Abstract
MicroRNAs (miRNAs) are a class of small non-coding RNAs that regulate cancer progression, especially the processes of invasion and metastasis. Although earlier studies in metastasis primarily focused on the impact miRNAs had on the intrinsic properties of cancer cells, recent reports reveal that miRNAs also shape interactions between cancer cells and their associated stroma. In this review, we discuss current mechanisms by which miRNAs execute their microenvironmental regulation of cancer metastasis, including regulating expression of cell membrane-bound and secreted proteins or directly transmitting mature miRNAs between different cell types. The significance of miRNA-mediated tumor-stroma interactions in regulating metastasis suggests miRNAs may be a potential therapeutic target.
Keywords: tumor microenvironment, metastasis, circulating microRNA, tumor-stroma interaction
MicroRNAs in cancer progression: focus on metastasis
MicroRNAs (miRNAs) are a class of small non-coding RNAs that regulate a wide range of biological processes through changing the expression and translation of their target mRNA genes [1]. Shortly after the identification of miRNAs, reports began surfacing regarding their dysregulation in a variety of cancer types [2–4]. Accumulating literature shows that abnormal expression of miRNAs in tumors has significant pathological consequences: they can either enhance oncogene expression to facilitate tumorigenesis [5], or reduce the expression of tumor suppressors resulting in enhanced overexpression of oncogene products [6, 7]. In addition to promoting tumor growth, many miRNAs also participate in the metastatic process, which accounts for the mortality of approximately 90% of cancer patients [8, 9].
Cancer metastasis is a complex and inefficient process. To form metastases at sites distant from the initial primary tumor location, cancer cells need to invade through the basement membrane, intravasate into the blood stream, disseminate through the circulation, extrasavate to distal tissues/organs, and adapt to a new environment at the secondary site for survival and proliferation [10, 11]. During each step, cancer cells closely interact with their surrounding microenvironment consisting of the extracellular matrix (ECM) and stromal cells including immune cells, fibroblasts, endothelial cells, bone marrow derived cells (BMDCs) and stem/progenitor cells. Indeed, it is now widely accepted that the metastatic potential of cancer cells is determined by both their intrinsic and extrinsic properties and the role of miRNAs has been implicated in regulating both to promote metastasis [12]. miRNAs were shown to regulate metastasis by altering the intrinsic properties of cancer cells such as cell proliferation, migration, apoptosis, cellular senescence and DNA damage responses [13–17]. In the past five years, several miRNAs have been shown to be actively involved in the formation and function of different microenvironments encountered during tumor dissemination. Through modulation of the tumor microenvironment, those miRNAs can regulate cancer cell interactions and their metastatic potential.
In this review, we focus on the effects of miRNAs on the microenvironmental regulation of cancer metastasis. We also discuss the context-dependent nature of miRNA regulation and its impact on understanding the role of miRNAs in metastasis. Since the majority of life-threatening cancers occur in epithelial tissues [18], this review primarily focuses on the metastatic process of carcinomas. However, due to similarities in metastatic routes with some other cancer types, many of these miRNAs may also play a role in the metastatic process of certain sarcomas or even lymphomas.
Primary Tumor Microenvironment
Oncogenesis initiates at the primary tumor site. Although still controversial, many believe the primary tumor site is also the location where cancer cells obtain their metastatic potential [11]. This hypothesis is supported by the similarities in gene expression signatures observed between metastases and their corresponding primary tumors in several different types of cancer including breast, pancreatic, colorectal, and prostate cancer [19–23]. These findings strongly suggest the significance of analyzing the pathological properties of primary tumors when defining mechanisms of cancer metastasis.
The primary tumor is largely composed of a population of cancer cells; however, a population of stromal cells also exists, which directly and indirectly interact with cancer cells and influence the process of tumor development. Cancer cells adapt to and utilize the primary tumor microenvironment to initiate metastasis through two complementary strategies: they change their own gene expression pattern to take advantage of the signaling input from the stroma and invade/migrate to a favorable place; alternatively, they actively recruit specific stromal cell types to the primary tumor site to facilitate metastasis. In this scenario, the interactions between cancer cells and stroma co-evolve as the tumor develops. Recent studies have shown that miRNAs can participate in both of these processes. Here, we review the mechanisms by which the dysregulation of miRNAs inside cancer cells can facilitate the adaptation and modulation of the primary tumor microenvironment to promote metastasis.
