Abstract
Background
Success in treating hepatitis B virus (HBV) infection with nucleoside analogues drugs is limited by the emergence of drug-resistant viral strains upon prolonged therapy. In addition to mutation patterns in the viral polymerase gene, host factors are assumed to contribute to failure of treatment in chronic HBV infections. The aim of this study was to analyze the correlation between efficacy of antiviral therapy and the prevalence of HBV pretreatment drug-resistant variants. We also analyzed the role of heterogeneity in the promoter region of the IL-10 on the HBV pol/s gene polymorphisms and efficacy of analogues-driven therapy.
Material/Methods
HBV DNA was extracted from 54 serum samples from chronic hepatitis B (CHB) patients. Drug-resistance mutations were analyzed using MALDI-TOF mass spectrometry technology (MALDI-TOF MS) and Multi-temperature single-strand conformation polymorphism (MSSCP). IL-10 gene promoter region polymorphisms at positions −1082, −819, and −592 were determined in allele-specific PCR reactions (AS-PCR).
Results
Drug-resistance mutations were detected in 74% of naïve and 93% of experienced patients, but the effect of pre-existence of drug-resistant HBV variants on antiviral therapy was not statistically significant (p=0.86). The role of polymorphisms at positions −1082 (p=0.88), −819 (p=0.26), and −592 (p=0.26) of IL-10 promoter region polymorphisms was excluded from the response-predicting factors. The main host factors predicting successful response to antiviral therapy were female sex (p=0.007) and young age (p=0.013).
Conclusions
The presence of drug-resistant HBV variants in baseline is not a viral predictor of good response to nucleoside/nucleotide analogues therapy. Only low HBV viral load predicted positive response to antiviral therapy. The ideal candidate for antiviral therapy is an immunocompetent, young female with low HBV viral load and elevated ALT activity.
Keywords: Polymorphism, Genetic, Drug Resistance, Interleukin-10, Mass Spectrometry, Hepatitis B, Chronic
Background
Hepatitis B virus (HBV) infection, affecting about 2 billion people worldwide, is still a global health problem. Despite the existence of a highly effective vaccine, 240–350 million chronic HBV carriers could develop chronic hepatitis B (CHB) with high risk of liver cirrhosis and hepatocellular carcinoma (HCC) [1–4]. The primary treatment goal in CHB patients is the prevention of liver disease progression. Life-long suppression of HBV replication seems to be the best antiviral strategy [4–6]. However, the existence of covalently closed circular DNA (cccDNA) in infected hepatocytes probably makes the eradication of HBV infection impossible [7,8].
Nucleotide/nucleoside analogues (NAs) and peg-interferon alpha (PegIFNα2a) are 2 major classes of antiviral drugs approved for CHB treatment. NAs agents (e.g., lamivudine, telbivudine, entecavir, adefovir, and tenofovir) can suppress viral load, but long-term therapy can lead to the selection of drug-resistant HBV variants [4,9]. Recent studies showed that there is a possibility of the pre-existence of natural resistance mutations in treatment-naïve patients [10–12]. Viral quasispecies evolve over time under selective pressure (e.g., antiviral therapy). These variants are more viable and spread more rapidly in the liver than does wild-type HBV [1,6,13].
Early detection of drug-resistant HBV variants is crucial for patients treated with low genetic barrier drugs [14,15]. Several methods are currently available to monitor HBV drug resistance (direct sequencing of PCR products, restriction fragment length polymorphism – RFLP, and mutation-specific real-time PCR) but reverse hybridization – Line Probe Assay is the most popular. Among these methods, MSSCP assay (multi-temperature single-strand conformation polymorphism) and MALDI-TOF MS (matrix-assisted laser desorption ionization time of flight mass spectrometry) are the most sensitive, being able to detect HBV mutants, which constitute only 1% of the viral population [16–19].
Development of HBV mutants is also related to the persistence of cccDNA and host factors affecting immune response [13,20]. Polymorphic sites within the IL-10 gene promoter region at positions −1082, −819, and −592 have been described [21–23]. It was published that certain polymorphisms within the IL-10 gene promoter region are associated with the development of chronic HBV infection and effects of IFNα therapy [21,24]. However, the impact of these polymorphisms on the development on drug-resistant HBV strains and response to NAs therapy has not been determined.
The aim of this study was to analyze the correlation between efficacy of antiviral therapy and the prevalence of HBV pretreatment drug-resistant variants. Moreover, the role of heterogeneity in the promoter region of the IL-10 on the HBV pol/s gene polymorphisms and efficacy of analogues-driven therapy was analyzed.
