Abstract
Mitochondrial DNA (mtDNA) exists in multiple copies per cell and is essential for oxidative phosphorylation. Depleted or mutated mtDNA promotes numerous human diseases and may contribute to aging. Reduced TORC1 signaling in the budding yeast, Saccharomyces cerevisiae, extends chronological lifespan (CLS) in part by generating a mitochondrial ROS (mtROS) signal that epigenetically alters nuclear gene expression. To address the potential requirement for mtDNA maintenance in this response, we analyzed strains lacking the mitochondrial base-excision repair enzyme Ntg1p. Extension of CLS by mtROS signaling and reduced TORC1 activity, but not caloric restriction, was abrogated in ntg1Δ strains that exhibited mtDNA depletion without defects in respiration. The DNA damage response (DDR) kinase Rad53p, which transduces pro-longevity mtROS signals, is also activated in ntg1Δ strains. Restoring mtDNA copy number alleviated Rad53p activation and re-established CLS extension mtROS-mediated longevity signaling, indicating that Rad53p senses mtDNA depletion directly. Finally, DDR kinases regulate nucleus-mitochondria localization dynamics of Ntg1p. From these results, we conclude that the DDR pathway senses mtDNA instability and regulates Ntg1p in response. Furthermore, Rad53p senses multiple mitochondrial stresses in a hierarchical manner to elicit specific physiological outcomes, exemplified by mtDNA depletion overriding the ability of Rad53p to transduce an adaptive mtROS longevity signal.
Keywords: chronological lifespan, DNA damage response, mtDNA, Rad53p, reactive oxygen species
1. Introduction
Mitochondrial DNA (mtDNA) encodes essential proteins of the oxidative phosphorylation system and the non-coding RNAs needed for their translation in the mitochondrial matrix, making it critical for cellular energy production, redox homeostasis, and signaling (Greaves et al., 2012; Wallace, 2005). The mitochondrial genome is present in multiple copies in most eukaryotic cells, with some human cells containing up to 10,000 mtDNA molecules (Robin and Wong, 1988). The importance of this component of the human genome is manifest by the numerous diseases resulting from mtDNA point mutations, deletions, or depletion (Greaves et al., 2012; Wallace, 2005). Furthermore, single cells can harbor mixtures of wild-type and mutant mtDNA, and the relative amount of each (degree of heteroplasmy) influences disease phenotypes (Schon et al., 2012). The accumulation of mtDNA mutations and reduced copy number is also strongly associated with age and often precedes functional decline in cells and tissues that accompanies aging (Clay Montier et al., 2009; Cortopassi and Arnheim, 1990; Trifunovic et al., 2004). Cellular mechanisms that maintain mtDNA integrity may therefore represent key regulators of longevity and healthspan.
Maintenance of mtDNA represents a balance between replication, repair, segregation to daughter cells, and degradation. Mitochondria of the budding yeast, Saccharomyces cerevisiae, have a base-excision repair (BER) pathway and other overlapping systems for mtDNA maintenance (O'Rourke et al., 2002). Full yeast chronological lifespan, defined as viability in post-diauxic and stationary phases of growth (Longo et al., 2012), requires a fully intact mitochondrial BER system under stress conditions, highlighting the importance of mtDNA repair during yeast aging (Maclean et al., 2003). Several modes of yeast mtDNA replication have been proposed (Lipinski et al., 2010). During transcription-dependent replication, short transcripts derived from replication origins serve as primers for initiation by mtDNA polymerase γ, or Mip1p (Baldacci and Bernardi, 1982). Alternately, formation of a double-strand breaks (DSB) and subsequent DNA resection generates short single-stranded tails, which can hybridize within double-stranded regions and have been proposed to initiate rolling circle or recombination-mediated replication (Ling et al., 2013; Maleszka et al., 1991). Importantly, these later mechanisms faithfully duplicate the parental mtDNA into concatamers, promoting propagation of mtDNA molecules with identical sequence to decrease heteroplasmy (Ling et al., 2013). Although all of the above modes of replication contribute to maintenance of normal mtDNA copy number, conditions or signaling pathways that might promote one form of replication over others and consequences of this selection on mtDNA stability, mitochondria function, and aging remain largely unknown.
Several studies indicate that proteins involved in the nuclear DNA damage response (DDR) sense mtDNA stability and regulate mtDNA maintenance. During a canonical DDR in S. cerevisiae, the kinases Tel1p and Mec1p (homologs of ATM and ATR in mammals) initially sense DNA double-strand breaks or stalled replication forks and activate the effector kinase Rad53p (Chk2 in mammals) (Pellicioli and Foiani, 2005). Rad53p then initiates cell cycle delay, increased dNTP production, and expression of enzymes involved in DNA repair (Branzei and Foiani, 2006). In both budding yeast and mammalian cells, DDR kinases regulate mtDNA copy number and stability. For example, cultured fibroblasts from patients with Ataxia-Telangiectasia, a disease caused by mutations in the ATM gene, have reduced mtDNA copy number (Eaton et al., 2007). In contrast, activation of an ATM/CHK2 checkpoint increases mtDNA copy number but also increases the frequency of a common mtDNA deletion (Niu et al., 2012). In yeast, both cell cycle progression and dNTP levels, factors regulated by Rad53p, determine mtDNA copy number (Lebedeva and Shadel, 2007; Taylor et al., 2005). Furthermore, Mec1p regulates sumoylation of many proteins involved in DNA repair, which may influence their nuclear versus mitochondrial localization and repair activity (Cremona et al., 2012; Psakhye and Jentsch, 2012). Finally, loss of mtDNA activates a Rad53p-dependent cell cycle checkpoint and phosphorylation of Rad53p target proteins (Crider et al., 2012), indicating that communication between the mitochondrial genome and the DDR pathway is bi-directional.