Epithelial-Mesenchymal Transition (EMT) and Extracellular Matrix Remodeling
During carcinoma metastasis, the first step towards reaching a distant site is to breach the basement membrane, which is a specialized type of ECM that separates epithelial cells from multiple layers of stroma. In a normal situation, epithelial cells form a strong connection with each other through epithelial adheren-junctions involving the protein E-cadherin, limiting their migratory ability. To acquire motility and invasiveness, carcinoma cells undergo a morphological alternation known as the epithelial-mesenchymal transition (EMT), to repress the expression of E-cadherin and thus detach from the epithelial sheets [24]. In addition to changes in shape, cancer cells undergoing EMT display fundamental differences in their gene expression profiles and exhibit some properties of stem cells [25]. These stem-like carcinoma cells are able to bypass cellular senescence induced by oncogene activation and maintain their self-renewal capacity at distant sites [25, 26]. Thus, the EMT process is not only a migratory strategy but also a crucial process for the establishment and maintenance of cancer cell stemness. Recent evidence suggests that multiple miRNAs are involved in regulating the EMT process.
The miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) and miR-205 were the first group of miRNAs reported to regulate EMT. MiR-200 family and miR-205 were shown to be down-regulated in a variety of tumors to facilitate EMT progression. Loss of these miRNAs leads to increased expression of the ZEB family of transcription factors, which regulate the expression of EMT-related genes such as E-cadherin inside carcinoma cells [27–29]. Since this discovery, additional miRNAs have been found to modulate the EMT process when intrinsically dysregulated in cancer cells by post-transcriptionally regulating EMT-related transcription factors. For example, miR-34, miR-9 and miR-30a regulate the expression of Snail1 [30–32], whereas miR-124, miR-203 and miR-204/211 target Snail2 [33–35]; miR-214, miR-580 and Let-7d regulate Twist1 [36–38]; miR-138, miR-215 and miR-708 suppress Zeb2 [39–41]. A detailed review on the EMT-related transcription factors and miRNAs regulating those factors can be found elsewhere [42].
In order to invade adjacent cell layers, cancer cells also need to excavate a passage through the ECM by remodeling the nearby tissue environment. In this process, matrix metalloproteinases (MMPs) are probably the most important effectors [43]. Although stromal cells, such as macrophages, mast cells and fibroblasts, are generally believed to be the main producers of MMPs, certain epithelial cells, under the appropriate stimuli, can also exhibit MMP production [44]. Several miRNAs have been shown to affect the expression of MMPs. For example, when miR-29b is induced by the transcription factor GATA3 in breast cancer cells, it suppresses the expression of a group of pro-metastatic regulators including MMP2 and MMP9, consequently re-shaping the tumor microenvironment and suppressing metastasis by altering collagen remodeling, angiogenesis and proteolysis [45]. Different from miR-29b, up-regulation of miR-21 has been reported to associate with increased expression of multiple MMPs including MMP2 and MMP9. It regulates MMP expression through suppressing the expression of both the phosphatase PTEN in hepatocellular carcinoma cells or MMP inhibitors RECK and TIMP3 in glioblastoma cells [46, 47]. In addition, down-regulation of miR-138 was shown to induce RhoC expression and consequently increase production of MMP2/MMP9 in cholangiocarcinoma [48]. Similarly, miR-24 was recently reported to enhance EGFR signaling, which regulates the expression and activity of several MMPs as its downstream targets, and thus increase the expression of MMP2 and MMP11 [49]. Collectively, these findings demonstrate the involvement of many microRNAs in EMT and extracellular matrix remodeling, two important processes whereby carcinoma cells adapt to and utilize the primary tumor microenvironment to facilitate their migration and local invasion.
Stromal Cell Recruitment
In general, invasion through the basement membrane places cancer cells in a favorable position for completing subsequent steps of metastasis. However, although cancer cells gain direct access to blood and lymphatic vessels located on the stromal side of the basement membrane, interactions between cancer cells and stroma may initially suppress the disease progression [12]. For example, co-culture of prostatic epithelial cancer cells with normal fibroblasts, but not cancer-associated fibroblasts (CAFs), suppressed the proliferation of cancer cells [50, 51]. To shift the microenvironment to a metastasis-promoting state, cancer cells need to either transform the resident stroma cells, such as normal fibroblasts, to facilitate their growth/invasion, or recruit other metastasis-promoting stromal cells to remodel the microenvironment. Although it is still largely unknown how normal resident stroma cells are transformed, recent evidence suggests that miRNAs can regulate metastasis by influencing the recruitment of stromal cells in the primary tumor.
miR-126 and its complementary “passenger strand” miR-126* was recently found to suppress metastasis by modulating the composition of primary tumor stroma [52]. Using the 4T1-Balb/c xenograft model, in which adaptive immunity functions as a contributor to metastasis, miR-126/126* was found to significantly inhibit lung metastasis when the 4T1 murine mammary tumor cells were implanted in the mammary gland (primary tumor site), but not when injected into the tail vein (metastatic site). This result indicates that this pair of miRNAs suppresses metastasis by altering the primary tumor microenvironment before the tumor cells enter circulation. Mechanistically, miR-126/126* directly inhibits the expression of the chemokine Sdf-1α in cancer cells and indirectly suppresses the production of chemokine Ccl2 in an Sdf-1α- and stroma-dependent manner. Both of these chemokines have been reported to promote breast cancer metastasis by recruiting additional stromal cell types. As a result, suppression of these two miRNAs in breast cancer cells leads to the increased recruitment of mesenchymal stem cells (MSCs) and inflammatory monocytes, two important metastasis-promoting stromal cell types [53, 54], into the primary tumor microenvironment.
