, H2 and H3 helical segments in the assembled hexamer, showing that the i and i + 3 monomers form a symmetric pair in the hexamer. (B) Pore-lining elements of the p7 channel. Left panel: cutaway view of the channel showing the pore-lining residues, with residues in red being strongly conserved. The numbers next to the helical segments represent the monomers to which the helices belong. Right panel: the view of the N-terminal opening of the channel showing the carboxamide ring formed with Asn9 sidechains.
Although having a minimalist architecture compared to other Ca2 + or Mg2 + channels, the p7 hexamer appears to possess essential features of ion channels: 1) pore elements that support selective ion dehydration; 2) a gate or constriction that prevents non-specific permeation, but can open in response to regulating factor such as pH, voltage, ligand, or the ion of selection itself [64], [65], [66]. The channel interior of p7 has a number of strongly conserved residues that are likely candidates to serve the above functions. The most obvious one is Asn9, which forms a ring of carboxamide near the narrow end of the channel (Fig. 2B). Residue 9 is asparagine in all strains except being substituted with histidine in genotype 2 viruses. Formation of a ring of carboxylates or carboxamides has been a recurring theme in prokaryotic and eukaryotic channels that have selectivity for divalent cations. For examples, central to the CorA Mg2 + channel is the pentameric ring of asparagines [67]. Moreover, the recent structure of the calcium release-activated calcium (CRAC) channel Orai shows a hexameric ring of aspartic acids [68]. These channels all have strong selectivity for divalent cations, although they can also conduct monovalent cations such as Na+ and K+.
Proximal to the Asn9 ring of p7 is the hydrophobic ring formed by Ile6, which defines the narrowest point of the channel. Residue 6 is isoleucine or valine throughout HCV genotypes and subtypes. Therefore, a narrow hydrophobic ring at this position likely serves as a hydrophobic constriction which prevents water from freely passing through. A mechanism of cation conduction can be envisaged from the structure. The role of the Asn9 ring is to provide ion selectivity by recruiting and dehydrating cations near the funnel exit, and the unhydrated cations can pass the hydrophobic ring. In this regard, the Asn9 has the feature of being a broad selectivity filter.
In addition to what appears to be the cation selectivity ring near the narrow, N-terminal exit of the channel, the wider, C-terminal entrance of the channel is decorated with a conserved ring of arginines or lysines. Placement of a positively charged ring at the entrance was incomprehensible to us initially because it may repel cations. But an earlier study reported that a designed TM barrel with internal arginine–histidine dyads forms efficient cation selective channels [69] because the immobile arginines can recruit mobile anions, which in turn facilitate cations to diffuse through the pore. It is interesting to note that a highly basic region containing Arg155, Lys159 and Lys163 in the pore was also found in the CRAC Orai structure [68]. One possible role of these basic residues in cation selective channels is binding and obstructing anions while allowing cations to diffuse into the pore. This mechanism would be consistent with the observation that replacing Arg35 with negatively charged aspartic acid largely abrogated conductance [63].
Finally, an unresolved issue with the NMR structure is that the size of the Asn9 ring (inner diameter ~ 7 Å) is significantly larger than those in, e.g., the CRAC and CorA crystal structures. At this size, the carboxamide ring clearly cannot provide tight coordination of Ca2 +, and this raises a number of questions that need to be addressed in the future. Does the NMR structure, solved in the absence of Ca2 + and inhibitors, represent the open or closed state of the channel (if the two states exist)? Is Ca2 + selectivity of p7 as strong as the highly regulated Ca2 + channel such as CRAC? Or was p7 developed as a general, unspecific cation channel for the purpose of dissipating membrane potential? Addressing these questions would require further structural investigation of p7 under different conditions and more extensive characterization of the channel conductance properties by single-channel measurements. From structural perspective, the funnel-like architecture is formed with multiple helical segments connected by hinges and short loops and we believe that this flexibility can afford the dynamic opening and closing of the tip of the channel.
2.3. Other viroporins
In addition to the above viroporin structures, structural characterization has been conducted for several other viroporins. Combined use of solid-state NMR PISEMA experiment and solution NMR first identified the membrane-associated region of the Vpu protein from HIV-1 including a TM helix and a juxtamembrane amphipathic helix parallel to the membrane [16]. The same research group obtained a more detailed structure of the TM helix and its tilt angle relative to the lipid bilayer, and in combination with computational modeling, provided a homopentameric model of the Vpu channel [70]. A somewhat different model of the pentameric Vpu was recently developed using inter-monomer distance restraints from magic angle spinning (MAS) NMR experiments, suggesting that Vpu, like M2 and p7, also adopts substantial structural variability [71]. In addition to Vpu, the oligomeric state analyses have been performed for the Kcv protein from PBCV-1 [22], and the 3a and E proteins from SARS-CoV [72], [73]. These studies suggested that Kcv and 3a form homotetramers while the E protein forms a stable pentamer. We believe that these viroporins may adopt interesting architectures as well to transport ions.