Material and Methods
Study population and sample design
We enrolled into the study 54 consecutive CHB (29 male and 25 female) patients, mean age 48.7±2 yrs, qualified between January 2011 and June 2012 to antiviral therapy at the Department of Infectious Diseases, Medical University of Gdansk. They were qualified and received treatment accordingly to recommendations of the Polish National Health Service (NFZ). These recommendations require the use of peg-IFNα2a or lamivudine (when cytokines are contradicted) as a primary therapy in HBeAg-negative subjects. A pretreatment HBV drug resistance test is required in analogues therapy, and the detection of lamivudine-resistant strains allows use of entecavir, adefovir, or tenofovir. In HBeAg-reactive patients, entecavir or tenofovir is recommended as a first-line therapy.
The majority of subjects received lamivudine – 30/54 (55%), entecavir 16/54 (30%), tenofovir 7/54 (13%), and combined therapy (lamivudine with adefovir) in 1/54 (2%). The therapy effect was assessed 48 weeks after NAs administration, accordingly to the NFZ recommendation. Patients were classified into 2 groups: those with detectable HBV DNA (non-responders) and those with undetectable HBV DNA (responders). Patients receiving peg-IFNα2a; HCV or HIV co-infected individuals were excluded from the study.
Whole blood and serum samples for analyzing HBV viral load, HBV pol/S region, and IL-10 promoter region polymorphisms were collected before antiviral therapy (baseline). Serum samples taken at week 12, 24, and 48 were stored for HBV DNA quantitative testing. At baseline, biochemical and serological tests and liver biopsy assessments were part of routine analyses.
The study protocol was approved by the local ethics committee and informed consent for participation was obtained from all enrolled subjects.
HBV viral load and pol/S region analysis
Whole blood samples were collected into Vacutainer tubes without anticoagulant and then incubated in an upright position for 30–45 min to allow clotting. The clot was then removed by centrifuging at 3500 × g for 15 min and serum was immediately transferred into a clean Eppendorf tube. All samples were stored at −20°C until further analysis. Viral DNA was extracted from 200 μl of serum samples using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics, Germany) with a slightly modified manufacturer’s protocol: the incubation time with Proteinase K was 1 h instead of 10 min and the final elution volume was 30 μl. Quality and quantity of the DNA was measured using a spectrophotometer (NanoDrop™ ND-1000, Thermo Scientific, USA). The Roche Cobas Amplicor HBV Monitor Assay (Roche Diagnostics, Pleasanton, USA) was used to quantify the level of HBV DNA in serum (sensitivity 34.36 IU/ml) using the manufacturer’s protocol.
Multi-temperature single-strand conformation polymorphism (MSSCP)
The MSSCP assay was done by means of the DNA Pointer System (Biovectis, Poland). To increase the sensitivity of HBV YMDD (rt204) variants detection, nested PCR was performed. The reaction mixture for the first step (1st PCR) contained (25 μl): 2 μl of HBV DNA, 1× chelating buffer, 1.2 mM Mg(OAc)2, 0.2 mM dNTP, 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Germany), 0.1 mg/ml of casein, 0.01% (v/v) formamide, and 0.125 μM of primers: 1F and 1R (1F: 5′-GACTCGTGGTGGACTTCTCTC-3′, 1R: 5′-TGATCCTGTGGCAAAGTTCC-3′). The reaction mixture for the second step (2nd PCR) consisted of (25 μl): 1× chelating buffer, 1.2 mM MgCl2, 0.5 mM dNTP, 1 U of AmpliTaq Gold polymerase, 0.25% (v/v) glycerol, 0.4% (v/v) BSA, 0.125 μM of primers: 2F [17] and 2R (5′-TAACAGCGGTATAAAGGGCCT-3′), and 2 μl of the first amplification step product. PCR conditions for each step are shown in Table 1. Positive and negative control was used at each step (HBV DNA External Quality Control, PeliSpy™PRO; AcroMetrix, USA). To determine the detection limit, serial dilutions of Quantification Standard QS HBV Real Star® (Altona Diagnostics, Germany) were used: 104 copies/μl (1718 IU/mL), 103 copies/μl (171.8 IU/mL), 102 copies/μl (17.18 IU/mL), 10 copies/μl (1.718 IU/mL), 5 copies/μl (0.859 IU/mL), 2 copies/μl (0.3436 IU/mL), and 1 copy/μl (0.1718 IU/mL).
Table 1.
PCR thermal cycling parameters.