Immune Surveillance
The immune system is another important component of the tumor stroma. Accumulating evidence shows that the immune system is able to distinguish cancer cells from normal cells and eliminate neoplastic cells in many situations [55, 56]. However, if cancer cells obtain the ability to escape immune surveillance, they can take advantage of immune responses that promote a pro-metastatic environment permissive for their proliferation and invasion [57]. To evade immune detection, cancer cells can either inhibit the expression and display of tumor antigens on the cell surface or increase the expression of certain surface immunomodulatory inhibitors such as certain glycoproteins that disrupt immune recognition. Although it remains to be determined how important miRNAs are in modulating this process, some reports demonstrated that dysregulation of certain miRNAs in cancer cells could contribute to immune evasion. For example, enhancer of zeste homolog 2 (EZH2), a recently identified tumor-associated antigen in prostate cancer [58], can be suppressed by miR-138, partially explaining the up-regulation of this miRNA in high-grade prostate tumors [59]. As an alternative immune-evasive strategy, miR-9 is able to suppress the expression of MHC class I gene transcription and thus disturb the display of antigens at the cell surface [60]. Different from these miRNAs, miR-29 modulates immunity against cancer cells through a distinct process. Down-regulation of miR-29 in cancer cells leads to the up-regulation of B7-H3, a cell surface immunomodulatory glycoprotein that attenuates attack from natural killer cells and cytotoxic T cells. As a result, cancer cells expressing lower levels of miR-29 may escape detection by the immune system [61].
In addition to intrinsic changes occurring within cancer cells, several miRNAs can modulate immune surveillance by regulating the secretion of proteins from cancer cells needed to attract immunosuppressive T-regulatory cells (Tregs) or launch counterattacks on cytotoxic lymphocytes. For example, miR-30d promoted the metastatic behavior of melanoma cells by directly suppressing the GalNac transferase GALNT7. By repressing GALNT7, miR-30d stimulated the expression of the immunosuppressive cytokine IL-10, which has strong cytostatic effects on T lymphocytes and prevents their expression of MHC type II molecules [62]. In addition, miR-34a, which is down-regulated by TGF-β in liver cancer cells, suppressed the expression of chemokine CCL22 resulting in an attenuated ability of cancer cells to attract Tregs. In hepatocellular carcinoma, elevated TGF-β activity leads to reduced miR-34a expression and increased Treg infiltration. As a result, this immune-subversive microenvironment facilitates the colonization of metastatic tumor cells to the major branches of the portal vein, with the clinical symptom termed portal vein tumor thrombus (PVTT). PVTT is strongly correlated with a poor prognosis for liver cancer patients [63].
It should be noted that up to now we have focused our discussion on how dysregulation of miRNAs in cancer cells can contribute to immune evasion. Since many miRNAs also play important roles in mediating the normal function of immune responses, abnormal expression of those miRNAs in immune cells, through certain stimuli, may also affect cancer cell immune escape. In addition, recent studies suggest that certain secreted miRNAs can serve as a paracrine ligand for Toll-like receptors (TLRs) on immune cells and induce the secretion of pro-metastatic inflammatory cytokines from those cells [64]. For cancer cells that can already escape the immune attack, this strategy can further modulate the tumor microenvironment to favor the proliferation and invasion of cancer cells.
Hypoxia and Angiogenesis
Inside the primary tumor, the rapid proliferation of cancer cells frequently outgrows its blood supply, leaving many tumor cells in regions where the oxygen concentration is significantly lower compared with normal tissues. To adapt to and survive in this hypoxic microenvironment, cancer cells need to change their intrinsic gene expression pattern and concomitantly induce angiogenesis to provide enough nutrients and oxygen. Hypoxia-inducible factors (HIFs) are the most important transcription factors that respond to hypoxia. HIFs orchestrate a signaling cascade to promote tumor cell survival and the formation of neovasculature [65]. Recent studies have delineated the role of several microRNAs that coordinate with HIF signaling to regulate oxygen homeostasis inside tumors.