In addition to sensing nuclear DNA damage and mtDNA maintenance, Rad53p transduces a mitochondrial ROS (mtROS) signal that can extend yeast chronological lifespan (CLS) (Schroeder et al., 2013). CLS measures viability in post-diauxic and stationary phases of yeast growth and models post-mitotic cellular aging in higher eukaryotes (Longo et al., 2012). Mitochondrial ROS adaptation is also a key aspect by which reduced signaling through the conserved Target of Rapamycin (TOR) pathway extends yeast CLS (Pan et al., 2011). Treatment with a sub-lethal dose of the redox-cycling compound menadione during the exponential growth phase generates mitochondrial matrix superoxide that initiates mtROS signaling and mimics the effects of tor1Δ on lifespan and ROS adaptation (Pan et al., 2011; Schroeder et al., 2013). One outcome of mtROS pro-longevity signaling is repression of subtelomeric gene expression mediated by the histone 3 lysine 36 (H3K36) demethylase Rph1p, which enhances heterochromatin formation at subtelomeres. These Rad53p-dependent epigenetic changes occur in the absence of canonical DNA damage response signaling. Rad53p therefore transduces both beneficial (mtROS) and detrimental (lack of mtDNA) mitochondrial signals to elicit either lifespan extension or cell cycle arrest, but how multiple signals might be integrated remains unknown. Additionally, given that complete loss of mtDNA represents a physiologically extreme circumstance that also induces extensive metabolic and transcriptional reprograming (Butow and Avadhani, 2004; Traven et al., 2001), it is not known how less severe mtDNA instability influences longevity. In this study, we used strains lacking the mitochondrial BER enzyme Ntg1p to examine the involvement of mtDNA copy number and stability in aging of budding yeast and potential intersections of different mitochondrial and nuclear signaling modes to Rad53p.
2. Materials and Methods
2.1. Yeast growth and chronological lifespan measurement
All yeast strains used in this study are derivatives of DBY2006 (MATa his3-Δ200 leu2-3,-112 ura3-52 trp1-Δ1 ade2-1) or BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and are listed in Table S1. Deletion strains were generated via gene replacement with URA3 or KanMX6 cassettes and transformed using the lithium acetate method. Ntg1p was tagged at the C-terminus with either GFP-KanMX6 or HA-KanMX6. Strains overexpressing Rnr1p were transformed with the plasmid pBAD71-RNR1 (2μ, URA3) (Lebedeva and Shadel, 2007) or pRS316 (CEN, URA3) as a control empty vector. Chronological lifespan was determined as described (Schroeder et al., 2013). For adaptive menadione treatment, saturated cultures from single colonies were diluted to an OD600 of 0.01 in 50 mL fresh minimal media and grown at 30°C, 200 RPM until the OD600 reached 0.5. Either menadione (50 μM final concentration for DBY2006 background, 80 μM final concentration for BY4742 background) or an equivalent volume of ethanol (no treatment control) was added, and cultures were grown until the OD600 reached 2.0, approximately 24 hours after inoculation. At this point, cells were pelleted and resuspended in media from an untreated parallel culture. For lifespan measurement under caloric restriction (CR), cells were grown in minimal media supplemented with essential amino acids and 0.5% glucose. CLS measurements were initiated 24 hours (day 1 of CLS) or 48 hours (day 2 of CLS) following adaptive menadione treatment and were analyzed in triplicate for all strains.
2.2. ROS measurements and oxygen consumption
Measurements of cellular superoxide were performed as described (Bonawitz et al., 2007; Bonawitz et al., 2006; Pan et al., 2011). Oxygen consumption was measured using a Clark electrode (SI130) paired with an oxygen meter (model 782, Stathkelvin Instruments). Respiration of one mL of cells at either OD600=0.5 or collected at day one of CLS (48 hours after inoculation) was measured as %oxygen/minute/OD600 for biological triplicate samples and normalized to the wild-type rate, which was set to one.
2.3. Microscopy
Yeast expressing NTG1-GFP at OD600=0.5 were visualized with an Olympus IX-71 inverted microscope. Prior to image acquisition, one mL of cells were stained with MitoTracker Red and DAPI for 15 minutes at 30°C at final concentrations of 100 nM and 5 ng/mL, respectively. Three microliters of 0.2% low melting point agarose in SD medium warmed to 50°C were pipetted onto glass slides and allowed to cool briefly before 3 μL of stained cells were added, mixed by pipetting, and covered with a glass coverslip. All images were acquired within 30 minutes of slide preparation using Metamorph software. Merged images were generated in Adobe Photoshop.
2.4. Subcellular fractionation
Mitochondria and nuclei were isolated from exponentially growing cells (OD600=0.5) as described (Daum et al., 1982). The crude nuclear pellet, isolated after the second round of dounce homogenization, was purified further by resuspension in Ficoll buffer and centrifugation as described (Mosley et al., 2009).
2.5. Western blotting
Yeast whole-cell extracts were prepared from cells at OD600=0.5 treated with 0.01% MMS or water (vehicle control) for 30 minutes (Keogh et al., 2006). Proteins were separated in 6% SDS-PAGE gels containing 100 μM PhosTAG (Wako Chemicals) and 100 μM MnCl2 to resolve phosphorylated forms of Rad53p or 15-8% gradient gels and transferred to PVDF membranes (Millipore Immobilon). Blocking and antibody incubations were carried out in TBS-T (10 mM Tris [pH 8.0], 150 mM NaCl, 0.05% Tween 20) containing 5% nonfat dry milk. Antibodies used were anti-Rad53p (Santa Cruz sc-6749, 1:500), anti-HA (Abcam ab9110, 1:1000), anti-histone 3 (Abcam ab1791, 1:10,000), and anti-porin (Invitrogen 459500, 1:1000). Primary antibodies were detected with appropriate HRP-conjugated secondary antibodies. GAPDH conjugated to HRP (Abcam ab9385, 1:1000) was used as a loading control. Immunoblotting was performed on protein isolated from two independent experiments. For Rad53p blots, the degree of the Rad53p band shift was approximated using ImageJ as described (Schroeder et al., 2013).
2.6. Mitochondrial DNA copy number determination
Total DNA was isolated from cells at OD600=0.5 and at day 1 of CLS (48 hours after inoculation) using the “smash and grab”method. DNA pellets were resuspended in 100 μL TE with 10μg/mL RNase and incubated at 65°C for one hour. DNA was then diluted 1:200 in water. Five microliters of diluted DNA was used in each qPCR reaction with 4.6 μL SYBR Green (Applied Biosystems) and 0.2 μL of each primer listed below (from 25 mM primer stocks):
Cox1 F: 5′-CTACAGATACAGCATTTCCAAGA-3′
Cox1 R: 5′-GTGCCTGAATAGATGATAATGGT-3′
Actin F 5′-GTATGTGTAAAGCCGGTTTTG-3′
Actin R 5′-CATGATACCTTGGTGTCTTGG-3′
PCR was performed using a Bio-Rad C1000 thermocycler/CFX96 RT system used the following program: 1 cycle of 3 min at 95°C, followed by 39 cycles of 30 seconds at 95°C, 30 seconds at 53°C, and 30 seconds at 72°C. Ct values were obtained with Bio-Rad CFX manger software.