3. Drug inhibition of viroporins
One of the motivations for investigating viroporins is their anticipated structural difference with human channels and thus the potential as antiviral drug targets. A successful example of antiviral drug that targets viroporin is amantadine (Symadine) or rimantadine (Flumadine), which inhibits proton conduction of the M2 channel [74], [75] and was the first licensed drug for treating influenza infections [76]. Rimantadine has also been shown to inhibit the HCV p7 channel [18], [77]. Since the first successful drug targeting a viroporin, researches on viral membrane proteins have identified several more viroporin inhibitors with varying efficacies, including hexamethylene amiloride that inhibits HIV-1 Vpu [17], HCV p7 [59], [60] and E from SARS-CoV [78], long-alkyl-chain iminosugar derivatives that inhibit HCV p7 [19], as well as a new compound, BIT225, that inhibits HIV-1 Vpu [79] and HCV p7 [80].
3.1. Amantadine inhibition of M2 channel
The mechanism of how amantadine/rimantadine inhibit M2 channel has been elusive until very recently. Previous confusion came mainly from the multiple binding sites that have been observed experimentally. In a crystallographic study of the M2 TM domain (residues 24–46) in the presence of amantadine, the drug density was found inside the channel near residue Ser31, but at structural resolution of 3.5 Å, it was difficult to confirm the position of amantadine binding [49]. At the same time, however, a solution NMR study of a longer version of M2 (residues 18–60) showed that rimantadine binds to an external, lipid-facing pocket around residue Asp44 between adjacent TM helices [48]. The mechanistic implications of the two binding sites are obviously different. One is the drug directly blocking the channel passage, and the other is an allosteric mechanism by which drug binding to the external site favors the closed state of the channel. Subsequent solid-state NMR measurements of the TM domain in lipid bilayer showed that rimantadine binds to both sites, with higher affinity for the pore site [53]. Functional studies using an AM2–BM2 chimera protein provided the strongest evidence that the pore binding site is the primary site of drug inhibition [81], [82]. In this study, the authors used a chimera of M2 variants from influenza A and B viruses that contains only the pore-binding site which showed that the chimera channel is still sensitive to amantadine or rimantadine inhibition, indicating that the pore-binding site is the primary site of drug inhibition. Later, the structure of the TM domain of the chimera in complex with rimantadine was determined by NMR, providing a precise view of the drug binding inside the channel pore [83].
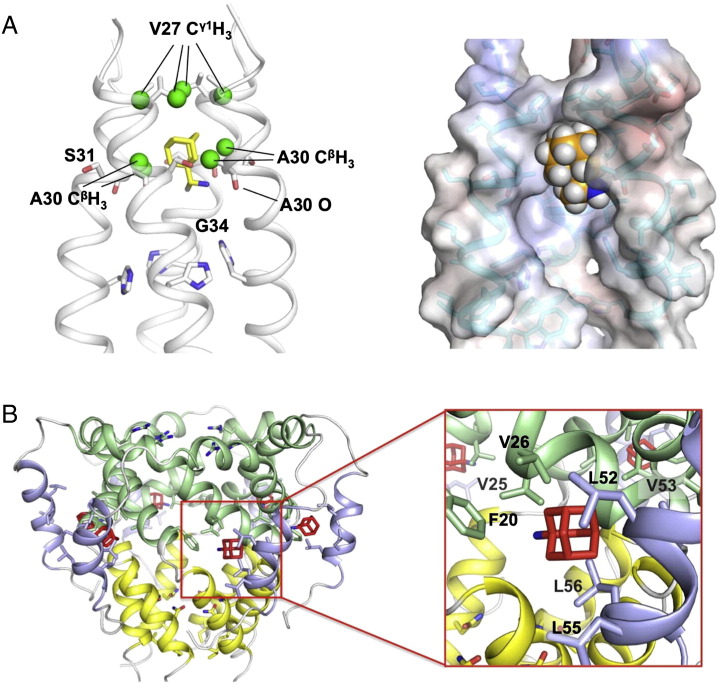
The structure shows that eight methyl groups of the M2 tetramer (two from each subunit: Val27 Cγ1H3 and Ala30 CβH3) together form a deep internal pocket that completely wraps the adamantane cage of the drug (Fig. 3A). The nitrogen of the rimantadine amino group is on average 2.8 Å from the backbone carbonyl oxygen of Ala30 of one of the four subunits, probably forming a hydrogen bond. The terminal methyl group of the rimantadine is in the middle of the pore, facing the open space in the channel around the Gly34 position. The structure also shows that rimantadine binding is slightly tilted: its vertical axis is on average ~ 20° from the C4 symmetry axis of the channel. The tilt angle is consistent with the amantadine tilt in the M2 channel observed with solid-state NMR spectroscopy [53].
Fig. 3.