| Step | Temperature/duration | ||||||
|---|---|---|---|---|---|---|---|
| 1st PCR | 2nd PCR | HBV genotyping | rs1082 | rs819 | rs592 | ||
| No. of amplification cycles | 35 | 35 | 35 | 33 | 33 | 33 | |
| Initial inactivation/denaturation | 95°C/10 min | 95°C/10 min | 95°C/10 min | 95°C/3 min | 95°C/3 min | 95°C/3 min | |
| Parameters of amplification | Denaturation | 94°C/30 sec | 94°C/30 sec | 94°C/30 sec | 95°C/30 sec | 95°C/30 sec | 95°C/30 sec |
| Annealing | 56°C/30sec | 57°C/30 sec | 57°C/30 sec | 57.9°C/30 sec | 61.4°C/30 sec | 53.7°C/30 sec | |
| Extension | 72°C/30 sec | 72°C/30 sec | 72°C/90 sec | 72°C/60 sec | 72°C/60 sec | 72°C/60 sec | |
| Final extension | 72°C/10 min | 72°C/10 min | 72°C/10 min | 72°C/1 min | 72°C/1 min | 72°C/1 min | |
| Hold | 4°C | 4°C | 4°C | 4°C | 4°C | 4°C | |
One μl of denatured PCR products (159 bp) containing HBV mutant and wild-type (WT) sequences at the codons of interest were loaded onto a 11% polyacrylamide gel (29:1 acrylamide: bisacrylamide) with addition of 5% glycerol. The electrophoresis was run under sequentially changed gel temperature (15°C, 10°C, and 5°C for 900 Vxh each) at a constant voltage of 40V in 0.5×TBE. After silver staining, specific band patterns characteristic for each HBV variant were cut out from the gel, purified using the QIAquick Gel Extraction Kit (Qiagen, Germany), and then sequenced.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS)
The MALDI-TOF MS-based HBV genotyping assay started with amplification of 754 bp DNA fragment containing 12 codons: 80, 169, 173, 180, 181, 184, 194, 202, 204, 233, 236, and 250 [27–35]. The PCR reaction mixture contained (25 μl): 200 nmol/l of each primer [25], 5 μl of HBV DNA, 1x chelating buffer, 1.2 mM Mg(OAc)2, 0.2 mM dNTP and 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Germany). Amplification conditions of HBV pol/S region analysis are shown in Table 1.
PCR products were then purified from non-incorporated dNTPs by treating with shrimp alkaline phosphatase (SAP) solution (40 min at 37°C and 5 min at 85°C). The iPLEX Gold assay was done in 4 separate primer-extension reactions on the Mass Array genotyping platform (Sequenom Inc., USA) with a standard procedure following the iPLEX kit protocol (Sequenom Inc., USA). To desalt the iPLEX reaction products, a resin kit was used (SpectroCLEAN resin, Sequenom Inc., USA), then cleaned extension products were dispensed onto a 384-element SpectroChip using the Nanodispenser, and mass differences were detected with MALDI-TOF MS (matrix-assisted laser desorption/ionization time-of-flight mass spectrometry). All data analyses were carried out with TyperAnalyzer Application, version 4 (Sequenom Inc., USA).
IL-10 promoter region polymorphism
Interleukin 10 promoter region polymorphism DNA was extracted from 500 μl of whole blood using the QIAamp® DNA Blood Mini Kit (Qiagen, Germany). DNA was eluted in 50 μl of AE buffer. To determine 3 biallelic polymorphisms at positions −1082, −819, and −592 in the IL-10 gene promoter region, allele-specific PCR reactions (AS-PCR) were performed. Each polymorphism (rs) was assessed in 2 separate reactions. While the common primer was the same for both reactions (rs1082: 5′-AAGCTTCTGTGGCTGGAGTC-3′; rs819: 5′-GGCACATGTTTCCACCTCTTC-3′; rs519: 5′-GGGGTCATGGTGAGCACTAC-3′), 2 different primers were designed for each allele (rs1082: A: 5′-AACACTACTAAGGCTTCTTTGGGTA-3′ and G: 5′-AACACTACTAAGGCTTCTTTGGGTG-3′; rs819: C: 5′-TACCCTTGTACAGGTGATGTACC-3′ and T: 5′-TACCCTTGTACAGGTGATGTACT-3′; rs519: C: 5′-TCCAGAGACTGGCTTCCTACAAG-3′). The Human β-globin primers were used as an experimental control (5′-TTGGACCCAGAGGTTCTTTG-3′, 5′-GAGCCAGGCCATCACTAAAG-3′). The PCR reaction mixture contained (25 μl): 1× Taq buffer +NH4SO4 −MgCl2 (Thermo Scientific, USA), 2 mM MgCl2 (Thermo Scientific, USA), 0.2 mM dNTPs (Thermo Scientific, USA), 0.4 μM of each primer, 5 μl of BSA (1 mg/μl), 1 U DreamTaq (Thermo Scientific, USA), and ~20 ng/μl DNA. The PCR conditions are shown in Table 1.
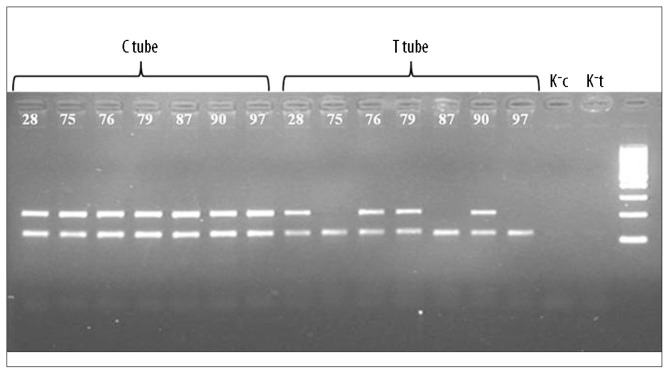
Further analysis involved agarose gel (2%) electrophoresis of PCR products (40 min, 100V). Based on the presence or absence of a PCR product, the patient’s genotype was determined. For example, if a product was seen in a “G” tube line without a product in an “A” tube line (rs1082), the patient was a GG homozygote. If PCR products were seen in both lines, the patient was a GA heterozygote (Figure 1).