miR-210 is probably the most prominent microRNA upregulated by hypoxia via HIF transcription. In breast cancer, melanoma, pancreatic cancer and ovarian cancer, elevated expression of miR-210 under hypoxia has been reported to correlate with metastasis [66–68]. Mechanistically, miR-210 targets a MYC antagonist MAX dimerization protein (MNY) which allows cancer cells to bypass the hypoxia-induced cell cycle arrest [69]. It also regulates the mitochondrial metabolism of cancer cells by upregulating glycolysis, a metabolic pathway that is more adaptable for low oxygen conditions, through suppressing the expression of iron-sulfur cluster scaffold homolog (ISCU) and cytochrome c oxidase assembly protein (COX10) [70]. Interestingly, hypoxia stimulation or direct transmission from cancer cells via miRNA-containing microvesicles can enhance miR-210 expression in endothelial cells (the latter mechanism will be discussed later). In endothelial cells, elevated expression of miR-210 suppresses the production of Ephrin-A3 (EFNA3), a member of the ephrin family ligand that plays a crucial role in the development of the cardiovascular system and in vascular remodeling. This suppression via miR-210 stimulates the formation of capillary-like structures and cell migration [71]. In addition to miR-210, miR-424 is also induced by hypoxia to suppress a scaffolding protein cullin 2 (CUL2), which is critical to the assembly of the ubiquitin ligase system, resulting in the stabilization of HIF-α and increased angiogenesis [72].
Despite these findings, current understanding of the role of microRNAs in the hypoxic microenvironment is still limited. For example, it is unclear what the molecular nature of the regulatory process is for many hypoxia-induced or -suppressed microRNAs that are not direct targets of HIF transcription factors [73]. It also remains to be determined whether microRNAs are involved in other hypoxia-related processes such as the maintenance of a cancer stem cell niche [74].
Invasive and Metastatic Tumor Microenvironment
En route to form metastases, cancer cells must travel from the primary site through blood or lymphatic vessels and colonize at the secondary site. During this process, cancer cells face at least two distinct microenvironments from that found at the primary tumor site. Many of the aforementioned miRNA-related strategies used to promote metastasis from the primary site, such as immune evasion, are also applicable in these situations. However, there are also several miRNA-related mechanisms that specifically influence these later steps of metastasis.
A recent study provided mechanistic evidence for how the miR-200 family promotes cancer cell colonization independent from their EMT regulatory function [75]. Combining xenograft models and clinical correlation studies, the miR-200 family was found to suppress the expression of Sec23a, an essential component of COPII vesicles that transports proteins from the ER to Golgi apparatus, and thus induce a global reduction in protein secretion in breast cancer cells. The miR-200-Sec23a axis specifically functions at the colonization step, in which cancer cells initiate micrometastases formation at a distant organ, by decreasing the secretion of metastasis-suppressive proteins including insulin-like growth factor binding protein 4 (IGFBP4) and tubulointerstitial nephritis antigen-like 1 (Tinagl1). Although it remains to be determined how these two molecules contribute to the formation of a permissive environment at the distant site, this study suggests a novel mechanism by which miR-200 family promotes metastatic colonization by influencing the tumor cell secretome. Similar to miR-200 family, miR-17/20 cluster has been reported to regulate neighboring cell migration/invasion, in addition to regulating the intrinsic properties of cancer cells, to inhibit breast cancer cell invasion. High expression of miR-17/20 in breast cancer cells inhibits expression of a subset of metastasis-promoting cytokines, such as IL-8 and CXCL1, while concomitantly suppressing secreted plasminogen activators cytokeratin 8 and α-enolase that facilitate cell migration/invasion once recognized by nearby cells [76].
In addition to its role at the primary tumor site, miR-126 is also known to promote late stages of metastasis. Using human MDA-MB-231 cell series and immune-deficient SCID mice, it was shown that miR-126 attenuates the metastatic colonization of breast cancer cells by suppressing pro-angiogenic factors, including insulin-like growth factor binding protein 2 (IGFBP2), phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1) and c-mer proto-oncogene tyrosine kinase (MERTK) [77]. Consequently, this miRNA inhibits the recruitment of endothelial cells and angiogenesis at the metastatic site. These two studies on miR-126 suggest miRNA function is highly dependent on tumor cell intrinsic and extrinsic properties. This context-dependent nature of miRNAs will be further discussed in this review.
Direct Transmission of microRNAs between Cancer Cells and Associated Stromal Cells
miRNAs have been detected in the serum of cancer patients and can serve as circulating biomarkers for several different cancer types such as breast, pancreatic, colon, and lung cancer [78–82]. Circulating miRNAs can originate either from cancer cell debris generated through apoptosis or necrosis, or from miRNA-containing microvesicles that are actively produced by cancer cells. While it has been speculated that circulating miRNAs, once absorbed by another cell, can change the gene expression pattern of its recipients, the experimental evidence to support this hypothesis has just emerged. A recent study demonstrated that mast-cell-secreted exosomes contained mRNAs and miRNAs and these RNAs were transferable to and functional in other cells [83]. Recent studies provided mechanistic evidence that certain miRNAs in this form could directly regulate the cancer metastasis process by modulating cancer cell-stroma interactions.