The amantadine and rimantadine binding sites in the M2 and p7 channels. (A) The precise NMR structure of rimantadine bound to the internal pocket of the AM2–BM2 chimeric channel determined in DHPC micelles and at pH 7.5 [83]. Left panel: detailed illustration of the methyl groups (in green) that interact with the adamantane cage. Right panel: surface representation for showing the hydrophobic pocket that fits the drug snuggly. One of the four subunits is omitted for drug visibility. (B) The amantadine or rimantadine binding site of the p7 channel determined by NMR in DPC micelles and at pH 6.5 [63]. Left panel: the drug binds to six equivalent hydrophobic pockets of the p7 channel. Right panel: a close view of amantadine docked into the binding pocket as determined using NMR NOE restraints.
Although amantadine or rimantadine has been widely used as anti-influenza drug for decades, drug resistant variants are widespread, with resistance now > 90% [84]. The reported mutations that confer resistance include V27A, A30T, S31N, and G34E, among which S31N and V27A account for the vast majority of resistant strains [85], [86]. It is obvious from the rimantadine-bound NMR structure that V27A and A30T will change the size and/or hydrophobicity of the adamantane binding pocket. The G34E mutation is likely to cause steric collision with the amino and methyl groups of the drug. Moreover, introducing four negative charges inside the small pore could alter the channel conformation. The S31N mutation is less straightforward to explain because Ser31 faces the helix–helix interface and does not appear to be involved in any direct interactions with the drug. Previous NMR analysis of the S31N mutant suggested that replacing Ser31 with the much bulkier asparagine has the effect of destabilizing helical packing [51]. We believe that the S31N mutation abrogates drug binding allosterically by disrupting or loosening the internal binding pocket. Modifying the adamantane compound to overcome this mutation is clearly not easy, but a recent study reported that conjugation of a —CH2-heteroaryl group to the amine of amantadine leads to significant inhibition of the S31N mutant [87].
3.2. Inhibition of the p7 channel by the adamantane compounds
In addition to blocking the M2 channel, the adamantane derivatives such as amantadine and rimantadine have also been reported to pose some inhibitory effects on the p7 channel conductance [18], [88]. The therapeutic relevance of this finding was however controversial in clinical trials that investigated p7 amino acid variations in response to antiviral therapy with amantadine [89]. When treating HCV genotype 1a/b infected patients with amantadine in the clinical study, no significant correlation of overall amino acid variations in p7 with response to the treatment was found, although a p7 mutation, L20F, was observed more often in the drug resistant patients. Another in vitro study using single-channel recording technique showed that the activity of p7 from genotype 1b is not sensitive to amantadine [60]. The above discrepancies around amantadine inhibition could be explained by the sequence variations of p7 among the different genotypes. Indeed, a systematic investigation using multiple functional readouts including viral entry and release demonstrated substantial genotype-dependent and subtype-dependent sensitivity to several p7 inhibitors, including amantadine and rimantadine [77], but the sequence differences are not sufficiently localized to enable elucidation of the drug binding location.
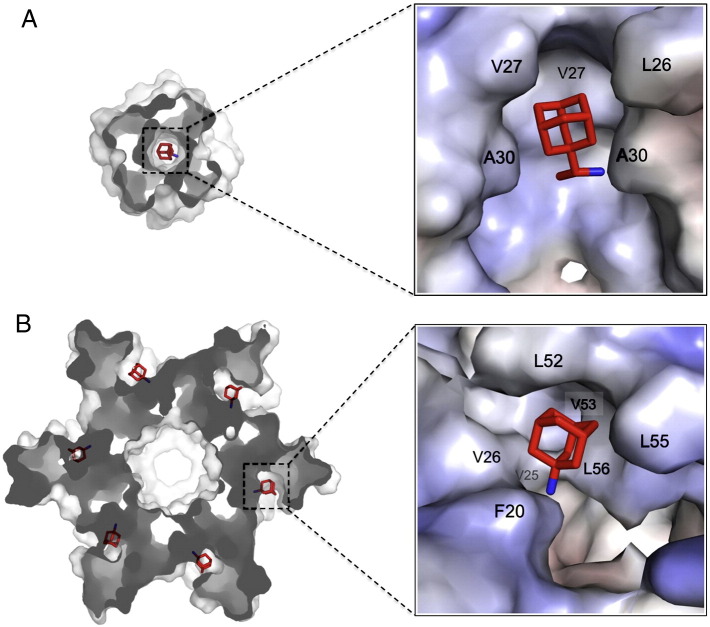
The most recent structural characterization of HCV p7 provided a detailed view of the binding site of amantadine or rimantadine [63]. NMR measurements of the channel-drug complex revealed that amantadine or rimantadine binds to six equivalent hydrophobic pockets (due to the six-fold symmetry of the p7 channel) between the pore-forming and peripheral helices (Fig. 3B). In each site, Leu52, Leu53, and Leu56 from H3 of the i monomer, Val25 and Val26 from H2 of the i + 2 monomer, and Phe20 from H2 of the i + 3 monomer together form a deep hydrophobic pocket that wraps around the adamantane cage of the drug. The amino group of amantadine or rimantadine is facing the largely hydrophilic channel lumen. An important property of the drug binding site is that it consists of elements from different helical segments and from different monomers. As rationalized above, permeation of cations through the p7 channel may depend on opening the narrow end of the funnel, which in turn depends on the reorientation of the helical segments. The binding of adamantane derivatives to the pocket may inhibit channel activity allosterically by causing the channel to close.