Figure 1.
An example of results for 7 patients (28, 75, 76, 87, 90, 97) obtained for rs871. K−c and K−t are negative controls for C and T tubes.
Statistical analysis
Statistical analysis was done using STATISTICA data analysis software, version 8.0 (StatSoft Inc., USA). All statistical data are presented as a mean ± standard error of means (±SE), or median value. Standard error was used because distributions of data were skewed. Analysis of differences between variations was done using nonparametric statistics: Mann-Whitney’s U test, chi-squared test, and Kruskal-Wallis one-way analysis of variance. Relations between variables were estimated by univariate and multivariate logistic regressions. P-value less then 0.05 was considered statistically significant.
Results
The studied group of 54 consecutive CHB patients consisted of 27 naïve and 27 NAs-experienced individuals. The naïve patients had lower prevalence of HBV drug-resistant strains (p=0.019) and were more frequently treated with lamivudine 23/27 (85%) (χ2=20.5, p=0.0001) (Table 2). Entecavir was administrated in 4/27 (15%) subjects. Undetectable HBV DNA at week 24 and at week 48 was similar (p=0.7 and p=0.98, respectively).
Table 2.
Demographic and clinical characteristics in responder v/s nonresponder and naïve v/s NAs-experienced CHB patients.
| Analogues antiviral therapy | Significance p<0.05) | Analogues antiviral therapy | Significance p<0.05) | ||||
|---|---|---|---|---|---|---|---|
| responder (n=28) | nonresponder (n=26) | Naïve (n=27) | NAs experienced (n=27) | ||||
| Gender (male/female) | 10/18 | 19/7 | 0.0066 | 11/16 | 18/9 | 0.10 | |
| Age (years) | 43.4±1.98 | 54.4±3.51 | 0.011 | 47.67±3.3 | 49.70±2.59 | 0.62 | |
| ALT (IU/l) | 136±41 | 76±11 | 0.75 | 120±40 | 95±20 | 0.74 | |
| Baseline viral load (kIU/ml) | 13702±8385 | 57857±15205 | 0.0035 | 33989±12765 | 35935±12814 | 0.98 | |
| HBe Ag (reactive/nonreactive) | 8/20 | 15/11 | 0.033 | 9/18 | 14/14 | 0.25 | |
| HBe Ab (reactive/nonreactive) | 21/7 | 12/14 | 0.032 | 19/8 | 14/13 | 0.25 | |
| Baseline HBV drug-resistant variants* (Yes/No) | 19/9 | 17/9 | 0.86 | 13/14 | 23/4 | 0.019 | |
| Immunocompromised (Yes/No) | 10/17 | 8/17 | 0.71 | 8/19 | 10/15 | 0.52 | |
| NAs therapy** (naïve/experienced) | 13/15 | 14/12 | 0.60 | NA$ | NA## | NA## | |
| Antiviral therapy | Lamivudine | 16 | 14 | 0.029 | 23 | 7 | 0.0001 |
| Entecavir | 6 | 10 | 4 | 12 | |||
| Tenofovir | 6 | 1 | 0 | 7 | |||
| lamivudine + adefovir | 0 | 1 | 0 | 1 | |||
| Liver biopsy# | inflammation grade | 2 | 2 | 0.50 | 1.75 | 2 | 0.70 |
| fibrosis stage | 1.5 | 1.5 | 0.99 | 1.5 | 1.5 | 0.42 | |
Data are presented as a mean value ± standard error (SE);
data presented as a median value;
the presence of HBV drug-resistant minor variant before NAs therapy;
nucleoside/nucleotide analogues therapy;
not applicable.