2.7. Quantitative reverse transcription PCR
RNA was extracted using acid phenol and purified with Qiagen RNeasy columns as described (Bonawitz et al., 2008). RNA was then converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Agilent) according to the manufacturer's instructions. cDNA was diluted 1:10, and PCR was performed as described for copy number measurement using previously published primers (Schroeder et al., 2013).
2.8. Cell cycle analysis
Analysis of DNA content by propidium iodide staining and flow cytometry was performed as described (Zhang and Siede, 2004). Briefly, cultures were grown to early exponential growth phase (OD600=0.3) and 2 mL of cells were pelleted, washed once in water, and fixed in 1 mL 100% ethanol. Cells were then treated with RNase A and Proteinase K in sodium citrate buffer prior to staining with propidium iodide. Fluorescence in the FL2-A channel was immediately analyzed using a FACS Calibur. The percent of cells with 1N or 2N DNA content was determined using FloJo software.
2.9. Statistical analyses
All data points represent the mean of biological triplicate samples, and error bars represent the standard error of the mean. Statistical significance was determined using Student's t test. In all figures, “*” represents p ≤ 0.05, “**” represents p ≤ 0.01, and “n.s.”indicates “not significant.”
3. Results
3.1. Lack of the base-excision repair enzyme Ntg1p specifically prevents lifespan extension by adaptive mtROS signaling independently of changes to cellular respiration
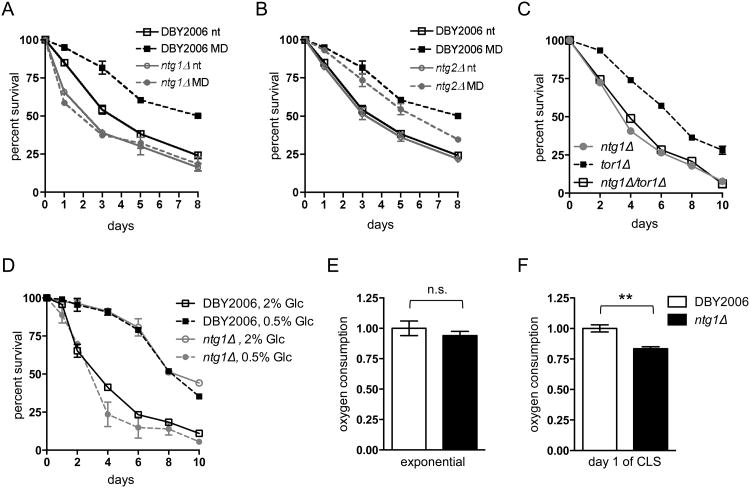
In order to examine the contribution of mtDNA maintenance to mtROS-mediated longevity, we generated yeast strains lacking the homologous DNA repair enzymes Ntg1p and Ntg2p. Both proteins are involved in base-excision repair (BER), the primary repair pathway for oxidative DNA damage (Alseth et al., 1999). Ntg1p and Ntg2p are bi-functional enzymes that excise damaged bases via N-glycosylase activity and process the resulting apurinic/apyrmidinic (AP) site, allowing further processing by a 3′ phosphodiesterase before DNA polymerase and DNA ligase complete BER (Eide et al., 1996; Meadows et al., 2003; You et al., 1999). Although Ntg1p and Ntg2p catalyze similar activities, Ntg1p contains both a mitochondrial-targeting sequence and a nuclear-localization signal, allowing it to function in both mitochondrial and nuclear DNA repair (Griffiths et al., 2009; Swartzlander et al., 2010). Ntg2p contains only a nuclear localization signal (Griffiths et al., 2009; Swartzlander et al., 2010). Importantly, strains lacking Ntg1p are respiratory competent and do not exhibit increased petite formation (a measure of mtDNA instability) compared to wild-type strains (Doudican et al., 2005; O'Rourke et al., 2002), allowing us to analyze the effects of mtDNA mutation or instability on lifespan independently of major changes to cellular respiration. We observed that an ntg1Δ strain exhibited a slightly reduced chronological lifespan (CLS) relative to wild-type (DBY2006) when grown in minimal media, in accordance with previously published CLS assays in which yeast were aged in water at elevated temperatures after growth in rich media (Maclean et al., 2003). Furthermore, the ntg1Δ strain did not exhibit extended CLS in response to elevated mtROS induced by menadione treatment (Fig. 1A). However, loss of NTG2 did not shorten lifespan or affect CLS extension following menadione treatment (Fig. 1B), suggesting that mitochondrial functions of Ntg1p are required to observe an adaptive mtROS longevity response. The requirement for NTG1 in mtROS adaptation was also observed in the BY4742 background (Fig. S1A), and was not influenced by the inability of the ntg1Δ strains to produce ROS in response to menadione treatment or by differences in basal ROS levels during exponential growth (Fig. S1B) (Doudican et al., 2005). Lifespan extension when TORC1 activity is reduced also requires mtROS signaling (Pan et al., 2011). Accordingly, we found that ntg1Δ completely eliminated CLS extension observed in a tor1Δ mutant strain (Fig. 1C). However, the ntg1Δ strain exhibited extended CLS under caloric restriction (CR, 0.5% glucose, Fig. 1D), demonstrating that loss of NTG1 does not generally limit the ability of lifespan to be extended. Given the different requirements for Ntg1p in mtROS- versus CR-mediated lifespan extension, we examined the effects of combined CR and menadione treatment on CLS. Menadione treatment did not further extend the median CLS of cells grown in 0.5% glucose, but combined treatments slightly reduced survival at late time points (Fig. S1C).
Figure 1. Lack of Ntg1p prevents adaptive mtROS-mediated lifespan extension and elicits mild and conditional effects on respiration.
A. Chronological lifespan (CLS) analysis of wild-type DBY2006 and ntg1Δ following treatment with 50 μM menadione (MD) or ethanol (nt) during exponential growth. In all graphs, data points represent the average viability of three biological replicates and error bars represent the standard error of the mean. B. CLS of wild-type and ntg2Δ as in A. C. CLS of wild-type, tor1Δ, and ntg1Δ/tor1Δ. D. CLS of wild-type and ntg1Δ grown in 20% glucose (nt) or 0.5% glucose (CR). E-F. Analysis of mitochondrial oxygen consumption in wild-type DBY2006 and ntg1Δ strains during exponential growth (OD=0.5, E) and at day one of CLS (48 hours after inoculation, F). Oxygen consumption was measured as %O2/min/OD600, and the wild-type values were set to one. In all graphs, data points represent the mean of three biological replicates and error bars the SEM.