For example, miR-210 can be released from metastatic breast cancer cells via an nSMase2-dependent exosomal secretion pathway and transported to endothelial cells. Once inside, this miRNA increased cell migration and capillary formation thereby promoting angiogenesis and metastasis [84]. Alternatively, miRNAs can also been transmitted from stroma cells to cancer cells. A recent study showed that miR-223, which is highly expressed in IL-4 activated tumor-associated macrophages (TAMs) but not in breast cancer cells, can be transmitted from TAMs to co-cultured cancer cells. After entering breast cancer cells, miR-223 suppressed the expression of Myocyte-specific enhancer factor 2C (Mef2c), whose reduction has been linked to nuclear accumulation of β-catenin and the promotion of cell migration [85]. This is a novel mechanism in which TAMs promote breast cancer cell invasion and metastasis by secreting miRNA-containing microvesicles [86]. Furthermore, the secreted miRNAs that function as ligands for TLRs also belong to this type of regulation [64]. The transmission of miRNAs-containing microvesicles between different cell types provides an additional mechanism of microenvironmental regulation of metastasis beyond alterations in the expression of membrane and secreted proteins.
Beyond the surface: why the effects of microRNAs seem so complex
Based on previous discussions, it is obvious that miRNAs are widely involved in the interactions between cancer cells and their associated stroma. However, it has been difficult to fully understand the regulatory network of miRNAs in the microenvironmental regulation of metastasis given the complexity of tumor-stroma interaction and the context-dependent nature of miRNA functions. It is quite common to notice some seemingly contradictory observations about miRNAs in cancer metastasis: some miRNAs have different effects at the same stage for a specific cancer type; others regulate multiple steps throughout the metastasis process. To better understand this phenomenon, there are at least three issues that should be noted:
First, several miRNAs function as master regulators of cancer metastasis that also regulate more than one step in the invasion-metastasis cascade. For example, oncogenic miR-21 is involved in multiple aspects of cancer oncogenesis and metastasis [14]. miR-21 is overexpressed in most tumor types analyzed and is known to promote cancer cell survival, proliferation, migration and invasion [47, 87–89]. It is also involved in remodeling the ECM and facilitating angiogenesis [46, 90]. A recent study used the Cre- and Tet-off technique to generate mice that conditionally express miR-21 and demonstrated that tumors could become addicted to miR-21 similar to other oncogene addictions [91]. Similarly, miR-31 is also found to have pleiotropic functions in cancer metastasis. By coordinating repression of a cohort of metastasis-promoting genes, miR-31 inhibits several steps of metastasis including local invasion, extravasation, initial survival and metastatic colonization [92–94]. Given the strong effects of these master miRNAs in cancer progression, the prospect of utilizing these miRNAs as therapeutic targets in cancer treatment is exciting.
Second, although a single miRNA can usually regulate hundreds of different mRNA genes, the ability to select different target genes may be based on a specific cellular context, even within the same step of metastasis. As previously mentioned, miR-126/126* regulates the primary tumor microenvironment by recruiting MSCs and inflammatory monocytes [52]. In contrast, it changes the metastatic tumor microenvironment by suppressing endothelial cells infiltration [77]. Since the infiltration of the different types of stromal cells should not be restricted by the physical locations, it is more likely that the different phenotypes observed in these studies are due to different experimental settings or different cellular contexts. Analysis of potential targets of miR-126 revealed that the key targets in the MDA-MB-231 system, IGFBP2, PITPNC1 and MERTK were not the same crucial miR-126/126* targets in the 4T1 model. It is possible that MDA-MD-231 and 4T1 cells represent different breast cancer cell populations with distinct expression profiles of potential miR-126/126* targets and/or RNA binding proteins that modulate the accessibility of the putative targets to these microRNAs [52]. As a result, these two miRNAs may suppress the metastatic process through different mechanisms in different subsets of breast cancer patients. The complexity of miRNA in mediating cancer metastasis requires additional studies on the impact of the cellular context. These studies, however, also suggest the advantage of developing miRNAs that regulate multiple steps of cancer progression as therapeutic targets.
Third, determined by the specific cellular contexts in different stages of metastasis, miRNAs may target the same gene but result in different phenotypic outcomes. For example, miR-200 is well established as a breast tumor suppressor at the primary tumor site and promotes epithelial characteristics by suppressing the EMT-regulatory transcription factor ZEB1/2 [95]. However, the same miR-200-ZEB1/2 axis has been shown to promote macroscopic metastases in a mouse xenograft model whereby tumor colonization might be enhanced by a mesenchymal to epithelial cell transition (MET) [96]. These studies again demonstrate the context-dependent nature of miRNAs in metastasis regulation.