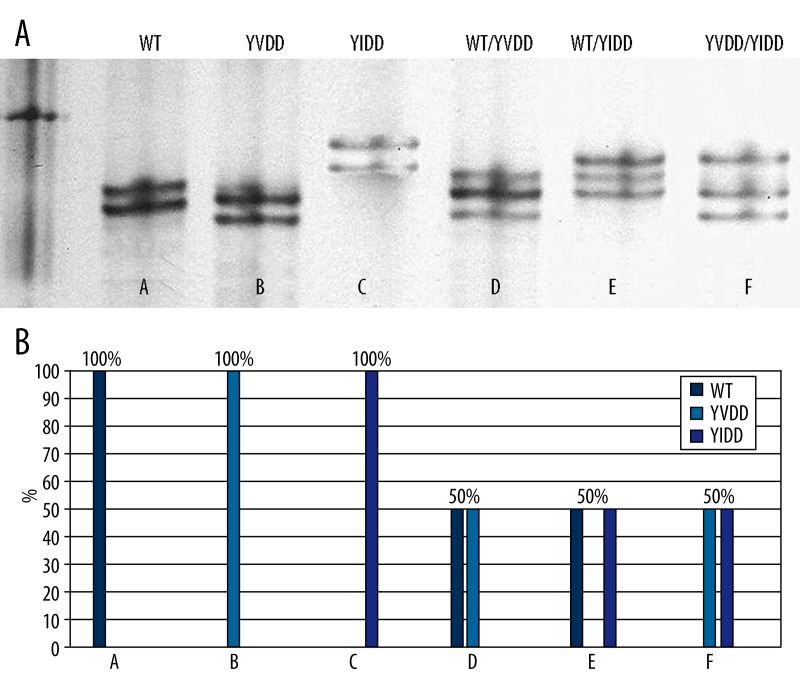
Some HBV drug-resistant variants were detected in 20/27 (74%) naïve and 25/27 (93%) NAs-experienced patients in MSSCP assay, confirmed in randomly selected cases by MALDI-TOF MS (Figure 2A, 2B). The HBV drug-resistant variant could affect prescribed drug activity in 13/27 (48%) naïve and 23/27 (85%) NAs-experienced subjects. Commercially available tests (Line Probe Assay – INNOLiPA HBV DR) did not detect drug-resistant strains in naïve patients qualified to NAs therapy [26]. The National Health Service (NFZ) requires pretreatment drug resistance tests in all patients (naïve and experienced) during qualification to NAs therapy.
Figure 2.
HBV YMDD variants obtained for 7 patients (A–F) during MSSCP (A) and MALDI-TOF MS analysis (B).
The efficacy of antiviral therapy in our study did not depend on liver biopsy inflammation activity (p=0.5), fibrosis stage (p=0.99), polymorphisms promoter IL-10 gene (Table 2), or preexistence of HBV drug-resistant variants (p=0.86). The strongest factor predicting the response to antiviral therapy was baseline viral load, which was significantly lower in the responder then the non-responder group (Table 2). The antiviral therapy was significantly different in responder and non-responder groups (p=0.03), but the frequency of lamivudine administration was similar in both groups (Table 2).
The highest mean viral load (kIU/ml) was detected in patients receiving entecavir (66,138±20,800). Significantly lower concentration of HBV DNA was measured in lamivudine (19,378±9,379) and tenofovir (11,769±11,539) groups (p=0.03). HBeAg reactive was significantly more frequent in the entecavir 11/16 (67%) and tenofovir 4/7 (57%) than in the lamivudine 7/23 (30%) groups (χ2=10.98, p=0.0118).
The univariate and multivariate comparison of variables between patients with good and weak response to antiviral therapy are reported in Table 4. Univariate logistic regression analysis showed the strongest influence of female sex (OR 4.88, 95% CI 1.49–16.04, p=0.007) on the response to NAs therapy. Multivariate logistic regression analysis revealed significant correlations (χ2=25.95, p=0.00009) between the following independent variables predicting better response to NAs therapy: younger age (OR 0.90, 95% CI 0.83–0.98, p=0.011), female sex (OR 10.3, 95% CI 1.60–65.81, p=0.011), lower baseline viral load (OR 0.999961, 95% CI 0.99993–0.99999, p=0.008), higher baseline ALT activity (OR 1.01, 95% CI 0.99–1.02, p=0.178), and patient’s immunocompetence (OR 0.09, 95% CI 0.009–0.93, p=0.038).
Table 4.
Univariate and multivariate logistic regression analyses of host and viral predicting NAs therapy response factors in CHB patients.
| Analogues antiviral therapy | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Responder (n=28) | Nonresponder (n=26) | OR (95% Cl) | p value | OR (95% Cl) | p value | |
| Age (year) | 43.4±1.98 | 54.4±3.51 | 0.95 (0.91–0.99) | 0.013 | 0.90 (0.83–0.98) | 0.011 |
| Female gender | 18 | 7 | 4.88 (1.49–16.04) | 0.007 | 10.3 (1.60–65.81) | 0.011 |
| Baseline HBV viral load (kIU/ml) | 13.702±8.385 | 57.857±15.205 | 0.999987 (0.99997–0.999998) | 0.019 | 0.999961 (0.99993–0.99999) | 0.008 |
| Baseline HBeAg (reactive) | 8 | 15 | 0.29 (0.09–0.90) | 0.033 | ||
| Baseline ALT (IU/l) | 136±41 | 76±11 | 1.003 (0.998–1.009) | 0.22 | 1.01 (0.99–1.02) | 0.178 |
| Previous NAs* therapy | 15 | 12 | 1.35 (0.45–4.03) | 0.586 | ||
| Baseline drug resistance presence ** | 19 | 17 | 1.12 (0.36–3.47) | 0.85 | ||
| Liver biopsy inflammation activity# | 2 | 2 | 1.42 (0.60–3.35) | 0.41 | ||
| Liver biopsy fibrosis stage# | 1.5 | 1.5 | 0.93 (0.46–1.93) | 0.86 | ||
| ATA IL-10 haplotype | 8 | 11 | 0.54 (0.18–1.69) | 0.29 | ||
| Immunocompetent patient | 17 | 17 | 0.8 (0.25–2.59) | 0.7 | 0.09 (0.009–0.93) | 0.038 |
Data are presented as a mean value ± standard error (SE);
data presented as a median value;
nucleoside/nucleotide analogues;
the presence of HBV drug-resistant minor variant before NAs therapy.