Since we showed previously that CLS extension in tor1Δ strains requires mitochondrial respiration (Bonawitz et al., 2007), we speculated that ntg1Δ might be blocking lifespan extension by altering respiratory rates. Although loss of NTG1 promotes oxidative damage to mtDNA and slightly increases point mutation frequency, ntg1Δ does not enhance petite formation, suggesting that sufficient copies of wild-type mtDNA are present to support respiration (Doudican et al., 2005; O'Rourke et al., 2002). Consistent with this, we observed no significant growth differences between wild-type and ntg1Δ strains in respiratory (glycerol) medium, using the yeast strain lacking ABF2, a protein required for mtDNA packaging, inheritance, and expression as a positive control for slow growth due to respiration deficiency (Zelenaya-Troitskaya et al., 1998) (Fig. S1 D and E). Furthermore, loss of NTG1 did not affect cellular oxygen consumption during exponential growth under the CLS conditions used in our laboratory (Fig. 1E), and only reduced oxygen consumption at day one of CLS to 80% of the wild-type respiration rate (Fig. 1F). The observed respiration rate in ntg1Δ in stationary phase is well above the approximate 40% threshold documented to influence yeast CLS (Ocampo et al., 2012), suggesting that these subtle alterations in mitochondrial respiration do not account for the inability of ntg1Δ strains to extend CLS.
3.2. Depletion of mtDNA in ntg1Δ strains activates Rad53p checkpoint signaling
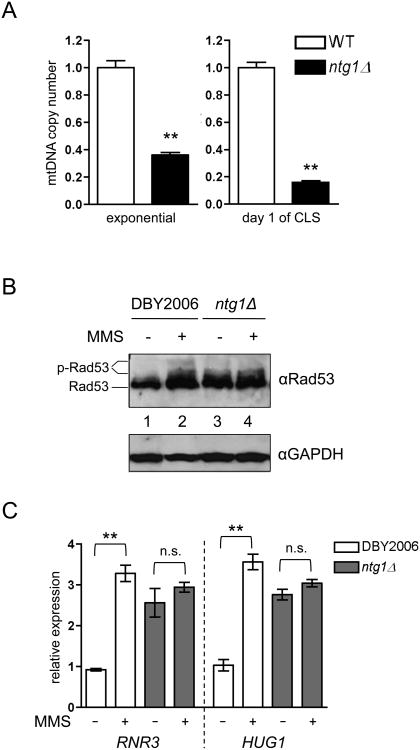
Given the minor effects on mitochondrial respiration, we sought alternate consequences of ntg1Δ that might account for the observed deficiencies in lifespan extension. In addition to its function in BER, Ntg1p has been proposed to initiate rolling-circle mtDNA replication by inducing double-strand breaks at sites of oxidative DNA damage within the replication origin ori5, and is required for mtDNA copy number elevation following hydrogen peroxide treatment (Hori et al., 2009). Consistent with a role in mtDNA replication or maintenance, we observed that ntg1Δ strains had reduced mtDNA copy number during exponential growth, which was exacerbated as cells underwent chronological aging (Fig. 2A). However, the degree of mtDNA depletion observed in ntg1Δ does not promote significant changes in cellular respiration (Fig. 1 E-F), suggesting that an alternate event downstream of mtDNA depletion is blocking mtROS-mediated CLS extension. The complete absence of mtDNA was recently shown to induce Rad53p-dependent cell cycle arrest and phosphorylation of downstream targets (Crider et al., 2012). Adaptive mtROS also induce phosphorylation and activation of Rad53p to elicit outcomes distinct from those associated with a DNA damage response (Schroeder et al., 2013). To examine potential crosstalk between these signals, we examined Rad53p phosphorylation by western blot in wild-type and ntg1Δ strains with and without exposure to the DNA-damaging agent methylmethane sulfanote (MMS). In ntg1Δ strains, Rad53p phosphorylation was amplified basally and did not exhibit additional phosphorylation upon MMS treatment (Fig. 2B and S2A). Additionally, Rad53p was phosphorylated in strains lacking ABF2, which results in mtDNA depletion and an increased propensity to form rho° petite cells (Lebedeva and Shadel, 2007) (Fig. S2 B and C). Strains lacking ABF2 also fail to extend CLS following menadione-induced adaptive mtROS signaling (Fig. S2D). DNA damage activates Rad53p to induce transcription of many genes required for cell cycle regulation, nucleotide biosynthesis, and DNA repair (Branzei and Foiani, 2006). Accordingly, MMS strongly induced expression of the Rad53p-regulated genes RNR3 and CLN2 (Huang et al., 1998; Travesa et al., 2012) in wild-type cells (Fig. 2C). However, RNR3 and CNL2 transcripts were elevated basally in an ntg1Δ strain and were not elevated further upon MMS treatment, mirroring Rad53p phosphorylation in this strain.
Figure 2. Lack of Ntg1p causes mtDNA depletion and Rad53p activation.
A. Analysis of mtDNA copy number in wild-type DBY2006 and ntg1Δ during exponential growth (OD=0.5) and at day one of CLS (48 hours after inoculation). The copy number of the wild-type samples was set to one. B. Western blot of Rad53p phosphorylation in wild-type DBY2006 and ntg1Δ strains treated with 0.01% methylmethane sulfate (MMS, +) or water (-) for 30 minutes during exponential growth. C. Expression of two Rad53p-regulated genes in wild-type DBY2006 and ntg1Δ strains treated with MMS as described in B.