Concluding remarks
In summary, there are many miRNAs involved in the regulation of tumor-stroma interactions (Table 1). These miRNAs execute their microenvironmental regulation of cancer metastasis through several different mechanisms, including regulating expression of cell membrane proteins and secreted proteins, or directly transmitting mature miRNAs between different cell types (Fig. 1). Although our current understanding in the field is still in its nascent stage, the significance of miRNA-mediated tumor-stroma interaction in metastasis implies the possibility to develop such miRNAs as therapeutic targets. Since the genomes of stroma cells are not as mutable as cancer cells, it is difficult for tumors to develop resistance to treatments targeting the tumor microenvironment. However, the complexity of such regulation requires further basic and pre-clinical studies to examine the upstream regulation of these miRNAs, the mechanisms underlying how the cellular context determines their regulation, as well as the potential adverse effects of such treatments.
Table 1.
microRNAs regulate metastasis through modulating tumor-stroma interactions
| microRNA | Function | Direct Targets | Refs |
|---|---|---|---|
| let-7d | EMT | Twist1 | 38 |
| miR-124 | EMT | Snail2 | 33 |
| miR-126/126* | Stromal Cell Recruitment Colonization |
Sdf-1 IGFBP2, PITPNC1, MERTK |
52 77 |
| miR-138 | EMT ECM Remodeling Immune Escape |
ZEB2 RhoC EZH2 |
39 48 58, 59 |
| miR-17/20 | Migration/Invasion of Neighboring Cells | IL-8, CXCL1, CK8, α-ENO | 76 |
| miR-200 family | EMT Colonization |
ZEB1/2 Sec23a |
27–29, 95 75 |
| miR-203 | EMT | Snail2 | 34 |
| miR-204/211 | EMT | Snail2 | 35 |
| miR-205 | EMT | ZEB1/2 | 27–29 |
| miR-21 | ECM Remodeling Proliferation, Migration/Invasion, Apoptosis, Angiogenesis |
PTEN, RECK, TIMP3 PDCD4, TPM1 |
46, 47 87–90 |
| miR-210 | Angiogenesis Survival in Hypoxic Microenvironment |
Ephrin-A3 MNY, ISCU, COX10 |
71, 84 69, 70 |
| miR-214 | EMT | Twist1 | 36 |
| miR-215 | EMT | ZEB2 | 40 |
| miR-223 | Migration/Invasion | Mef2c | 86 |
| miR-24 | ECM Remodeling | PTNP9, PTPRF | 49 |
| miR-29b | ECM Remodeling Immune Escape |
MMP2, MMP9 B7-H3 |
45 61 |
| miR-30a | EMT | Snail1 | 32 |
| miR-30d | Immune Escape | GALNT7 | 62 |
| miR-31 | Migration/Invasion, Extravasation, Colonization | ITGA5, RDX, and RhoA | 92–94 |
| miR-34a | EMT Immune Escape, Stromal Cel Recruitment |
Snail1 CCL22 |
30 63 |
| miR-424 | Angiogenesis | CUL2 | 72 |
| miR-580 | EMT | Twist1 | 37 |
| miR-708 | EMT | ZEB2 | 41 |
| miR-9 | EMT Immune Escape |
Snail1 MHC class I genes |
31 60 |
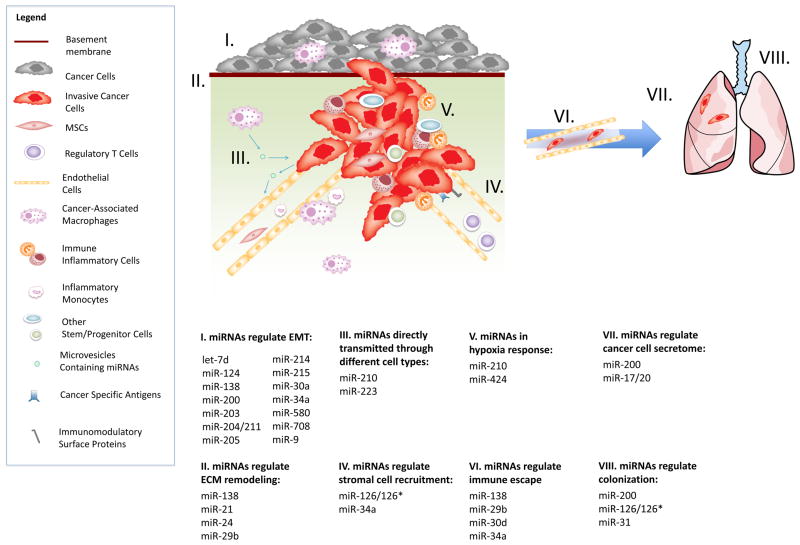
Figure 1. The mechanisms by which miRNAs execute their microenvironmental regulation of cancer metastasis.