Discussion
We collected blood samples only from patients randomly qualified to NAs therapy in the Therapeutical Programs of the National Health Service (NFZ). Criteria for qualification to antiviral therapy was different from the EASL CHB statement and probably contributed to the slightly higher rate of therapeutic success of NAs in experienced compared to naïve patients [4,14]. NFZ qualification criteria still promotes lamivudine therapy in subjects without detectable HBV drug-resistant strains, and explains the overrepresentation of these patients in naïve and NAs-experienced individuals. A relatively weak HBV DNA suppression rate in entecavir-treated subjects have an effect on entecavir proscribing in lamivudine-resistant or high viremic HBeAg-reactive patients. These criteria selected patients in the entecavir-treated group who were more difficult to treat.
It was interesting to discover drug-resistant HBV variants in 13/27 (48%) treatment-naïve CHB patients, but without a significant influence on the therapy success rate (Table 1). We found drug-resistant HBV variants significantly more frequently in NAs-experienced than among naïve patients. However, some HBV variants seemed to be non-correlated with previous antiviral therapy and could have appeared spontaneously. Recently a natural prevalence of HBV reverse transcriptase amino acid substitutions in treatment-naïve patients with CHB has been shown [10–12,36,37]. However, these mutations in our subjects were more common. MSSCP and MALDI-TOF MS techniques used in our study had superior sensitivity than did commercially available HBV drug-resistance assays [26].
In our study there were no significant differences in frequencies of genotypes and alleles of IL-10 gene promoter region at position −1082 G/A, −819 T/C, −592 A/C and the initial response of HBV infection to treatment with NAs. Although IL-10 has been shown to have an important role in chronic inflammation and fibrogenesis, as well as in IFN-α therapy of hepatitis B patients, it cannot become a predictive factor of response to NAs in chronic HBV infection. Moreover, in contrast to IFN therapy, no differential haplotype distribution responsible for IL-10 expression was observed between NAs responders and non-responding patients [21]. This suggests that the association of remaining putative functional single nucleotide polymorphisms (SNPs) (e.g., 2753 A/C and 3575 T/A) in IL-10 and other host genetic factors should be investigated. It is well known that the actions of cytokines may be profoundly conditioned by the presence of other cytokines. Thus, there is a need to identify new predictors for treatment outcome in these patients. New studies of particular combinations of IL-10 SNPs and other host predictors could reveal the relation with susceptibility to nucleos(t)ide analogues and may help in making appropriate treatment decisions [20–23].
In the presented study, female sex and young age were the main host factors predicting successful response to antiviral therapy. The presence of drug-resistant HBV variants at baseline was not a viral predictor of good response to NAs therapy. Only low HBV viral load was useful in identifying the likelihood of response to antiviral therapy. Multivariate analysis connected essential host and viral factors predicting response to NAs drugs, investigated in univariate analysis, with higher ALT activity and absence of immunosuppressive therapy or past bone marrow transplantation. The ideal candidate for antiviral therapy is an immunocompetent, young, female with low HBV viral load and higher ALT activity, Similar observations were previously reported [4,6,12,22].
Conclusions
We did not confirm the influence of presence drug-resistant HBV variants on NAs treatment efficacy in naïve patients. The Polish National Health Service requirement of a pre-treatment drug resistance test during qualification to antiviral therapy has no justification in our data or in commonly accepted knowledge. We did not find any host- or virus-related causes of HBV drug-resistant variants except for previous insufficient NAs therapy. The main HBV-related predicting response factor was low viral load. Host-related factors of good antiviral response predictors were young age, female sex, high ALT activity, and immunocompetence. Our study did not confirm the role of IL-10 promoter region polymorphisms in efficacy of NAs therapy in CHB patients. Natural polymorphism of the host immune system genes seems to be an essential factor affecting elimination of varied pathogens and needs further investigation.
Table 3.