3.3. The same drug, different viroporins, and different modes of inhibition
Comparing the amantadine or rimantadine binding mode of HCV p7 to that of influenza M2 shows two fundamentally different mechanisms of drug inhibition. In the case of M2, one drug binds to one channel. Drug binding inhibits proton transport by directly blocking the channel passage; it also prevents channel from opening. In the case of p7, amantadine and rimantadine are clearly too small to block the channel. They instead bind to six equivalent sites outside of the channel cavity, which can afford up to six drugs per channel. If rimantadine binding to this site is relevant to inhibition, as crudely suggested by previous functional mutagenesis, drug binding to these sites inhibits cation conduction with an allosteric mechanism, possibly by stabilizing the closed state of the channel.
Although the mechanisms of drug inhibition may be completely different, the structural bases that govern drug-binding affinity for M2 and p7 are actually similar, and they involve hydrophobicity and size of the pocket, and position of the drug amino group. In the case of the M2 tetramer, the drug adamantane cage fits snuggly in a hydrophobic pocket formed by eight methyl groups from Val27 and Ala30 (two from each subunit), while the drug amino group forms polar contact with the backbone oxygen of Ala30 and points to the polar region of the channel cavity (Fig. 4A). For the p7 channel, the adamantane cage is in contact with ten methyl groups and an aromatic group from the protein. These hydrophobic groups form a deep hydrophobic pocket that also matches closely the size of the adamantane cage (Fig. 4B). The amino group of amantadine or rimantadine points to the channel lumen; it is in position to form polar contacts with the electronegative groups such as backbone carbonyl of residues 15–17.
Fig. 4.
Comparison between adamantane binding sites of influenza M2 and HCV p7 channels. (A) The internal pocket that wraps around the adamantane cage of rimantadine determined earlier for the AM2–BM2 chimeric channel [83]. The AM2–BM2 chimeric channel is a well-behaved model system for studying influenza M2 channels; its N-terminal half is from influenza A M2 protein (sensitive to amantadine or rimantadine inhibition) and its C-terminal half is from influenza B M2 protein (insensitive to amantadine or rimantadine). One subunit of the tetrameric complex is removed to unveil the channel interior. (B) The peripheral pocket between the H2 and H3 helices to which amantadine binds [63]; it is a representative pocket among six equivalent pockets in the p7 hexamer.
The similarity in amantadine or rimantadine binding between p7 and M2 allude to similar mechanism of drug resistance. Known drug resistance mutations of M2 such as V27A and A30T confer resistance by decreasing the hydrophobicity of the adamantane binding pocket, and this is conceptually consistent with the observed mutations that confer drug resistance in p7. In the case of p7, our functional assay showed that single mutations L20F and F26A as well as double mutation L53T/A56S also significantly reduced rimantadine inhibition (unpublished data), and these mutations all decrease hydrophobicity of the adamantane binding pocket. Thus variations in the hydrophobicity of the adamantane binding pocket among the p7 variants explain the large differences in drug efficacies observed between different HCV genotypes.
4. Conclusion
Viruses have developed membrane-embedded viroporins for transport of ions and solutes during viral life cycle. The name “porin” seems to imply leaky pores, similar to the β-barrel porins in bacteria. But, the recent structural data show that these viroporins are more sophisticated than leaky pores. Although viroporins generally adopt minimalist architecture, they have the essential structural elements compatible with selective transport of protons and/or cations. These viral channels may not be as functionally robust or specifically regulated as some of their counterparts in prokaryotes and eukaryotes, because viruses often do not need intricate regulation of channel activities during their infection cycles. Nonetheless, the unique structures of viroporins can provide unique opportunities for developing novel antiviral therapeutics. We believe, with multiple biophysical and biochemical tools being established for structural studies of membrane proteins, new viroporin architectures will be discovered at a faster pace. Despite the progress in structure determination, major challenges lie ahead in viroporin research. The precise functional roles of many viroporins are still unclear. Moreover, due to the lack of robust single-channel recording setups suitable for viroporins, the ion conductance properties of most viroporins have not been fully characterized. Therefore, better definition of the functional aspects of viroporins will certainly draw greater enthusiasm for developing therapeutics that target this interesting family of membrane channels.
Acknowledgements
This work was supported by NIH Grant GM094608 (to J.J.C.).
Footnotes
This article is part of a Special Issue entitled: Viral Membrane Proteins—Channels for Cellular Networking.