Through many different mechanisms, miRNAs regulate the tumor-stroma interactions and the metastasis process. These mechanisms include: I. miRNAs regulate epithelial-mesenchymal transition by suppressing EMT related transcription factors; II. miRNAs regulate extracellular matrix remodeling through modulating certain matrix metalloproteinase; III. miRNAs are able to function as direct transmitters that exchange information between cancer cells and stromal cells and thus facilitate metastasis; IV. miRNAs regulate recruitment of several stromal cell types including endothelial cells, MSCs, inflammatory monocytes and immune cells via changing the expression level of specific chemokines; V. miRNAs can coordinate with HIF transcription factors to regulate the oxygen homeostasis in hypoxic microenvironment; VI. miRNAs facilitate immune escape through either changing the expression of cancer specific antigens and surface immunomodulatory proteins on cancer cell membrane or regulating the infiltration of immune regulatory cell types such as regulatory T cells; VII. miRNAs regulate cancer cell secretome to affect neighboring cells’ metastatic ability or to form a pro-metastatic microenvironment; VIII. miRNAs promote colonization through increasing angiogenesis, promoting cancer cell proliferation or forming a pro-metastatic environment at the distant site.
Highlights.
miRNAs are able to regulate metastasis through modulating tumor microenvironment
Mechanistically, miRNAs regulate tumor cell membrane and secreted proteins
Mature miRNAs can also be transmitted between tumor cells and stromal cells
miRNAs regulating tumor-stroma interaction may serve as new therapeutic targets
Acknowledgments
We apologize to researchers whose studies we were unable to cite due to the space limitation of this review. We thank G. Markowitz and X. Xu for providing insightful advises. Our research is supported by the NIH grant CA151541 to X. W.
Footnotes
Conflict of Interest
The authors declare no conflict of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Hammond SM. MicroRNAs as tumor suppressors. Nature genetics. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- 7.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. The New England journal of medicine. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 8.Nicoloso MS, et al. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 9.Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends in genetics: TIG. 2008;24:448–456. doi: 10.1016/j.tig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen DX, et al. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 11.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu S, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell research. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 15.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tazawa H, et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pothof J, et al. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. The EMBO journal. 2009;28:2090–2099. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 19.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiery JP, et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ansieau S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 28.Korpal M, et al. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. The Journal of biological chemistry. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim NH, et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. The Journal of cell biology. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, et al. MicroRNA-9 up-regulates E-cadherin through inhibition of NF-kappaB1-Snail1 pathway in melanoma. The Journal of pathology. 2012;226:61–72. doi: 10.1002/path.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumarswamy R, et al. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. International journal of cancer. Journal international du cancer. 2012;130:2044–2053. doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 33.Xia H, et al. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. The Journal of biological chemistry. 2012;287:9962–9971. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. Epigenetic Silencing of miR-203 Upregulates SNAI2 and Contributes to the Invasiveness of Malignant Breast Cancer Cells. Genes & cancer. 2011;2:782–791. doi: 10.1177/1947601911429743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang FE, et al. MicroRNA-204/211 alters epithelial physiology. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2010;24:1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, et al. Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. The FEBS journal. 2012;279:2393–2398. doi: 10.1111/j.1742-4658.2012.08618.x. [DOI] [PubMed] [Google Scholar]
- 37.Nairismagi ML, et al. Translational control of TWIST1 expression in MCF-10A cell lines recapitulating breast cancer progression. Oncogene. 2012;31:4960–4966. doi: 10.1038/onc.2011.650. [DOI] [PubMed] [Google Scholar]
- 38.Chang CJ, et al. Let-7d functions as novel regulator of epithelial-mesenchymal transition and chemoresistant property in oral cancer. Oncol Rep. 2011;26:1003–1010. doi: 10.3892/or.2011.1360. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, et al. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. The Biochemical journal. 2011;440:23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White NM, et al. miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br J Cancer. 2011;105:1741–1749. doi: 10.1038/bjc.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saini S, et al. MicroRNA-708 induces apoptosis and suppresses tumorigenicity in renal cancer cells. Cancer Res. 2011;71:6208–6219. doi: 10.1158/0008-5472.CAN-11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng H, Kang Y. Multilayer control of the EMT master regulators. Oncogene. 2013 doi: 10.1038/onc.2013.128. [DOI] [PubMed] [Google Scholar]
- 43.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer metastasis reviews. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 44.Overall CM, Kleifeld O. Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nature reviews. Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 45.Chou J, et al. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2013;15:201–213. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gabriely G, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Molecular and cellular biology. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng F, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q, et al. Downregulation of microRNA-138 enhances the proliferation, migration and invasion of cholangiocarcinoma cells through the upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncology Reports. 2013;29:2046–2052. doi: 10.3892/or.2013.2304. [DOI] [PubMed] [Google Scholar]