Distribution of alleles promoter region IL-10 in CHB patients.
| IL-10 polymorphism | Analogues antiviral therapy | Significance p<0.05) | |
|---|---|---|---|
| Responder (n=28) | Nonresponder (n=26) | ||
| rs1800896 (1082) | |||
| GG | 4 | 4 | 0.88 |
| AA | 3 | 3 | |
| GA | 21 | 19 | |
| rs1800871 (819) | |||
| CC | 20 | 15 | 0.26 |
| TT | 0 | 1 | |
| CT | 8 | 10 | |
| rs1800872 (592) | |||
| CC | 19 | 13 | 0.26 |
| AA | 2 | 1 | |
| CA | 7 | 12 | |
| ATA haplotype IL-10 (present/absent) | 8 | 11 | 0.3 |
Acknowledgments
Magda Rybicka is the recipient of the European Social Fund in the framework of the project “InnoDoktorant” – Scholarships for PhD students, 5th Edition.
Footnotes
Source of support: Ministry of Science and Higher Education, grant no. N N302183639 and Medical University of Gdansk, grant no. ST79
Conflict of interest
None declared.
References
- 1.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielawski KP, Dybikowska A, Lisowska-Charmuszko U, et al. Distribution of HBV genotypes and mutants among hepatitis B infected patients from northern Poland. Int J Mol Med. 2004;14:301–4. [PubMed] [Google Scholar]
- 3.WHO HBV fact sheet No 204, Revised July 2012, World Health Organisation: http://www.who.int/mediacentre/factsheets/fs204/en/
- 4.European Association For The Study Of The Liver. EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–85. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Bielawski KP, Stalke P. Molecular epidemiology of chronic hepatitis B virus infection in northern Poland. J Clin Virol. 2005;34(Suppl 1):S63–69. doi: 10.1016/s1386-6532(05)80012-5. [DOI] [PubMed] [Google Scholar]
- 6.Chen CJ, Yang HI. Natural history of chronic hepatitis B REVEALed. J Gastroenterol Hepatol. 2011;26:628–38. doi: 10.1111/j.1440-1746.2011.06695.x. [DOI] [PubMed] [Google Scholar]
- 7.Cai D, Mills C, Yu W, et al. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrob Agents Chemother. 2012;5–6:4277–88. doi: 10.1128/AAC.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locarnini S, Mason WS. Cellular and virological mechanisms of HBV drug resistance. J Hepatol. 2006;44:422–31. doi: 10.1016/j.jhep.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 9.Lok AS, Zoulim F, Locarnini S, et al. Hepatitis B Virus Drug Resistance Working Group. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254–65. doi: 10.1002/hep.21698. [DOI] [PubMed] [Google Scholar]
- 10.Han Y, Huang LH, Liu CM, et al. Characterization of hepatitis B virus reverse transcriptase sequences in Chinese treatment naive patients. J Gastroenterol Hepatol. 2009;24:1417–23. doi: 10.1111/j.1440-1746.2009.05864.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu BM, Li T, Xu J, et al. Characterization of potential antiviral resistance mutations in hepatitis B virus reverse transcriptase sequences in treatment-naive Chinese patients. Antiviral Res. 2010;85:512–19. doi: 10.1016/j.antiviral.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen MH, Garcia RT, Trinh HN, et al. Prevalence of hepatitis B virus DNA polymerase mutations in treatment-naive patients with chronic hepatitis B. Aliment Pharmacol Ther. 2009;30:1150–58. doi: 10.1111/j.1365-2036.2009.04151.x. [DOI] [PubMed] [Google Scholar]
- 13.Rybicka M, Charmuszko U, Stalke P, Bielawski KP. The influence of hepatitis B virus polymorphism on the progression of chronic liver disease. Postepy Hig Med Dosw. 2011;65:244–54. doi: 10.5604/17322693.939665. [DOI] [PubMed] [Google Scholar]
- 14.Keffe EB, Dieterich DT, Pawlotsky JM, Benhamou Y. Chronic hepatitis B: preventing, detecting, and managing viral resistance. Clin Gastroenterol Hepatol. 2008;6:268–74. doi: 10.1016/j.cgh.2007.12.043. [DOI] [PubMed] [Google Scholar]
- 15.Paik YH, Han KH, Hong SP. The clinical impact of early detection of YMDD mutants on the outcomes of long-term lamivudine therapy in patients with chronic hepatitis B. Antivir Ther. 2006;11:447–55. [PubMed] [Google Scholar]
- 16.Han KH, Hong SP, Choi SH, et al. Comparison of multiplex restriction fragment mass polymorphism and sequencing analyses for detecting entecavir resistance in chronic hepatitis B. Antivir Ther. 2011;16:77–87. doi: 10.3851/IMP1702. [DOI] [PubMed] [Google Scholar]
- 17.Bielawski KP, Al-Soud WA, Stalke P, et al. Determination of lamivudine-resistant variants of hepatitis B virus by denaturing gradient gel electrophoresis: A novel approach to monitoring drug resistance. Med Sci Monit. 2008;14(5):CR281–85. [PubMed] [Google Scholar]