References
- 1.Fischer W.B., Sansom M.S. Viral ion channels: structure and function. Biochim. Biophys. Acta. 2002;1561:27–45. doi: 10.1016/s0304-4157(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 2.Wang K., Xie S., Sun B. Viral proteins function as ion channels. Biochim. Biophys. Acta. 2011;1808:510–515. doi: 10.1016/j.bbamem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nieva J.L., Madan V., Carrasco L. Viroporins: structure and biological functions. Nat. Rev. Microbiol. 2012;10:563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer W.B., Hsu H.J. Viral channel forming proteins—modeling the target. Biochim. Biophys. Acta. 2011;1808:561–571. doi: 10.1016/j.bbamem.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madan V., Sanchez-Martinez S., Vedovato N., Rispoli G., Carrasco L., Nieva J.L. Plasma membrane-porating domain in poliovirus 2B protein. A short peptide mimics viroporin activity. J. Mol. Biol. 2007;374:951–964. doi: 10.1016/j.jmb.2007.09.058. [DOI] [PubMed] [Google Scholar]
- 6.Madan V., Redondo N., Carrasco L. Cell permeabilization by poliovirus 2B viroporin triggers bystander permeabilization in neighbouring cells through a mechanism involving gap junctions. Cell. Microbiol. 2010;12:1144–1157. doi: 10.1111/j.1462-5822.2010.01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agirre A., Lorizate M., Nir S., Nieva J.L. Poliovirus 2b insertion into lipid monolayers and pore formation in vesicles modulated by anionic phospholipids. Biochim. Biophys. Acta. 2008;1778:2621–2626. doi: 10.1016/j.bbamem.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Melton J.V., Ewart G.D., Weir R.C., Board P.G., Lee E., Gage P.W. Alphavirus 6K proteins form ion channels. J. Biol. Chem. 2002;277:46923–46931. doi: 10.1074/jbc.M207847200. [DOI] [PubMed] [Google Scholar]
- 9.Sanz M.A., Perez L., Carrasco L. Semliki Forest virus 6K protein modifies membrane permeability after inducible expression in Escherichia coli cells. J. Biol. Chem. 1994;269:12106–12110. [PubMed] [Google Scholar]
- 10.Firth A.E., Chung B.Y., Fleeton M.N., Atkins J.F. Discovery of frameshifting in alphavirus 6K resolves a 20-year enigma. Virol. J. 2008;5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoine A.F., Montpellier C., Cailliau K., Browaeys-Poly E., Vilain J.P., Dubuisson J. The alphavirus 6K protein activates endogenous ionic conductances when expressed in Xenopus oocytes. J. Membr. Biol. 2007;215:37–48. doi: 10.1007/s00232-007-9003-6. [DOI] [PubMed] [Google Scholar]
- 12.Pielak R.M., Chou J.J. Influenza M2 proton channels. Biochim. Biophys. Acta. 2011;1808:522–529. doi: 10.1016/j.bbamem.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., Qiu J.X., Soto C., DeGrado W.F. Structural and dynamic mechanisms for the function and inhibition of the M2 proton channel from influenza A virus. Curr. Opin. Struct. Biol. 2011;21:68–80. doi: 10.1016/j.sbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross T.A., Dong H., Sharma M., Busath D.D., Zhou H.X. M2 protein from influenza A: from multiple structures to biophysical and functional insights. Curr. Opin. Virol. 2012;2:128–133. doi: 10.1016/j.coviro.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schubert U., Ferrer-Montiel A.V., Oblatt-Montal M., Henklein P., Strebel K., Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398:12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- 16.Marassi F.M., Ma C., Gratkowski H., Straus S.K., Strebel K., Oblatt-Montal M., Montal M., Opella S.J. Correlation of the structural and functional domains in the membrane protein Vpu from HIV-1. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14336–14341. doi: 10.1073/pnas.96.25.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romer W., Lam Y.H., Fischer D., Watts A., Fischer W.B., Goring P., Wehrspohn R.B., Gosele U., Steinem C. Channel activity of a viral transmembrane peptide in micro-BLMs: Vpu(1–32) from HIV-1. J. Am. Chem. Soc. 2004;126:16267–16274. doi: 10.1021/ja0451970. [DOI] [PubMed] [Google Scholar]
- 18.Griffin S.D., Beales L.P., Clarke D.S., Worsfold O., Evans S.D., Jaeger J., Harris M.P., Rowlands D.J. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 2003;535:34–38. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 19.Pavlovic D., Neville D.C., Argaud O., Blumberg B., Dwek R.A., Fischer W.B., Zitzmann N. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mould J.A., Paterson R.G., Takeda M., Ohigashi Y., Venkataraman P., Lamb R.A., Pinto L.H. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell. 2003;5:175–184. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 21.Plugge B., Gazzarrini S., Nelson M., Cerana R., Van Etten J.L., Derst C., DiFrancesco D., Moroni A., Thiel G. A potassium channel protein encoded by chlorella virus PBCV-1. Science. 2000;287:1641–1644. doi: 10.1126/science.287.5458.1641. [DOI] [PubMed] [Google Scholar]
- 22.Gazzarrini S., Severino M., Lombardi M., Morandi M., DiFrancesco D., Van Etten J.L., Thiel G., Moroni A. The viral potassium channel Kcv: structural and functional features. FEBS Lett. 2003;552:12–16. doi: 10.1016/s0014-5793(03)00777-4. [DOI] [PubMed] [Google Scholar]