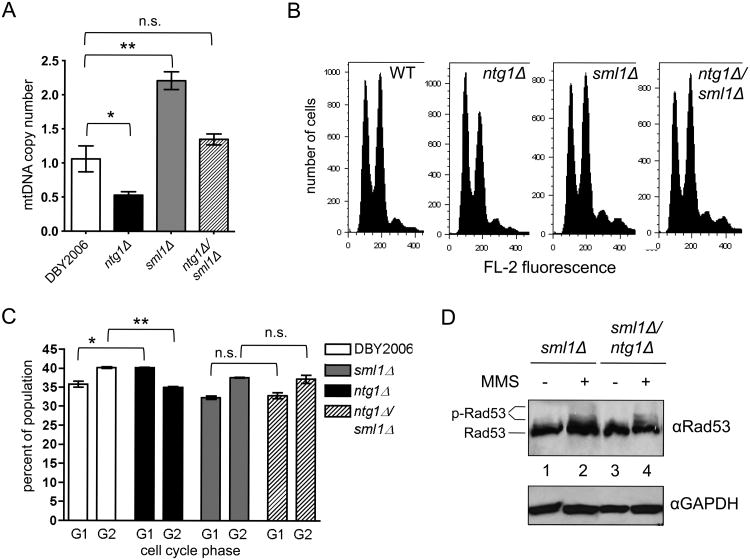
To determine if ntg1Δ was impacting Rad53p activity via mtDNA depletion, we restored mtDNA copy number in this strain by elevating free dNTP pools. Over-expression of RNR1, the large subunit of ribonucleotide reductase, or deletion of SML1, a repressor of RNR activity (Zhao et al., 1998), increased mtDNA copy number by about two fold in wild-type yeast and restored the low copy number of ntg1Δ strains to approximately wild-type levels (Fig. 3A and Fig. S3A) (Lebedeva and Shadel, 2007; Taylor et al., 2005). Analysis of cell cycle progression as a readout of active Rad53p signaling in the ntg1Δ strain revealed a slight G1 delay relative to the wild-type (Fig. 3B and C), consistent with the increased CLN2 expression observed in the ntg1Δ strain in Figure 2C. However, ntg1Δ/sml1Δ cells displayed cell cycle profiles similar to those of the wild-type and sml1Δ single-mutant strains, indicating that sml1Δ prevents Rad53p-dependent cell cycle checkpoint activation in the ntg1Δ strain. Finally, deletion of SML1 reduced the basal level of Rad53p phosphorylation in the ntg1Δ background and restored the phosphorylation response to MMS treatment (Fig. 3D and S3B). Together, these data indicate that mtDNA depletion in ntg1Δ strains induces Rad53p phosphorylation and downstream cell cycle arrest, while blocking DNA damage-dependent activation of Rad53p.
Figure 3. Restoring mtDNA copy number in ntg1Δ alleviates Rad53p activation.
A. Mitochondrial DNA copy number in wild-type DBY2006, ntg1Δ, sml1Δ, and ntg1Δ/sml1Δ measured at OD=0.5. B. Cell cycle analysis by FACS of asynchronous cultures in early exponential growth (OD=0.2-0.3). The first peak indicates cells in G1, and the second indicates cells in G2. Representative histograms of three biological replicates are shown C. Quantification of cell cycle profiles determined by FACS. D. Western blot of Rad53p phosphorylation in sml1Δ and ntg1Δ/sml1Δ strains treated with 0.01% methylmethane sulfate (MMS, +) or water (-) for 30 minutes during exponential growth.
3.3. A mtDNA copy number threshold supports adaptation to mtROS
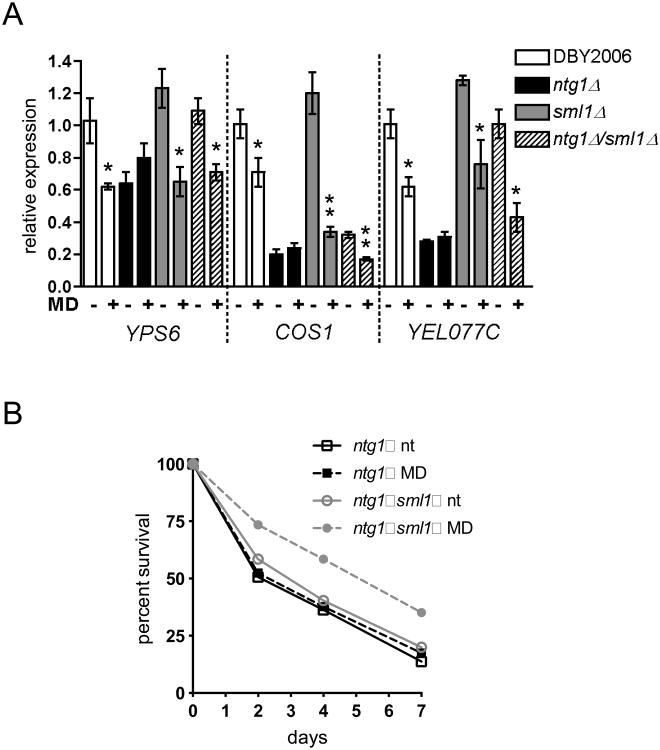
Since the results so far demonstrate mtDNA depletion activates Rad53p checkpoint signaling, we hypothesized that activation of this mode of signaling in ntg1Δ strains prevents Rad53p from transducing a pro-longevity mtROS signal. To test this hypothesis, we examined CLS extension and subtelomeric silencing (Schroeder et al., 2013) following mtROS signaling in ntg1Δ strains with or without mtDNA depletion restored by RNR manipulation. We measured transcriptional repression of three subtelomeric genes in wild-type, ntg1Δ, sml1Δ and ntg1Δ/sml1Δ twenty-four hours after menadione treatment during exponential growth. The ntg1Δ strain failed to repress transcription of all three genes in response to menadione treatment, and for COS1 and YEL077C, gene expression was reduced in the absence of menadione treatment (Fig. 4A). The sml1Δ strain exhibited a transcriptional response comparable to wild-type. For all genes examined, combined deletion of SML1 and NTG1 restored mtROS-dependent repression of subtelomeric transcripts, and in the case of YEL077C restored the basal expression to wild-type levels. Finally, both sml1Δ and RNR1 overexpression rescued CLS extension following menadione treatment in the ntg1Δ strain background (Fig. 4B and Fig. S3C). We therefore conclude that reduced mtDNA copy number blocks the ability of mtROS to extend CLS via convergent signaling to the DNA damage response kinase Rad53p.
Figure 4. An mtDNA copy number threshold supports mtROS adaptation and longevity.
A. RT-PCR analysis of subtelomeric genes at day one of CLS, or 24 hours after treatment with menadione (MD+) or ethanol (-) during exponential growth. Statistically significant changes between ethanol treated and menadione treated samples are indicated. B. CLS of ntg1Δ and ntg1Δ/sml1Δ following treatment with menadione (MD) or ethanol (nt) during exponential growth.