- 18.van Belkum A, Welker M, Erhard M, Chatellier S. Biomedical mass spectrometry in today’s and tomorrow’s clinical microbiology laboratories. J Clin Microbiol. 2012;50:1513–17. doi: 10.1128/JCM.00420-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tost J, Gut IG. Genotyping single nucleotide polymorphisms by MALDI mass spectrometry in clinical applications. Clin Biochem. 2005;38:335–50. doi: 10.1016/j.clinbiochem.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Zoulim F. Hepatitis B virus resistance to antiviral drugs: where are we going? Liver Int. 2011;31(Suppl 1):111–16. doi: 10.1111/j.1478-3231.2010.02399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Huang D, Sun S, et al. Interleukin-10 promoter polymorphism predicts initial response of chronic hepatitis B to interferon alfa. Virol J. 2011;20(8):28. doi: 10.1186/1743-422X-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao QJ, Liu DW, Zhang SY, et al. Polymorphisms of some cytokines and chronic hepatitis B and C virus infection. World J Gastroenterol. 2009;28(15):5610–19. doi: 10.3748/wjg.15.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Z, Tan W, Zhao W, et al. Regulatory polymorphisms in the IL-10 gene promoter and HBV-related acute liver failure in the Chinese population. J Viral Hepat. 2009;16:775–83. doi: 10.1111/j.1365-2893.2009.01139.x. [DOI] [PubMed] [Google Scholar]
- 24.Thursz M, Yee L, Khakoo S. Understanding the host genetics of chronic hepatitis B and C. Semin Liver Dis. 2011;31:115–27. doi: 10.1055/s-0031-1276642. [DOI] [PubMed] [Google Scholar]
- 25.Luan J, Yuan J, Li X, et al. Multiplex detection of 60 hepatitis B virus variants by maldi-tof mass spectrometry. Clin Chem. 2009;55:1503–9. doi: 10.1373/clinchem.2009.124859. [DOI] [PubMed] [Google Scholar]
- 26.Rybicka M, Stalke P, Dręczewski M, et al. High-throughput MALDI-TOF Mass Spectrometry as an Alternative Approach to HBV Drug Resistance Monitoring. J Clin Microbiol. 2013 doi: 10.1128/JCM.01891-13. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warner N, Locarnini S, Kuiper M, et al. The L80I substitution in the reverse transcriptase domain of the hepatitis B virus polymerase is associated with lamivudine resistance and enhanced viral replication in vitro. Antimicrob Agents Chemother. 2007;51:2285–92. doi: 10.1128/AAC.01499-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenney DJ, Levine SM, Rose RE, et al. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to Lamivudine. Antimicrob Agents Chemother. 2004;48:3498–507. doi: 10.1128/AAC.48.9.3498-3507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaney WE, Yang H, Westland CE, et al. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J Virol. 2003;77:11833–41. doi: 10.1128/JVI.77.21.11833-11841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono SK, Kato N, Shiratori Y, et al. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J Clin Invest. 2001;107:449–55. doi: 10.1172/JCI11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheldon J, Camino N, Rodes B, et al. Selection of hepatitis B virus polymerase mutations in HIV-coinfected patients treated with tenofovir. Antivir Ther. 2005;10:727–34. [PubMed] [Google Scholar]
- 32.Allen MI, Deslauriers M, Andrews CW, et al. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 1998;27:1670–77. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 33.Bozdayi AM, Uzunalimoglu O, Turkyilmaz AR, et al. YSDD: a novel mutation in HBV DNA polymerase confers clinical resistance to lamivudine. J Viral Hepat. 2003;10:256–65. doi: 10.1046/j.1365-2893.2003.00435.x. [DOI] [PubMed] [Google Scholar]
- 34.Schildgen O, Sirma H, Funk A, et al. Variant of hepatitis B virus with primary resistance to adefovir. N Engl J Med. 2006;354:1807–12. doi: 10.1056/NEJMoa051214. [DOI] [PubMed] [Google Scholar]
- 35.Angus P, Vaughan R, Xiong S, et al. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology. 2003;125:292–97. doi: 10.1016/s0016-5085(03)00939-9. [DOI] [PubMed] [Google Scholar]
- 36.Singla B, Chakraborti A, Sharma BK, et al. Hepatitis B virus reverse transcriptase mutations in treatment Naïve chronic hepatitis B patients. J Med Virol. 2013;85:1155–62. doi: 10.1002/jmv.23608. [DOI] [PubMed] [Google Scholar]
- 37.Du QW, Ding JG, Sun QF, et al. Combination lamivudine and adefovir versus entecavir for the treatment of naïve chronic hepatitis B patients: a pilot study. Med Sci Monit. 2014;20:751–56. doi: 10.12659/MSM.889443. [DOI] [PMC free article] [PubMed] [Google Scholar]