- 49.Du WW, et al. MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. Journal of cell science. 2013;126:1440–1453. doi: 10.1242/jcs.118299. [DOI] [PubMed] [Google Scholar]
- 50.Olumi AF, et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer research. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iacopino F, et al. Interactions Between Normal Human Fibroblasts and Human Prostate Cancer Cells in a Co-culture System. Anticancer Res. 2012;32:1579–1588. [PubMed] [Google Scholar]
- 52.Zhang Y, et al. miR-126 and miR-126*repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nature Cell Biology. 2013;15:284–294. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karnoub AE, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 54.Qian BZ, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smyth MJ, et al. Cancer immunosurveillance and immunoediting: the roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Advances in immunology. 2006;90:1–50. doi: 10.1016/S0065-2776(06)90001-7. [DOI] [PubMed] [Google Scholar]
- 56.Vesely MD, et al. Natural innate and adaptive immunity to cancer. Annual review of immunology. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 57.Zitvogel L, et al. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nature reviews. Immunology. 2006;6:715–727. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- 58.Itoh Y, et al. New peptides of the polycomb group protein enhancer of zeste homolog 2 with the potential to induce cancer-reactive cytotoxic T lymphocytes in human leukocyte antigen-A2+ prostate cancer patients. Oncology reports. 2007;18:1231–1237. [PubMed] [Google Scholar]
- 59.Walter BA, et al. Comprehensive microRNA Profiling of Prostate Cancer. Journal of Cancer. 2013;4:350–357. doi: 10.7150/jca.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao F, et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochemical and Biophysical Research Communications. 2013;431:610–616. doi: 10.1016/j.bbrc.2012.12.097. [DOI] [PubMed] [Google Scholar]
- 61.Xu H, et al. MicroRNA miR-29 Modulates Expression of Immunoinhibitory Molecule B7-H3: Potential Implications for Immune Based Therapy of Human Solid Tumors. Cancer Research. 2009;69:6275–6281. doi: 10.1158/0008-5472.CAN-08-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaziel-Sovran A, et al. miR-30b/30d Regulation of GaINAc Transferases Enhances Invasion and Immunosuppression during Metastasis. Cancer Cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang P, et al. TGF-beta-miR-34a-CCL22 Signaling-Induced Treg Cell Recruitment Promotes Venous Metastases of HBV-Positive Hepatocellular Carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho AS, et al. Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Translational Oncology. 2010;3:109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giannakakis A, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biology & Therapy. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Camps C, et al. hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clinical Cancer Research. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Z, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 70.Chen Z, et al. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 71.Fasanaro P, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. Journal of Biological Chemistry. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ghosh G, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. Journal of Clinical Investigation. 2010;120:4141–4154. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ivan M, et al. Hypoxia response and microRNAs: no longer two separate worlds. Journal of Cellular and Molecular Medicine. 2008;12:1426–1431. doi: 10.1111/j.1582-4934.2008.00398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Korpal M, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nature Medicine. 2011;17:1101–U1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu Z, et al. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci U S A. 2010;107:8231–8236. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Png KJ, et al. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 78.Roth C, et al. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Research. 2010:12. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kawaguchi T, et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. British Journal of Cancer. 2013;108:361–369. doi: 10.1038/bjc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheng H, et al. Circulating Plasma MiR-141 Is a Novel Biomarker for Metastatic Colon Cancer and Predicts Poor Prognosis. Plos One. 2011:6. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng D, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. International Journal of Clinical and Experimental Pathology. 2011;4:575–586. [PMC free article] [PubMed] [Google Scholar]
- 83.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 84.Kosaka N, et al. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. The Journal of biological chemistry. 2013;288:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vanpoucke G, et al. GATA-4 and MEF2C transcription factors control the tissue-specific expression of the alphaT-catenin gene CTNNA3. Nucleic Acids Res. 2004;32:4155–4165. doi: 10.1093/nar/gkh727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang M, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Molecular cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frankel LB, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. The Journal of biological chemistry. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 88.Zhu S, et al. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) The Journal of biological chemistry. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 89.Papagiannakopoulos T, et al. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer research. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 90.Liu LZ, et al. MiR-21 induced angiogenesis through AKT and ERK activation and HIF–1alpha expression. PLoS One. 2011;6:e19139. doi: 10.1371/journal.pone.0019139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Medina PP, et al. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 92.Valastyan S, et al. Activation of miR-31 function in already-established metastases elicits metastatic regression. Genes & development. 2011;25:646–659. doi: 10.1101/gad.2004211. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Valastyan S, et al. Concurrent suppression of integrin alpha5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer research. 2010;70:5147–5154. doi: 10.1158/0008-5472.CAN-10-0410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94.Valastyan S, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95.Bracken CP, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 96.Dykxhoorn DM, et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PloS one. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]