3.4. DNA damage response kinases regulate the subcellular localization of Ntg1p
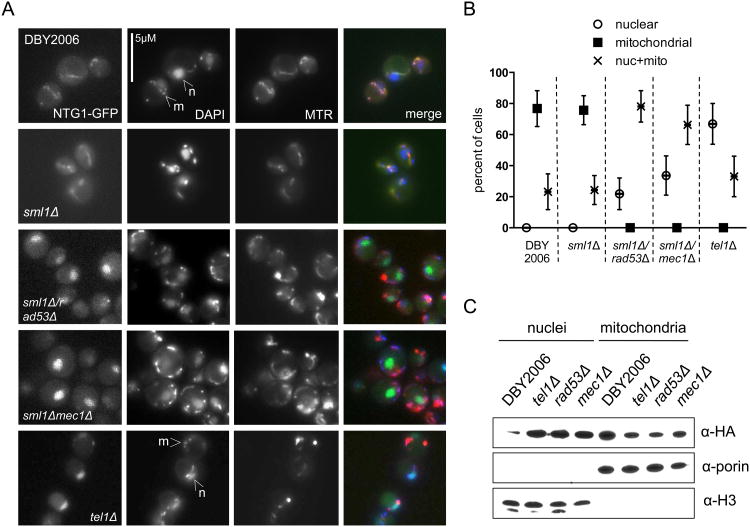
Multiple lines of evidence suggest that the DNA damage response senses mtDNA loss and depletion. However, this signaling pathway also regulates mtDNA copy number. Ntg1p can dynamically localize to both nuclei and mitochondria, presumably to facilitate BER where oxidative DNA damage is expected or experienced (Griffiths et al., 2009). We showed here that Ntg1p also maintains mtDNA copy number, and speculated that altered localization of this protein, possibly regulated by DDR kinases, might represent an additional level at which the DDR influences mtDNA copy number. To determine how the major DDR kinases affect the abundance of Ntg1p in mitochondria or nuclei, we examined localization of Ntg1p-GFP in strains lacking RAD53, MEC1, and TEL1. The rad53Δ and mec1Δ strains also lacked SML1 to suppress the lethality of these deletions (Zhao et al., 1998). During exponential growth in the wild-type and sml1Δ strains, Ntg1-GFP primarily co-localized with a mitochondrial marker. However, deletion of any DDR kinase increased the nuclear localization of Ntg1p (Fig. 5A and B). Similarly, subcellular fractionation of strains lacking DDR kinases demonstrated increased nuclear and decreased mitochondrial abundance of Ntg1p-HA relative to the wild-type control (Fig. 5C). Together, these results demonstrate that, in the absence of a DNA-damage signal, an intact DDR signaling pathway is required to promote mitochondrial localization of Ntg1p. The predominant nuclear localization of this protein in cells lacking Mec1p, Tel1p, or Rad53p may contribute the observed mitochondrial genomic instability in these backgrounds (Lebedeva and Shadel, 2007; Taylor et al., 2005).
Figure 5. DNA damage response kinases regulate Ntg1p localization.
A. Fluorescence microscopy of Ntg1p-GFP in strains lacking DNA damage response kinases during exponential growth. Mitochondria were visualized with MitoTracker Red (MTR), and DAPI staining identifies nuclear DNA (indicated by arrows labeled “n”) and mitochondrial DNA nucleoids (“m”). B. Quantification of Ntg1-GFP mitochondrial and nuclear co-localization. The percentage of cell with exclusively nuclear GFP (open circles), exclusively mitochondrial (filled squares), or mixed nuclear and mitochondrial (“X”) localization was determined by counting at least 100 cells for each condition from two experiments. Data points represent the mean and error bars the range of percentages. C. Western blot of nuclear and mitochondrial fractions of the indicated strains expressing Ntg1p-HA. Rad53Δ and mec1Δ are also lacking SML1. Porin and Histone 3 serve as mitochondrial and nuclear loading controls, respectively.
4. Discussion
Impaired mtDNA maintenance correlates with aging and may precipitate age-related disease onset and tissue functional decline (Greaves et al., 2012; Wallace, 2005). Although there is strong evidence that mtDNA maintenance supports healthy lifespan, few studies have investigated the role of mitochondrial genome integrity or copy number in longevity. Here, we show that the base excision repair enzyme Ntg1p, previously shown to initiate mtDNA replication following oxidative stress (Hori et al., 2009), maintains mtDNA copy number under normal laboratory growth conditions. Reduced mtDNA copy number in ntg1Δ strains blocks lifespan extension and adaptive responses to mtROS signaling (Fig. 4). Both mtROS signaling (Schroeder et al., 2013) and reduced copy number activate the DDR kinase Rad53p. However, our results show that Rad53p activation by mtDNA depletion blocks the ability of this kinase to respond to either nuclear DNA damage or mtROS signaling. Loss of NTG1 slightly decreases CLS in a wild-type strain background when analyzed in minimal media (Fig. 1A) or after initial growth in rich media followed by aging in water at elevated temperatures (Maclean et al., 2003). The modest effect of ntg1Δ on lifespan suggests that aberrant activation of Rad53p, rather than some significant functional consequence of mtDNA depletion, blocks mtROS-mediated lifespan extension. Rad53p is therefore a key convergence point for multiple mitochondrial and nuclear stress signals, and orchestrates hierarchical and specific cellular responses to these conditions.
Several mechanisms of mtDNA replication have been proposed, but little is known about the relative contribution of each mechanism under various growth or stress conditions. One exception is the recent finding that Ntg1p might initiate rolling-circle replication in response to oxidative mtDNA damage by inducing a DSB at the site of damage (Hori et al., 2009). Our findings suggest an important role for Ntg1p in copy number maintenance during normal growth and chronological aging. Due to its close proximity to the electron transport chain, mtDNA experiences a high volume of oxidative DNA damage (Richter et al., 1988). We propose that physiological amounts of ROS are sufficient to initiate Ntg1p-mediated replication to sustain mtDNA copy number throughout exponential growth and especially into stationary phase, where ROS levels increase and mitochondrial function becomes essential (Werner-Washburne et al., 1993). Alternately, lack of Ntg1p slightly increases mtDNA oxidative damage (Doudican et al., 2005), and damaged mtDNA molecules may be degraded in the mitochondrial matrix or removed by autophagy, resulting in the observed mtDNA depletion. Lifespan extension in tor1Δ strains requires elevated mtROS, and we show that ntg1Δ blocks CLS extension in this background (Fig. 1C). Although we believe this is largely due to Rad53p activation by mtDNA depletion, Ntg1p may also regulate mtDNA replication and mitochondria biogenesis downstream of elevated mtROS or another aspect of TORC1 signaling to support CLS extension (Bonawitz et al., 2007; Pan and Shadel, 2009). In contrast, ntg1Δ does not alter lifespan extension in response to caloric restriction, suggesting either that mtDNA copy number maintenance is less critical for CLS extension under these conditions or that copy number is regulated by an Ntg1p-independent mechanism. The effects of combined menadione and CR on yeast CLS (Fig. S1C) indicates potentially complex interactions between these two mechanisms of lifespan extension. Increased respiratory rates and ROS levels during exponential growth have been observed under CR (Goldberg et al., 2009; Lin et al., 2002) suggesting that mtROS signaling may contribute to CR-mediated lifespan extension. However, respiratory thresholds and altered metabolism, which are known to determine CLS extension under CR (Ocampo et al., 2012), may occur independent of mtROS signaling. Additional experiments will be required to fully understand the many facets of CR and mtROS signaling, including pathways that regulate mtDNA copy number, that influence longevity.