- 23.Strauss M., Levy H.C., Bostina M., Filman D.J., Hogle J.M. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J. Virol. 2013;87:3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin K., Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 25.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 26.Grambas S., Bennett M.S., Hay A.J. Influence of amantadine resistance mutations on the pH regulatory function of the M2 protein of influenza A viruses. Virology. 1992;191:541–549. doi: 10.1016/0042-6822(92)90229-i. [DOI] [PubMed] [Google Scholar]
- 27.Jones C.T., Murray C.L., Eastman D.K., Tassello J., Rice C.M. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinmann E., Penin F., Kallis S., Patel A.H., Bartenschlager R., Pietschmann T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3:e103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agarkova I., Dunigan D., Gurnon J., Greiner T., Barres J., Thiel G., Van Etten J.L. Chlorovirus-mediated membrane depolarization of Chlorella alters secondary active transport of solutes. J. Virol. 2008;82:12181–12190. doi: 10.1128/JVI.01687-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madan V., Castello A., Carrasco L. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell. Microbiol. 2008;10:437–451. doi: 10.1111/j.1462-5822.2007.01057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen B.J., Leser G.P., Jackson D., Lamb R.A. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 2008;82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imai M., Kawasaki K., Odagiri T. Cytoplasmic domain of influenza B virus BM2 protein plays critical roles in production of infectious virus. J. Virol. 2008;82:728–739. doi: 10.1128/JVI.01752-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Pielak R.M., McClintock M.A., Chou J.J. Solution structure and functional analysis of the influenza B proton channel. Nat. Struct. Mol. Biol. 2009;16:1267–1271. doi: 10.1038/nsmb.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gouklani H., Beyer C., Drummer H., Gowans E.J., Netter H.J., Haqshenas G. Identification of specific regions in hepatitis C virus core, NS2 and NS5A that genetically interact with p7 and co-ordinate infectious virus production. J. Viral Hepat. 2013;20:e66–71. doi: 10.1111/jvh.12004. [DOI] [PubMed] [Google Scholar]
- 35.Vieyres G., Brohm C., Friesland M., Gentzsch J., Wolk B., Roingeard P., Steinmann E., Pietschmann T. Subcellular localization and function of an epitope-tagged p7 viroporin in hepatitis C virus-producing cells. J. Virol. 2013;87:1664–1678. [Google Scholar]
- 36.Lamb R.A., Zebedee S.L., Richardson C.D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 37.Sugrue R.J., Hay A.J. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holsinger L.J., Lamb R.A. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 39.Paterson R.G., Takeda M., Ohigashi Y., Pinto L.H., Lamb R.A. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology. 2003;306:7–17. doi: 10.1016/s0042-6822(02)00083-1. [DOI] [PubMed] [Google Scholar]
- 40.Pinto L.H., Lamb R.A. The M2 proton channels of influenza A and B viruses. J. Biol. Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 41.Neirynck S., Deroo T., Saelens X., Vanlandschoot P., Jou W.M., Fiers W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999;5:1157–1163. doi: 10.1038/13484. [DOI] [PubMed] [Google Scholar]
- 42.Jegerlehner A., Schmitz N., Storni T., Bachmann M.F. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J. Immunol. 2004;172:5598–5605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- 43.De Filette M., Martens W., Roose K., Deroo T., Vervalle F., Bentahir M., Vandekerckhove J., Fiers W., Saelens X. An influenza A vaccine based on tetrameric ectodomain of matrix protein 2. J. Biol. Chem. 2008;283:11382–11387. doi: 10.1074/jbc.M800650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang B.Z., Gill H.S., Kang S.M., Wang L., Wang Y.C., Vassilieva E.V., Compans R.W. Enhanced influenza virus-like particle vaccines containing the extracellular domain of matrix protein 2 and a Toll-like receptor ligand. Clin. Vaccine Immunol. 2012;19:1119–1125. doi: 10.1128/CVI.00153-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim E.H., Lee J.H., Pascua P.N., Song M.S., Baek Y.H., Kwon H.I., Park S.J., Lim G.J., Decano A., Chowdhury M.Y., Seo S.K., Song M.K., Kim C.J., Choi Y.K. Prokaryote-expressed M2e protein improves H9N2 influenza vaccine efficacy and protection against lethal influenza A virus in mice. Virol. J. 2013;10:104. doi: 10.1186/1743-422X-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCown M.F., Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 2006;80:8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imai M., Watanabe S., Ninomiya A., Obuchi M., Odagiri T. Influenza B virus BM2 protein is a crucial component for incorporation of viral ribonucleoprotein complex into virions during virus assembly. J. Virol. 2004;78:11007–11015. doi: 10.1128/JVI.78.20.11007-11015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnell J.R., Chou J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stouffer A.L., Acharya R., Salom D., Levine A.S., Di Costanzo L., Soto C.S., Tereshko V., Nanda V., Stayrook S., DeGrado W.F. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Acharya R., Carnevale V., Fiorin G., Levine B.G., Polishchuk A.L., Balannik V., Samish I., Lamb R.A., Pinto L.H., DeGrado W.F., Klein M.L. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pielak R.M., Schnell J.R., Chou J.J. Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7379–7384. doi: 10.1073/pnas.0902548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pielak R.M., Chou J.J. Solution NMR structure of the V27A drug resistant mutant of influenza A M2 channel. Biochem. Biophys. Res. Commun. 2010;401:58–63. doi: 10.1016/j.bbrc.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cady S.D., Schmidt-Rohr K., Wang J., Soto C.S., Degrado W.F., Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma M., Yi M., Dong H., Qin H., Peterson E., Busath D.D., Zhou H.X., Cross T.A. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harbury P.B., Zhang T., Kim P.S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 56.Pielak R.M., Chou J.J. Kinetic analysis of the M2 proton conduction of the influenza virus. J. Am. Chem. Soc. 2010;132:17695–17697. doi: 10.1021/ja108458u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu F., Luo W., Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu J., Fu R., Nishimura K., Zhang L., Zhou H.X., Busath D.D., Vijayvergiya V., Cross T.A. Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Premkumar A., Wilson L., Ewart G.D., Gage P.W. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557:99–103. doi: 10.1016/s0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- 60.Montserret R., Saint N., Vanbelle C., Salvay A.G., Simorre J.P., Ebel C., Sapay N., Renisio J.G., Bockmann A., Steinmann E., Pietschmann T., Dubuisson J., Chipot C., Penin F. NMR structure and ion channel activity of the p7 protein from hepatitis C virus. J. Biol. Chem. 2010;285:31446–31461. doi: 10.1074/jbc.M110.122895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cook G.A., Opella S.J. Secondary structure, dynamics, and architecture of the p7 membrane protein from hepatitis C virus by NMR spectroscopy. Biochim. Biophys. Acta. 2011;1808:1448–1453. doi: 10.1016/j.bbamem.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luik P., Chew C., Aittoniemi J., Chang J., Wentworth P., Jr., Dwek R.A., Biggin P.C., Venien-Bryan C., Zitzmann N. The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12712–12716. doi: 10.1073/pnas.0905966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.OuYang B., Xie S., Berardi M.J., Zhao X., Dev J., Yu W., Sun B., Chou J.J. Unusual architecture of the p7 channel from hepatitis C virus. Nature. 2013;498:521–525. doi: 10.1038/nature12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gouaux E., Mackinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- 65.Cuello L.G., Jogini V., Cortes D.M., Pan A.C., Gagnon D.G., Dalmas O., Cordero-Morales J.F., Chakrapani S., Roux B., Perozo E. Structural basis for the coupling between activation and inactivation gates in K(+) channels. Nature. 2010;466:272–275. doi: 10.1038/nature09136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cuello L.G., Jogini V., Cortes D.M., Perozo E. Structural mechanism of C-type inactivation in K(+) channels. Nature. 2010;466:203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lunin V.V., Dobrovetsky E., Khutoreskaya G., Zhang R., Joachimiak A., Doyle D.A., Bochkarev A., Maguire M.E., Edwards A.M., Koth C.M. Crystal structure of the CorA Mg2 + transporter. Nature. 2006;440:833–837. doi: 10.1038/nature04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou X., Pedi L., Diver M.M., Long S.B. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakai N., Sorde N., Das G., Perrottet P., Gerard D., Matile S. Synthetic multifunctional pores: deletion and inversion of anion/cation selectivity using pM and pH. Org. Biomol. Chem. 2003;1:1226–1231. doi: 10.1039/b210604c. [DOI] [PubMed] [Google Scholar]
- 70.Park S.H., Mrse A.A., Nevzorov A.A., Mesleh M.F., Oblatt-Montal M., Montal M., Opella S.J. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J. Mol. Biol. 2003;333:409–424. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- 71.Lu J.X., Sharpe S., Ghirlando R., Yau W.M., Tycko R. Oligomerization state and supramolecular structure of the HIV-1 Vpu protein transmembrane segment in phospholipid bilayers. Protein Sci. 2010;19:1877–1896. doi: 10.1002/pro.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Torres J., Wang J., Parthasarathy K., Liu D.X. The transmembrane oligomers of coronavirus protein E. Biophys. J. 2005;88:1283–1290. doi: 10.1529/biophysj.104.051730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pinto L.H., Holsinger L.J., Lamb R.A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 75.Wang C., Takeuchi K., Pinto L.H., Lamb R.A. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davies W.L., Grunert R.R., Haff R.F., McGahen J.W., Neumayer E.M., Paulshock M., Watts J.C., Wood T.R., Hermann E.C., Hoffmann C.E. Antiviral activity of 1-adamantanamine (amantadine) Science. 1964;144:862–863. doi: 10.1126/science.144.3620.862. [DOI] [PubMed] [Google Scholar]