Complete lack of mtDNA was previously shown to activate Rad53p-dependent cell cycle arrest and protein phosphorylation (Crider et al., 2012), and our results demonstrate that Rad53p is also sensitive to less severe reductions in mtDNA copy number that do not impact mitochondrial respiration. The mechanism by which Rad53p senses mtDNA content remains unclear. During a canonical DNA-damage response, Tel1p and Mec1p sense nuclear DNA damage and activate Rad53p. Both upstream kinases have been identified in mitochondrial fractions in proteomic studies (Sickmann et al., 2003), suggesting that they may also communicate mitochondrial genome integrity to Rad53p. Alternately, these proteins may monitor mitochondrial processes that are sensitive to mtDNA levels. For example, mild mtDNA depletion may affect the efficiency of respiratory complex assembly or function, resulting in subtle effects on respiration that signal to sensor proteins. As the list of cytosolic and nuclear proteins involved in mitochondria signaling grows, it will be important to determine the initiating and intermediate mitochondrial outputs that directly regulate sensor protein activity or signaling.
Depletion of mtDNA, mtROS signaling, and nuclear DNA damage all activate Rad53p, but with different consequences for cellular longevity. Several hypotheses could explain how these diverse stimuli achieve unique outcomes despite converging on the same effector protein. First, different kinases upstream of Rad53p could sense specific stresses, as Tel1p activates Rad53p during mtROS signaling (Schroeder et al., 2013). Second, varying degrees of Rad53p phosphorylation under each condition suggests that unique phosphorylation sites or combinations of sites may be targeted in response to different stimuli. These different phosphorylation profiles may alter the ability of Rad53p to activate or inhibit downstream proteins, supporting the different outcomes of Rad53p signaling under mtDNA depletion, mtROS signaling, and nuclear DNA damage (Lee et al., 2003). The mammalian homolog of Rad53p, Chk2, is also phosphorylated at multiple residues in a DNA damage or stress-dependent manner to facilitate activation of specific downstream targets or binding to other checkpoint proteins (Stracker et al., 2009). Detailed characterization of stress-specific, post-translational modifications of Rad53p and Chk2 would provide insight into how these homologous proteins integrate diverse stimuli into specific responses that influence genomic stability, cellular transformation, and lifespan.
Although Rad53p is activated in response to mtDNA depletion in ntg1Δ strains, it and the DDR kinases Tel1p and Mec1p also regulate Ntg1p localization, with potential consequences on mtDNA replication or repair. How these proteins cooperate to determine Ntg1p localization is not yet clear. Many DNA repair proteins, including Ntg1p, are sumoylated following DNA damage, and absence of Mec1p causes these proteins to retain higher levels of sumoylation even in the absence of damage (Cremona et al., 2012). Furthermore, sumoylated Ntg1p accumulates in nuclear fractions (Griffiths et al., 2009). We propose that a functional DDR pathway maintains a pool of unmodified Ntg1p, promoting its mitochondrial localization. Additional characterization of Ntg1p modification in tel1Δ, rad53Δ, and mec1Δ strains would describe possible cooperation among the DDR kinases to regulate Ntg1p localization and mtDNA repair or replication activity.
Together, these findings support complex functions for DDR kinases as both sensors and regulators of mitochondrial genome integrity. In budding yeast, the DDR is at the nexus of mitochondria stress, mtDNA stability, and nuclear genome integrity, and coordinates these diverse signals into specific DNA repair, cell cycle delay, or pro-longevity outcomes. Future studies to investigate the conservation of these sensing and regulatory functions in the mammalian DDR should elucidate how this essential pathway integrates multiple stresses and signals to determine health, disease and lifespan.
Supplementary Material
Highlights.
Lack of the BER enzyme Ntg1p causes mtDNA depletion without respiration defects
mtDNA depletion induces Rad53p phosphorylation and cell cycle checkpoint activation
mtDNA depletion signaling to Rad53p overrides DNA damage and ROS longevity signaling
Rad53p senses and integrates mitochondrial and nuclear stress signals
Acknowledgments
This work was supported by grants W911NF-11-1-0376 from the Army Research Office and R01 AG047632 from the NIH to G.S.S, and NIH pre-doctoral fellowship F31 AG043242 to E.A.S. We thank Drs. Megan King, Paul Doetsch, and Daniel Swartzlander for insightful discussions and Dr. Patrick Lusk for reagents used in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alseth I, Eide L, Pirovano M, Rognes T, Seeberg E, Bjoras M. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Molecular and cellular biology. 1999;19:3779–3787. doi: 10.1128/mcb.19.5.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldacci G, Bernardi G. Replication origins are associated with transcription initiation sequences in the mitochondrial genome of yeast. The EMBO journal. 1982;1:987–994. doi: 10.1002/j.1460-2075.1982.tb01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Wearn CM, Shadel GS. Expression of the rDNA-encoded mitochondrial protein Tar1p is stringently controlled and responds differentially to mitochondrial respiratory demand and dysfunction. Curr Genet. 2008;54:83–94. doi: 10.1007/s00294-008-0203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Rodeheffer MS, Shadel GS. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Molecular and cellular biology. 2006;26:4818–4829. doi: 10.1128/MCB.02360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. The Rad53 signal transduction pathway: Replication fork stabilization, DNA repair, and adaptation. Exp Cell Res. 2006;312:2654–2659. doi: 10.1016/j.yexcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Clay Montier LL, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics. 2009;36:125–131. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, Zhao X. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol Cell. 2012;45:422–432. doi: 10.1016/j.molcel.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crider DG, Garcia-Rodriguez LJ, Srivastava P, Peraza-Reyes L, Upadhyaya K, Boldogh IR, Pon LA. Rad53 is essential for a mitochondrial DNA inheritance checkpoint regulating G1 to S progression. J Cell Biol. 2012;198:793–798. doi: 10.1083/jcb.201205193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Bohni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Doudican NA, Song B, Shadel GS, Doetsch PW. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Molecular and cellular biology. 2005;25:5196–5204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton JS, Lin ZP, Sartorelli AC, Bonawitz ND, Shadel GS. Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. J Clin Invest. 2007;117:2723–2734. doi: 10.1172/JCI31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide L, Bjoras M, Pirovano M, Alseth I, Berdal KG, Seeberg E. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. P Natl Acad Sci USA. 1996;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AA, Bourque SD, Kyryakov P, Gregg C, Boukh-Viner T, Beach A, Burstein MT, Machkalyan G, Richard V, Rampersad S, et al. Effect of calorie restriction on the metabolic history of chronologically aging yeast. Experimental gerontology. 2009;44:555–571. doi: 10.1016/j.exger.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Greaves LC, Reeve AK, Taylor RW, Turnbull DM. Mitochondrial DNA and disease. J Pathol. 2012;226:274–286. doi: 10.1002/path.3028. [DOI] [PubMed] [Google Scholar]
- Griffiths LM, Swartzlander D, Meadows KL, Wilkinson KD, Corbett AH, Doetsch PW. Dynamic compartmentalization of base excision repair proteins in response to nuclear and mitochondrial oxidative stress. Molecular and cellular biology. 2009;29:794–807. doi: 10.1128/MCB.01357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A, Yoshida M, Shibata T, Ling F. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res. 2009;37:749–761. doi: 10.1093/nar/gkn993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- Lebedeva MA, Shadel GS. Cell cycle- and ribonucleotide reductase-driven changes in mtDNA copy number influence mtDNA Inheritance without compromising mitochondrial gene expression. Cell Cycle. 2007;6:2048–2057. doi: 10.4161/cc.6.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Schwartz MF, Duong JK, Stern DF. Rad53 phosphorylation site clusters are important for Rad53 regulation and signaling. Molecular and cellular biology. 2003;23:6300–6314. doi: 10.1128/MCB.23.17.6300-6314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Ling F, Hori A, Yoshitani A, Niu R, Yoshida M, Shibata T. Din7 and Mhr1 expression levels regulate double-strand-break-induced replication and recombination of mtDNA at ori5 in yeast. Nucleic acids research. 2013;41:5799–5816. doi: 10.1093/nar/gkt273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski KA, Kaniak-Golik A, Golik P. Maintenance and expression of the S. cerevisiae mitochondrial genome--from genetics to evolution and systems biology. Biochimica et biophysica acta. 2010;1797:1086–1098. doi: 10.1016/j.bbabio.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and Chronological Aging in Saccharomyces cerevisiae. Cell Metab. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean MJ, Aamodt R, Harris N, Alseth I, Seeberg E, Bjoras M, Piper PW. Base excision repair activities required for yeast to attain a full chronological life span. Aging Cell. 2003;2:93–104. doi: 10.1046/j.1474-9728.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- Maleszka R, Skelly PJ, Clark-Walker GD. Rolling circle replication of DNA in yeast mitochondria. The EMBO journal. 1991;10:3923–3929. doi: 10.1002/j.1460-2075.1991.tb04962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows KL, Song B, Doetsch PW. Characterization of AP lyase activities of Saccharomyces cerevisiae Ntg1p and Ntg2p: implications for biological function. Nucleic Acids Res. 2003;31:5560–5567. doi: 10.1093/nar/gkg749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Florens L, Wen Z, Washburn MP. A label free quantitative proteomic analysis of the Saccharomyces cerevisiae nucleus. J Proteomics. 2009;72:110–120. doi: 10.1016/j.jprot.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu R, Yoshida M, Ling F. Increases in mitochondrial DNA content and 4977-bp deletion upon ATM/Chk2 checkpoint activation in HeLa cells. PLoS One. 2012;7:e40572. doi: 10.1371/journal.pone.0040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke TW, Doudican NA, Mackereth MD, Doetsch PW, Shadel GS. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Molecular and cellular biology. 2002;22:4086–4093. doi: 10.1128/MCB.22.12.4086-4093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell metabolism. 2012;16:55–67. doi: 10.1016/j.cmet.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Shadel GS. Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging. 2009;1:131–145. doi: 10.18632/aging.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Foiani M. Signal transduction: how rad53 kinase is activated. Curr Biol. 2005;15:R769–771. doi: 10.1016/j.cub.2005.08.057. [DOI] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. P Natl Acad Sci USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin ED, Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 1988;136:507–513. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nature reviews Genetics. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder EA, Raimundo N, Shadel GS. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 2013;17:954–964. doi: 10.1016/j.cmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Usui T, Petrini JH. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 2009;8:1047–1054. doi: 10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzlander DB, Griffiths LM, Lee J, Degtyareva NP, Doetsch PW, Corbett AH. Regulation of base excision repair: Ntg1 nuclear and mitochondrial dynamic localization in response to genotoxic stress. Nucleic Acids Res. 2010;38:3963–3974. doi: 10.1093/nar/gkq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SD, Zhang H, Eaton JS, Rodeheffer MS, Lebedeva MA, O'Rourke TW, Siede W, Shadel GS. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:3010–3018. doi: 10.1091/mbc.E05-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A, Wong JM, Xu D, Sopta M, Ingles CJ. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial dna mutant. The Journal of biological chemistry. 2001;276:4020–4027. doi: 10.1074/jbc.M006807200. [DOI] [PubMed] [Google Scholar]
- Travesa A, Kuo D, de Bruin RA, Kalashnikova TI, Guaderrama M, Thai K, Aslanian A, Smolka MB, Yates JR, 3rd, Ideker T, et al. DNA replication stress differentially regulates G1/S genes via Rad53-dependent inactivation of Nrm1. EMBO J. 2012;31:1811–1822. doi: 10.1038/emboj.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun E, Johnston GC, Singer RA. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You HJ, Swanson RL, Harrington C, Corbett AH, Jinks-Robertson S, Senturker S, Wallace SS, Boiteux S, Dizdaroglu M, Doetsch PW. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 1999;38:11298–11306. doi: 10.1021/bi991121i. [DOI] [PubMed] [Google Scholar]
- Zelenaya-Troitskaya O, Newman SM, Okamoto K, Perlman PS, Butow RA. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Siede W. Analysis of the budding yeast Saccharomyces cerevisiae cell cycle by morphological criteria and flow cytometry. Methods Mol Biol. 2004;241:77–91. doi: 10.1385/1-59259-646-0:77. [DOI] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.