- 77.Griffin S., Stgelais C., Owsianka A.M., Patel A.H., Rowlands D., Harris M. Genotype-dependent sensitivity of hepatitis C virus to inhibitors of the p7 ion channel. Hepatology. 2008;48:1779–1790. doi: 10.1002/hep.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilson L., Gage P., Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353:294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khoury G., Ewart G., Luscombe C., Miller M., Wilkinson J. Antiviral efficacy of the novel compound BIT225 against HIV-1 release from human macrophages. Antimicrob. Agents Chemother. 2010;54:835–845. doi: 10.1128/AAC.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luscombe C.A., Huang Z., Murray M.G., Miller M., Wilkinson J., Ewart G.D. A novel Hepatitis C virus p7 ion channel inhibitor, BIT225, inhibits bovine viral diarrhea virus in vitro and shows synergism with recombinant interferon-alpha-2b and nucleoside analogues. Antiviral Res. 2010;86:144–153. doi: 10.1016/j.antiviral.2010.02.312. [DOI] [PubMed] [Google Scholar]
- 81.Jing X., Ma C., Ohigashi Y., Oliveira F.A., Jardetzky T.S., Pinto L.H., Lamb R.A. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10967–10972. doi: 10.1073/pnas.0804958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohigashi Y., Ma C., Jing X., Balannick V., Pinto L.H., Lamb R.A. An amantadine-sensitive chimeric BM2 ion channel of influenza B virus has implications for the mechanism of drug inhibition. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18775–18779. doi: 10.1073/pnas.0910584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pielak R.M., Oxenoid K., Chou J.J. Structural investigation of rimantadine inhibition of the AM2–BM2 chimera channel of influenza viruses. Structure. 2011;19:1655–1663. doi: 10.1016/j.str.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bright R.A., Medina M.J., Xu X., Perez-Oronoz G., Wallis T.R., Davis X.M., Povinelli L., Cox N.J., Klimov A.I. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 85.Hay A.J., Wolstenholme A.J., Skehel J.J., Smith M.H. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bright R.A., Shay D.K., Shu B., Cox N.J., Klimov A.I. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 87.Wang J., Wu Y., Ma C., Fiorin G., Pinto L.H., Lamb R.A., Klein M.L., Degrado W.F. Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1315–1320. doi: 10.1073/pnas.1216526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Griffin S.D., Harvey R., Clarke D.S., Barclay W.S., Harris M., Rowlands D.J. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J. Gen. Virol. 2004;85:451–461. doi: 10.1099/vir.0.19634-0. [DOI] [PubMed] [Google Scholar]
- 89.Mihm U., Grigorian N., Welsch C., Herrmann E., Kronenberger B., Teuber G., von Wagner M., Hofmann W.P., Albrecht M., Lengauer T., Zeuzem S., Sarrazin C. Amino acid variations in hepatitis C virus p7 and sensitivity to antiviral combination therapy with amantadine in chronic hepatitis C. Antivir. Ther. 2006;11:507–519. [PubMed] [Google Scholar]
- 90.Gonzalez M.E., Carrasco L. The human immunodeficiency virus type 1 Vpu protein enhances membrane permeability. Biochemistry. 1998;37:13710–13719. doi: 10.1021/bi981527f. [DOI] [PubMed] [Google Scholar]
- 91.Verdia-Baguena C., Nieto-Torres J.L., Alcaraz A., Dediego M.L., Enjuanes L., Aguilella V.M. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim. Biophys. Acta. 2013;1828:2026–2031. doi: 10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Agirre A., Barco A., Carrasco L., Nieva J.L. Viroporin-mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem. 2002;277:40434–40441. doi: 10.1074/jbc.M205393200. [DOI] [PubMed] [Google Scholar]
- 93.Kabsch K., Alonso A. The human papillomavirus type 16 (HPV-16) E5 protein sensitizes human keratinocytes to apoptosis induced by osmotic stress. Oncogene. 2002;21:947–953. doi: 10.1038/sj.onc.1205147. [DOI] [PubMed] [Google Scholar]
- 94.Wetherill L.F., Holmes K.K., Verow M., Muller M., Howell G., Harris M., Fishwick C., Stonehouse N., Foster R., Blair G.E., Griffin S., Macdonald A. High-risk human papillomavirus E5 oncoprotein displays channel-forming activity sensitive to small-molecule inhibitors. J. Virol. 2012;86:5341–5351. doi: 10.1128/JVI.06243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Romani G., Piotrowski A., Hillmer S., Gurnon J., Vanetten J.L., Moroni A., Thiel G., Hertel B. Viral encoded potassium ion channel is a structural protein in the chlorovirus Paramecium bursaria chlorella virus-1 (PBCV-1) virion. J. Gen. Virol. 2013 doi: 10.1099/vir.0.055251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]